SUMMARY
During spermatogenesis, germ cells initially expand exponentially through mitoses. A majority of these cells are then eliminated through p53-mediated apoptosis to maintain germline homeostasis [1–4]. However, the activity of p53 must be precisely modulated, especially suppressed in postmitotic spermatogenic cells, to guarantee robustness of spermatogenesis. Currently, how the suppression is achieved is not understood. Here, we show that Pumilio 1, a posttranscriptional regulator, binds to mRNAs representing 1527 genes, with significant enrichment for mRNAs involved in pathways regulating p53, cell cycle, and MAPK signaling. Particularly, eight mRNAs encoding activators of p53 are repressed by Pumilio 1. Deleting Pumilio 1 results in strong activation of p53 and apoptosis mostly in spermatocytes, which disrupts sperm production and fertility. Removing p53 reduces apoptosis and rescues testicular hypotrophy in Pumilio 1-null mice. These results indicate that key components of the p53 pathway are coordinately regulated by Pumilio 1 at the posttranscriptional level, which may exemplify an RNA operon.
Keywords: Pumilio 1, RNA operon, spermatogenesis, apoptosis, p53, translational regulation
RESULTS AND DISCUSSION
Spermatogenesis in mammals is a complex process in which germline stem cells undergo 9–11 rounds of mitosis, followed by meiosis and a cellular morphogenic process called spermiogenesis that transforms round haploid spermatids into sperm. The multiple rounds of mitoses generate excess spermatogonia that must be eliminated to maintain the homeostasis of spermatogenesis. This is accomplished in part by p53-mediated apoptosis [1–4]. The temporally and spatially specific activation of p53 must be precisely controlled so that not only excess spermatogonia are eliminated, but enough germ cells must also survive the elimination to generate a sufficiently large number of sperm. However, how this control is achieved in mammals remains unknown. Here, we show that Pumilio 1 (Pum1), through coordinated posttranscriptional regulation of multiple factors in the p53 pathway, represses p53 activation and apoptosis after spermatogonial division.
Pum1 is one of the two members (Pum1 and Pum2) of the PUF (for Pumilio and FBF) protein family in the mouse. Its homolog in Drosophila, Pum, is a posttranscriptional regulator essential for a variety of germline processes, such as primordial germ cell proliferation [5], germline stem cell self-renewal [6, 7], ovarian morphogenesis, and oviposition [5]. Pum binds to a defined motif with a UGUAHAUA core in the 3′ untranslated region (3′ UTR) of its target mRNAs and mediates translational repression and/or mRNA decay [8, 9]. PUF proteins in C elegans are also known to be involved in multiple steps of germline development [10, 11]. In contrast to the critical roles of Pum proteins in the Drosophila and C. elegans germline, mouse Pum2 is not essential for spermatogenesis [12], calling for an understanding of Pum1 in the mouse.
Pum1 is expressed in the cytoplasm of spermatogenic cells
We first examined the expression of Pum1 in 11 mouse organs. The mRNA and protein levels were measured by quantitative reverse transcription PCR (qRT-PCR) and immunoblot analysis, respectively. Both analyses showed that Pum1 is highly expressed in the testis (Figures 1A and S1A). During the development of testis, Pum1 is expressed 2 days post partum (dpp) and then starts to increase at 14 dpp when pachytene spermatocytes first appear (Figure S1B). Immunofluorescence microscopy using an anti-Pum1 antibody further revealed that Pum1 is expressed in the cytoplasm of spermatocytes along with other germ cell types. (Figures 1B and S1C). To confirm the subcellular localization of Pum1, we separated adult testicular lysates into cytoplasmic and nuclear fractions. Pum1 was detected almost exclusively in the cytoplasm as indicated by immunoblot analysis (Figure 1C). This is consistent with the reported role of Drosophila and C. elegans PUF proteins in regulating the stability and translation of its target mRNAs [13–17].
Figure 1. Pum1 expression and function in the testis.
(A)qRT-PCR analysis of Pum1 mRNA levels in different organs and tissues, as normalized to Gapdh. ESCs: embryonic stem cells; BMW: bone marrow. Bars indicate SD (B) Immunofluorence microscopy of Pum1 in wild-type testis thin sections. Sg: spermatogonium; Sc: spermatocyte; Sd: spermatid. (C) Immunoblot analysis of Pum1 in testicular cytoplasmic and nuclear fractions. Nu: nucleus; Cy: cytoplasm. (D) Mature sperm counts in wild-type and Pum1 mutant males at different ages, with bars indicating SEM. (E) Size of litters sired by wild-type and Pum1 mutant males, with bars indicating SEM. (F and G) Testicular histology revealed by H&E staining in wild-type (F) and Pum1 null (G) mice. Arrow points to a spermatogenic cell undergoing cell death. (H) Quantification of apoptotic cells in adult wild-type and Pum1 mutant testes. Apoptosis was stained by TUNEL analysis. For each mouse analyzed, apoptotic cells were counted in 80 random seminiferous tubules sections and presented in (H) as numbers per 10 random tubules cross sections, with bars showing SD. *** indicates P<0.001 in (H). Scale bars in (B) and (F) both represent 50μm.
Pum1-null males show significantly reduced sperm counts and fertility
To understand the role of Pum1 in development, we generated Pum1-nullmice for phenotypic analysis (Figure S1D). The loss of Pum1 protein was confirmed by immunoblot analysis of testes from five Pum1−/− mice (Figure S1E). Pum1−/− mice are viable and grow to adulthood without apparent defects except that they are 18% (b.w., ±2.1% SEM, P=0.0003) smaller than wild-type mice at eight weeks of age (Figure S1F).
However, testicular hypoplasia was observed in all Pum1−/− males examined. The average testicular weight of Pum1−/− mice is 34% (±1.5% SEM, P=2.28×10−16) lower than that of wild-type mice at eight weeks of age (Figure S1G). In addition, the mature sperm count of Pum1−/− males is reduced by 80% (±2.0% SEM, P=1.40×10−16) at eight weeks of age and remains at this low level throughout adult life (Figure 1D). Consistently, the fertility of Pum1−/− males is reduced by 41% (±3.2% SEM, P=1.56×10−13) when compared to the wild-type level (Figure 1E). Taken together, these data indicate that spermatogenesis is compromised in Pum1−/− males.
Spermatocytes in Pum1-null testes display increased apoptosis
To understand which step of spermatogenesis is impaired in the Pum1−/− mice, we examined the testicular histology by hematoxylin and eosin (H&E) staining (Figures 1F and 1G) and periodic acid-Schiff (PAS) stain (Figures S2A and S2B). The global morphology of Pum1−/− testes looks normal, containing the complete lineage of germ cells. Acrosome morphogenesis is normal in Pum1−/− testes (Figures S2A and S2B), implicating normal spermiogenesis up to the round spermatid stage. Mature sperm are present (Figure 1D), indicating that spermatogenesis can reach completion for some of the Pum1−/− spermatogenic cells. In addition, Ki-67 immunohistochemistry analysis showed that the division rates of spermatogonia and spermatocytes are not affected in Pum1−/− testes (Figures S2C–S2E). This is confirmed by the normal level of phospho-Histone 3, another marker of cell division, in Pum1−/− testes, as revealed by immunoblot analysis (Figures S2F and S2G). Furthermore, serum testosterone level is unchanged in Pum1−/− mice, indicating normal steroidogenesis (Figure S2H). However, Pum1−/− spermatocytes more frequently contain enlarged nuclei and condensed chromatin, suggestive of apoptosis (Figures 1F and 1G). To investigate this defect further, we conducted a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay in 16 adult Pum1+/+, Pum1+/−, and Pum1−/− males. This analysis revealed that apoptosis in Pum1+/− and Pum1−/− testes is elevated by 2.7 fold (±0.29 SD, P=0.0002) and 6.8 fold (±1.02 SD, P=4.22×10−5), respectively (Figures1H and S2I–S2K). Moreover, Apoptosis is elevated mostly in primary spermatocytes (Figures S2L–S2N) in which Pum1 is normally expressed at high levels. Moreover, in the testes of Pum1−/flox; Vasa-cre+ mice where Pum1 is knocked out only in the germline, apoptosis is also elevated by 7.5 fold (±0.24 SD, P=1.98×10−7) from wild-type level (Figures 1H, S2O and S2P). It suggests that the apoptosis phenotype in Pum1 mutant testes is germline-autonomous.
Spermatocytes in Pum1-null testes are prematurely released into the epididymis
We then tracked the behavior of the apoptotic cells in the reproductive tract and found that the lumen of the epididymis in Pum1−/− mice contains unexpected round cells with aberrant morphology (Figures S3A–S3C). These cells are positive for Miwi (Figures S3D and S3E), a protein specifically expressed in spermatocytes and early spermatids. These cells also showed positive staining for TUNEL (Figures S3F and S3G). It indicates that these cells are apoptotic spermatocytes that are expelled from the testis rather than cells derived from the epididymis epithelium.
Pum1 binds to mRNAs representing 1,527 genes in the testis
To understand the mechanism of elevated apoptosis in the Pum1−/− testis, we wanted to know which mRNAs are targeted by Pum1 in the testis. To this end, we performed a genome-wide target identification of Pum1 using ribonucleoprotein-immunoprecipitation followed by microarray (RIP-Chip). The Pum1 ribonucleoprotein (RNP) complexes were precipitated from total testicular lysates with an anti-Pum1 antibody coupled to Protein A Sepharose beads. RNAs associated with Pum1 were extracted and analyzed by hybridization to a mouse cDNA microarray (Illumina Mouse Chip M6). To identify non-specifically precipitated RNAs, Pum1−/− testicular lysates were used in parallel experiments as negative controls.
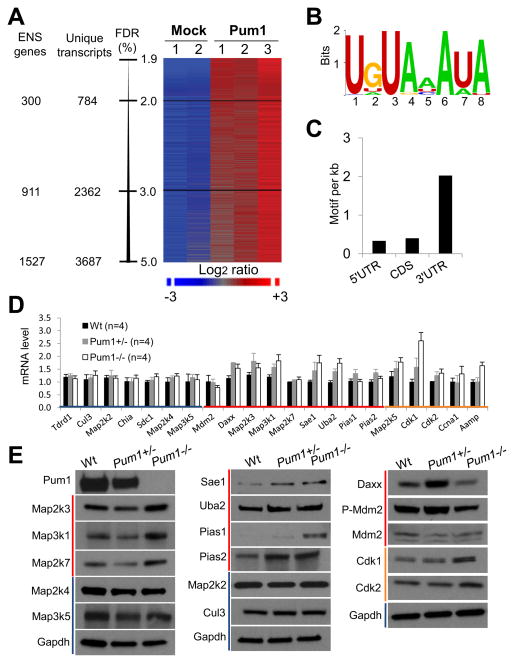
We identified 3,687 transcripts that are consistently associated with Pum1, representing 1,527 Ensembl genes (False Discovery Rate<5%; Figure 2A and Table S1). Similar studies performed on human PUM1 protein in Hela S3 cancer cells identified a similar number of mRNA targets [18, 19]. Multiple Expectation Maximization for Motif Elicitation (MEME) analysis revealed an eight nucleotide consensus sequence, UGUAHAUA, that exists among the Pum1 target mRNAs (Figure 2B). This motif predominantly resides in their 3′UTRs (Figure 2C), and is the same as those found in mRNA targets of yeast Puf3 [20], Drosophila Pum [9], and human PUM1 and PUM2 [18, 19], indicating that the Pum binding motif is highly conserved.
Figure 2. Pum1 binds to and represses mRNA targets via a defined binding motif.
(A) mRNAs associated with Pum1. Three independent experiments using wild-type testicular lysates and two mock experiments using Pum1−/− testicular lysates are shown. The Pum1 target mRNAs were co-immunoprecipitated by an anti-Pum1 antibody. mRNAs associated with Pum1 were analyzed by mouse cDNA microarrays. ENS: Ensembl; FDR: False Discovery Rate. (B) The consensus sequence among Pum1-associated mRNAs. A de novo sequence motif search was performed using the top 124 Pum1-assocated mRNAs by Multiple Expectation Maximization for Motif Elicitation (MEME). (C) Number of motifs per one kilobase of sequence in the 5′UTRs, CDS and 3′UTRs of Pum1 target mRNAs. (D and E) Expression levels of select Pum1-associated mRNAs and their protein levels in wild-type, Pum1+/− and Pum1−/− testes. Blue lines indicate non-Pum1 targets; red lines indicate Pum1 targets that are p53 regulators; and orange lines indicate Pum1 targets that are not p53 regulators. Bars in (D) indicate SEM.
Pathway analysis using MetaCore ™ (version 6.7 build 28822) was performed on 656 pathways to which genes on the Illumina mouse chip (M6) can be mapped. Pum1-associtated mRNAs are enriched in 11 pathways (P<0.001) that regulate p53 activation, cell cycle, and mitogen-activated protein kinase (MAPK) signaling (Table S2 and Figures S4A–S4C), which is also consistent with the pathways targeted by human PUM1 in Hela S3 cancer cells [18, 19]. In the p53-regulating pathways, nine mRNAs encoding factors that regulate p53-mediated apoptosis are Pum1 targets. Among them, Map3k1, Map2k3 and Daxx activate p38 MAPK which in turn activates p53 [21–25]; Map2k7 together with Map3k1 activate JNK which in turn activates p53 [22, 26]; Sae1, Uba2, Pias1 and Pias2 are sumoylation ligases that prime p53 for inducing apoptosis [27]; Mdm2 is a ubiquitin ligase for p53 that primes it for degradation.[28, 29]
Pum1 represses its target mRNAs in pathways that regulate p53 activity
Since Drosophila Pum promotes the decay of its target mRNAs and/or blocks their translation [13–15], we wanted to understand how murine Pum1 regulates its target mRNAs. To this end, we compared the expression levels of the nine regulators of p53 between wild-type, Pum1+/−, and Pum1−/− testes, along with another five Pum1 target mRNAs and seven non-target mRNAs as controls. qRT-PCR analysis revealed that all seven non-target mRNAs remain at wild-type levels in Pum1−/− testes; whereas 10 out of 14 Pum1 target mRNAs are slightly elevated in Pum1−/− testes, suggesting that Pum1 plays a minor role in reducing the stability of its target mRNAs (Figure 2D). More significantly, immunoblot analysis revealed the elevation of seven out of the nine Pum1 target genes that are p53 regulators at the protein level inPum1 −/− testis, except for Daxx and Mdm2 (Figures 2E). In addition, Cdk1 and Cdk2, two other Pum1 targets, are elevated at the protein level (Figure 2E). In contrast, the protein levels of Cul3, Map2k2, Map2k4 and Map3k5, which are not Pum1 targets, remain at the same levels in Pum1−/− testes (Figures 2E). The scale of elevation at the protein level (2–10 fold) is much larger than that at the mRNA level (mostly within 2 fold; Figure S4D), suggesting that Pum1 regulates its target mRNAs mostly via translational repression.
p53 is over-activated in spermatocytes in Pum1-null testes
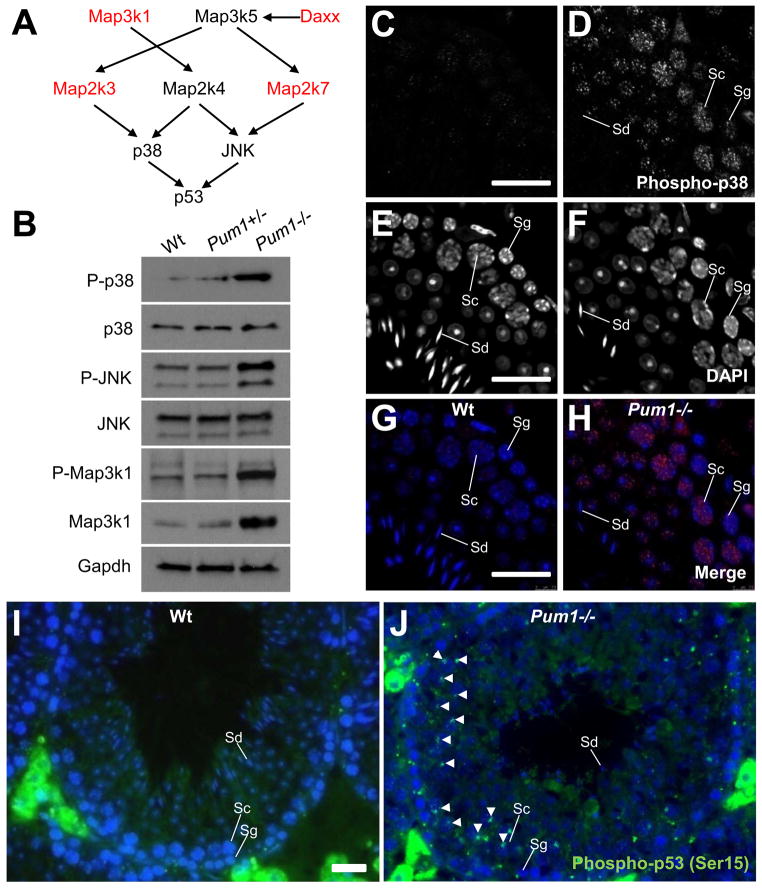
Pum1 represses at least two factors, Map2k3 and Map3k1, that activate p53 by activating the p38 MAPK (Figures 2E, 3A and 3B). We hypothesized that removing Pum1 would lead to elevated activation of p38. Indeed, immunoblot analysis showed an increase of phosphorylated p38 (Thr180/Tyr182) representing activated p38 in Pum1−/− testis; whereas its total protein level remains unchanged (Figure 3B). Similarly, the phosphorylated JNK (Thr183/Tyr185), representing activated JNK, is elevated when its upstream kinase, Map2k7, is increased due to Pum1 deficiency (Figures 3A and 3B).
Figure 3. Pum1 deficiency leads to activation of p38 MAPK, JNK and p53.
(A) Proteins encoded by Pum1 target mRNAs activate p53 through the MAPK signaling pathway. (B) Immunoblot analysis of total and activated levels of p38, JNK and Map3k1 in total testicular lysates. While the total protein levels of p38 and JNK remain unchanged, phosphorylated p38 (Thr180/Tyr182) and JNK (Thr183/Tyr185) are increased in Pum1−/− testes. Map3k1 is a Pum1 target that is upstream of both p38 and JNK. Both its total and phosphorylated (Thr1383) forms are elevated in Pum1−/− testes.( C–H) Immunofluorescence microscopy of activated p38 in testicular thin sections. DAPI staining (E and F) provides nucleus morphology that enables identification of spermatogonium (Sg), spermatocyte (Sc), and spermatid (Sd). The strongest elevation of p38 activation in Pum1−/− testes occurs in primary spermatocytes (C and D also G and F as merged images with DAPI). (I and J) Immunofluorescence microscopy of activated p53 (Serine 15) in testicular thin sections. White arrow heads indicate some of spermatocytes with activated p53. Extratubular green fluorescence in (I and J) is non-specific background from Leydig cells. Scale bars in (C), (E),(G) and (I) all represent 20 μm.
Immunofluorescence analysis showed that the strongest activation of p38 under Pum1 deficiency indeed occurs in primary spermatocytes (Figures 3C–3H). This is in agreement with our observations that Pum1 is most abundant in primary spermatocytes and that apoptosis is elevated in these cells under Pum1 deficiency.
The activated p38 phosphorylates p53 at Serine 15 to prime p53 for proapoptotic transcription [21, 23, 24]. Consistently, in wild-type testis, p53 is activated only in a small number of spermatogonia; however, in Pum1−/− testis, the activated form of p53 is extensively present in a large number of spermatocytes (Figures 3I and 3J). To confirm the activation of p53, the transcription of four p53 target genes were examined via qRT-PCR in total testicular lysates. Among them, Bax, Caspase 6, and Noxa showed increased mRNA level in Pum1−/− testes (Figure S4E). The over-activation of p53 can be partially rescued by an inhibitor of p38 (SB239063, i.v. at 15 μg/ml blood), suggesting p53 is over-activated in part via p38 in the Pum1−/− testis.
To further confirm the regulation of p53 by Pum1, we crossed the Pum1−/− mice with a Trp53 mutant line [30]. At 15 dpp, Pum1−/− testes contain significantly smaller seminiferous tubules than wild-type testes (Figures 4D and 4E), presumably due to an overkill of germ cells. Removing p53 partially restores this morphological defect (Figures 4F). TUNEL analysis of17 mice at 15 dpp revealed that removing p53 effectively reduces apoptosis in Pum1 mutant testes in a dose-dependent manner (Figure 4G). Moreover, removing p53 also rescues the testis hypotrophy in Pum1−/− testes (Figure 4H). These data indicate that p53 is a major mediator of the Pum1 function in maintaining the homeostasis of the mouse testicular germline.
Figure 4. Inhibition of p38 reduces p53 activation and removing p53 rescues Pum1 phenotype.
(A–C) Effects of p38 inhibition on p53 over-activation in Pum1−/− testes (D–F) H&E staining of thin-sectioned testes from wild-type (D), Pum1−/− (E) and Pum1−/− ;Trp53−/− testes at 15 dpp (F). (G) Removing p53 reduces apoptosis in Pum1−/− testes at 15 dpp. Apoptotic cells were counted in 80 random seminiferous tubules sections and presented in (D) as numbers per 10 random tubules cross sections, with bars showing SEM. (H) Removing p53 rescues the testis hypotrophy in Pum1−/− mice at 15 dpp. Bars indicate SEM. (I) A model illustrating that Pum1 represses eight mRNAs that encode activators of p53. Pum1 binds to these different mRNAs through a conserved binding motif in their 3′UTRs. Scale bars in (C) and (D) represent 5 and 50 μm for (A–C), and (D–F), respetively.
Conclusions
Previous studies have shown that p53-mediated apoptosis is used by mammals to eliminate excessive spermatogonia in order to maintain germline homeostasis [1–4]. Our data suggest that when a normal number of spermatogonia differentiate into spermatocytes, p53-mediated apoptosis must be suppressed to avoid the overkill of spermatocytes, and that this is achieved at least in part by Pum1 and its coordinated posttranscriptional suppression of multiple activators of p53 (Figure 4I).
In addition, Pum1 binds to mRNAs from 1527 genes. Most of these mRNAs may be true targets of Pum1 since 9 out of 11 Pum1 target mRNAs are elevated at the protein level under Pum1 deficiency (Figures 2E). The biological effects of Pum1-mediated regulation on these numerous targets await further investigation; why Pum1−/− mice are 18% smaller than wild-type mice (Figure S1D) may find an answer in the many Pum1 target mRNAs.
Furthermore, our data provide in vivo evidence in support of the RNA operon hypothesis proposed by Keene and colleagues [31, 32]. Unlike DNA operons, cistrons in an RNA operon (mRNAs) are not physically linked. Instead, each mRNA cistron carries a common sequence motif that is recognized by a shared regulator. On one hand, this allows efficient regulation of a pathway since multiple factors in it can be targeted by a single regulator, as demonstrated by Pum1 and its repression of eight activators of p53. On the other hand, each mRNA may carry multiple motifs recognized by different regulators and thus participate in more than one RNA operons. Lastly, even a single regulator can exert differential regulation towards different mRNA targets within an operon [33, 34]. These combined regulatory effects offer enormous capacity for precise regulation of many genes with biologically relevant functions.
Supplementary Material
HIGHLIGHTS.
Pum1-null males have significantly reduced sperm count and fertility.
Pum1-null testes display elevated apoptosis in spermatocytes.
Among its other target mRNAs, Pum1 binds to and represses multiple activators of p53.
Removing Pum1 in testes over-activates p53 in spermatocytes; whereas p53 deficiency rescues Pum1 phenotype.
Acknowledgments
We thank Drs. Sean Christenson, Celina Juliano, and Mona Nolde for critical reading of the manuscript. This work is supported by NIH grant R01HD42012 and a G. Harold & Leica Y. Mathers Award to H.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yin Y, Stahl BC, DeWolf WC, Morgentaler A. P53 and Fas are sequential mechanisms of testicular germ cell apoptosis. J Androl. 2002;23:64–70. doi: 10.1002/jand.2002.23.1.64. [DOI] [PubMed] [Google Scholar]
- 2.Russell LD, Chiarini-Garcia H, Korsmeyer SJ, Knudson CM. Bax-dependent spermatogonia apoptosis is required for testicular development and spermatogenesis. Biol Reprod. 2002;66:950–958. doi: 10.1095/biolreprod66.4.950. [DOI] [PubMed] [Google Scholar]
- 3.Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 4.Beumer TL, Roepers-Gajadien HL, Gademan IS, van Buul PP, Gil-Gomez G, Rutgers DH, de Rooij DG. The role of the tumor suppressor p53 in spermatogenesis. Cell Death Differ. 1998;5:669–677. doi: 10.1038/sj.cdd.4400396. [DOI] [PubMed] [Google Scholar]
- 5.Parisi M, Lin H. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics. 1999;153:235–250. doi: 10.1093/genetics/153.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin H, Spradling A. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 7.Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, McLachlan J, Zamore P, Hall T. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 9.Zamore P, Williamson J, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 10.Parisi M, Lin H. Translational repression: a duet of Nanos and Pumilio. Curr Biol. 2000;10:R81–83. doi: 10.1016/s0960-9822(00)00283-9. [DOI] [PubMed] [Google Scholar]
- 11.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Xu E, Chang R, Salmon N, Reijo Pera R. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- 13.Wreden C, Verrotti A, Schisa J, Lieberfarb M, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 14.Chagnovich D, Lehmann R. Poly(A)-independent regulation of maternal hunchback translation in the Drosophila embryo. Proc Natl Acad Sci U S A. 2001;98:11359–11364. doi: 10.1073/pnas.201284398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadyrova L, Habara Y, Lee T, Wharton R. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- 16.Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris A, Mukherjee N, Keene J. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol. 2008;28:4093–4103. doi: 10.1128/MCB.00155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber A. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3:e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerber A, Herschlag D, Brown P. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18:6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 23.She QB, Chen N, Dong Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J Biol Chem. 2000;275:20444–20449. doi: 10.1074/jbc.M001020200. [DOI] [PubMed] [Google Scholar]
- 24.Lin T, Mak NK, Yang MS. MAPK regulate p53-dependent cell death induced by benzo[a]pyrene: involvement of p53 phosphorylation and acetylation. Toxicology. 2008;247:145–153. doi: 10.1016/j.tox.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Chang HY, Nishitoh H, Yang X, Ichijo H, Baltimore D. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 1998;281:1860–1863. doi: 10.1126/science.281.5384.1860. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Lin A. Role of JNK activation in apoptosis: a double-edged sword. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- 27.Stehmeier P, Muller S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amst) 2009;8:491–498. doi: 10.1016/j.dnarep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 30.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Keene J. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 32.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 33.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Tuschl T, Ohler U, Keene JD. Integrative Regulatory Mapping Indicates that the RNA-Binding Protein HuR Couples Pre-mRNA Processing and mRNA Stability. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.