Abstract
Transgenesis is a cornerstone of molecular biology. The ability to integrate a specifically engineered piece of DNA into the genome of a living system is fundamental to our efforts to understand life and exploit its implications for medicine, nanotechnology and bioprospecting. However, transgenesis has been hampered by position effects and multi-copy integration problems, which are mainly due to the use of small, plasmid-based transgenes. Large transgenes based on native genomic regions cloned into bacterial artificial chromosomes (BACs) circumvent these problems but are prone to fragmentation. Herein, we report that contrary to widely held notions, large BAC-sized constructs do not prohibit transposition. We also report the first reliable method for BAC transgenesis in human embryonic stem cells (hESCs). The PiggyBac or Sleeping Beauty transposon inverted repeats were integrated into BAC vectors by recombineering, followed by co-lipofection with the corresponding transposase in hESCs to generate robust fluorescent protein reporter lines for OCT4, NANOG, GATA4 and PAX6. BAC transposition delivers several advantages, including increased frequencies of single-copy, full-length integration, which will be useful in all transgenic systems but especially in difficult venues like hESCs.
INTRODUCTION
Early work on transgenesis in animals and cell lines invariably used small transgenes, which only rarely achieved the intended expression pattern due mainly to position effects exerted by the genomic integration site or concatamerization. These major problems have been circumvented by the use of large transgenes such as bacterial artificial chromosomes (BACs), which carry intact genomic regions and often deliver the expected expression pattern precisely (1).
Due to their large size, BACs can accommodate complete genes including all cis-regulatory elements in their native configuration. Consequently, most BAC transgenes are indifferent to position effects and often deliver expression levels in proportion to the transgene copy number. Many BAC libraries have been annotated onto genome browsers and are readily available from genome resource providers such as CHORI (www.chori.org). Furthermore, BACs can be readily modified and mutated using recombineering (2–5). These advantages have promoted BACs to the forefront as transgenic tools and now BAC transgenesis has been successfully applied to produce a variety of transgenic animals, such as mice, rats, zebrafish and flies (6–9), as well as for studies of gene function, molecular complementation of mutations, identification of distant regulatory elements and analysis of gene dosage, among other applications (1,10–13). Because they often recapitulate expression patterns precisely, BAC transgenes are also widely used to create gene expression reporters for studies during development and differentiation.
Human embryonic stem cells (hESCs) (14) provide an essential venue for studies of human development and disease that complements work with model systems such as the mouse. Like mouse ESCs (mESCs), they can be differentiated in culture to recapitulate aspects of human embryology and to serve as paradigms for future medicine with cellular therapies. However, they are difficult to manipulate genetically, particularly for gene targeting (15–17).
The work reported here began with our efforts to create stable hESC reporter lines based on fluorescent protein expression driven by stage- and lineage-specific promoters. Although we were able to create an OCT4-GFP reporter line by gene targeting (data not shown), the efficiency of homologous recombination in hESCs is low (15,16) and our attempts to generate a knock-in for lineage-specific genes have not been successful. On the other hand, randomly integrated retroviral and small transgenes often undergo transcriptional silencing in hESCs (17–19). Consequently, we were attracted by the advantages of BAC transgenesis and used the only published method for BAC transgenesis in hESCs, which is based on nucleofection (20). Unexpectedly, transgene silencing was consistently observed, which we correlated with consistent failures to obtain integrations of full-length BAC transgenes. To solve the problem of BAC fragmentation, we explored the possibility that transposition could be used to integrate full-length BAC transgenes.
DNA transposons are mobile elements that contain inverted terminal repeats (ITRs), which are recognition sites for a transposase that cuts at the outside end of the inverted repeats and moves the excised DNA into a new site. Transposons have been used for insertional mutagenesis and gene transfer in many model organisms. However, applications in vertebrates were impeded due to the lack of active transposons until Tol2 was isolated from the Japanese Medaka fish Oryzias latipes (21,22) and Sleeping Beauty (SB) was reactivated from the salmon genome by the elimination of phylogenetically identified mutations (23,24). In 2005, PiggyBac transposon isolated from the cabbage looper moth Trichoplusia ni was reported to be active in mammalian cells including mouse and human (25). Consequently, several options for transposition in fish, mouse and human cells are now available. In particular, SB and PiggyBac appear most useful (26–31) and increased activity variants of both have been recently identified (32). Notably, transposase-mediated transgenesis has been used in cells that are difficult to transfect including human haematopoietic stem cells (32,33) and hESCs (34–36). Consequently, we were encouraged to examine whether BAC transgenesis in hESCs could be facilitated by transposition. However, transposons appear to have severe size limitations (37), which have limited their use for large transgenes.
During attempts to integrate large (up to 60 kb) transgenes into Myxococcus and Pseudomonas prokaryotic hosts, we encountered problems with fragmentation, which we solved by use of transposition (38). Furthermore, Tol2 transposition has been used to integrate a 66 kb transgene into zebrafish and mouse genomes (39). These studies indicate that fears about the size limitations of transposons may be misguided. Herein, we show that transposition can be applied to integrate full-length BACs larger than 150 kb into hESCs, which has implications for BAC transgenesis in general and particularly in systems that are difficult to work with.
MATERIALS AND METHODS
Generation of large reporter constructs and BAC reporters
The large constructs were made by subcloning from the respective BACs a region of 19 kb for hOCT4 gene and 25 kb for hNANOG into a plasmid with p15A origin of replication using recombineering technology (Supplementary Figure S1) (2,3). For the generation of large construct or BAC reporters, the green fluorescent protein (GFP) or Cherry cassettes were inserted directly after the initiating methionine (ATG) of the respective gene using recombineering. The PiggyBac or SB terminal repeats were inserted into different positions of the BAC backbone using a universal recombineering strategy applicable to most of the common used BAC vectors (Supplementary Figure S2). The recombineering details and list of oligos are presented in Supplementary Experimental Procedures.
hESC culturing
H7.S6 and H9 hESCs were cultured on mouse embryonic fibroblasts (MEFs) in DMEM/F12 medium supplemented with 20% Knockout Serum Replacement (Invitrogen) and 4 ng/ml basic fibroblast growth factor (bFGF) (Peprotech) and passaged using 1 mg/ml collagenase IV (Invitrogen) adding 10 µM Rho-associated kinase (ROCK) inhibitor Y-27632 (40). For transfections and differentiation assays, the cells were transferred to feeder-free conditions on Matrigel (BD Biosciences) in MEF-conditioned hESC medium, and propagated using TrypLE (Invitrogen).
Transfections of hESCs
Electroporation of large constructs into hESCs was performed according to the standard protocol at 320 V and 250 µF (15). BAC transfection was performed either by nucleofection (20) or lipofection. hOCT4-GFP, hNANOG-GFP, hPAX6-GFP and hGATA4-GFP BACs were prepared using Nucleobond BAC 100 kit (Macherey-Nagel). Nucleofection was done in 100 µl of solution V using program B-016 according to manufacturer protocol (Amaxa). 5 × 106 of cells were nucleofected with 5 µg of the BAC and 300 ng of the transposase expression or control vector.
For lipofection, hESCs were split to Matrigel-coated dishes in the ratio 1:3, 1 day before transfection. 3, 10, 30 or 50 µg of BAC and 3 or 10 µg of the transposase expression or control vector were used for lipofection of a 10 cm dish with hESCs using Lipofectamine LTX (Invitrogen) according to manufacturer protocol.
Selection with G418 (100 µg/ml; Invitrogen), puromycin (0.5 µg/ml; Sigma) or blasticidin (2 µg/ml; Invitrogen) started 2 days after transfection. After 14 days of selection, stable resistant clones were picked to 96-well plates and expanded.
Polymerase chain reaction analysis of hESC clones
Genomic DNA from the hESCs clones was prepared directly in 96-well plates and used for screening by polymerase chain reaction (PCR) for the presence of transposon inverted repeats and loss of ampicillin/spectinomycin cassette that occurs during transposition. The clones that contained a BAC integrated by transposase according to PCR analysis (PB5+ Amp− PB3+ or SB5+ Spec− SB3+) were checked for the BAC copy number by quantitative PCR (qPCR). Five to six pairs of primers were designed along each BAC randomly with the distance ∼30–40 kb between primer pairs (listed in Supplementary Experimental Procedures). The copy number was calculated by normalization of Ct-values for each primer pair to GAPDH gene relatively to wild-type cells, which contain two allelic copies of each genomic region.
Splinkerette PCR
The integration sites of BACs in the clones were determined using splinkerette PCR (41). Genomic DNA digested with ApoI or BstYI was ligated to 75 nM splinkerette adaptor (SPLK-A and SPLK-ApoI/BstYI). The 5′- and 3′-junctions were amplified using nested PCR and sequenced.
Differentiation of hESCs
For differentiation of H7.S6 hOCT4-GFP clones, bFGF was removed from the cultured medium for 10 days and then GFP expression was analysed by flow cytometry.
H7.S6 hPAX6-GFP clones and wild-type cells were differentiated to neural epithelial cells (42,43). Embryoid bodies (EBs) were formed in N2B27 medium with 10 µM transforming growth factor beta (TGFβ) receptor inhibitor SB431542 (Tocris). After 8 days, EBs were spread to tissue culture dishes coated with 100 µg/ml polyornithine and 10 µg/ml laminin (Sigma–Aldrich). Expression of PAX6 and GFP was checked by flow cytometry or immunocytochemistry on Day 14 of differentiation.
H7.S6 hGATA4-GFP clones and wild-type cells were differentiated to definitive endoderm (44). RPMI supplemented with B27 (1×), 1 mM sodium butyrate, 100 ng/ml activin A and 25 ng/ml Wnt3a (both from Peprotech) was used for the first day of differentiation. Next day, Wnt3a was omitted from the medium, and the cells were cultured further in RPMI with B27 (1×), 0.5 mM sodium butyrate and 100 ng/ml activin A. On Day 7, the cells were analysed for GFP and CXCR4 expression by flow cytometry, and for markers expression by qPCR.
Immunostaining and microscopy
The incubation with primary antibodies was for 1 h at room temperature with mouse anti-Oct4 (1:50, sc-5279; Santa Cruz), rabbit anti-Nanog (1:30, AB5731; Chemicon) or overnight at +4°C with mouse anti-Pax6 (1:30; Developmental Studies Hybridoma Bank). The cells were incubated with secondary antibodies diluted 1:500 (FITC goat anti-rabbit and TRITC goat anti-mouse; Jackson Immunoresearch Laboratories and Alexa633 goat anti-mouse; Molecular Probes, Invitrogen) for 1 h at room temperature. Fluorescence images were taken using Leica SP5 laser scanning confocal microscope.
Flow cytometry
The cells were dissociated and fixed in phosphate buffered saline (PBS) with 1% formaldehyde. For antibody staining, 106 of live cells were incubated with R-Phycoerythrin (PE)-conjugated anti-CD184 or immunoglobulin G (IgG) isotype control (BD Biosciences) diluted 1:100 in PBS with 2% fetal calf serum (FCS) for 30 min at +4°C. The cells were analysed with flow cytometer LSR II (Becton Dickinson) using FACSDiva software. The data were processed with FloJo software.
RESULTS
Generation of OCT4 and NANOG fluorescent reporter H7.S6 cells using large constructs
To study pluripotency, lineage commitment and differentiation pathways in hESCs, we aimed to create a panel of reporter cell lines based on the expression of fluorescent proteins under control of selected promoters. Using recombineering, genomic regions containing the OCT4 and NANOG genes (19 and 25 kb, respectively) were subcloned from BACs and GFP or mCherry IRES neomycin cassettes were inserted at the initiating methionine codon (Figure 1a and Supplementary Figure S1). Stable H7.S6 hESC clones were established after electroporation and G418 selection. However, we failed to obtain clones with uniform expression of the reporters, despite the fact that OCT4 and NANOG are expressed in undifferentiated hESCs. All clones (n = 8) showed mosaic expression (59.9–85.0% positive cells by flow cytometry), which was further reduced when G418 selection pressure was removed. However, the non-fluorescent cells were not differentiated, as shown by staining with OCT4 and NANOG antibodies (Figure 1b). We also generated double stable reporter lines after a second round of electroporation. The double reporter clones H7.S6 OCT4-mCherry/OCT4-GFP and NANOG-mCherry/NANOG-GFP also displayed mosaic expression of both fluorescent transgenes, which only partially overlapped (Figure 1c), indicating that even these relatively large transgenes undergo random silencing.
Figure 1.
H7.S6 OCT4 and NANOG reporter lines created by existing transgenic methods show mosaic expression, highlighting the need for BAC transgenes. (a) Stable H7.S6 hESC clones carrying 19 kb OCT4 or 25 kb NANOG reporter transgenes to express mCherry or GFP IRES neo reporter cassettes (see also Supplementary Figure S1), exhibited mosaic expression as determined by fluorescent imaging (left panels) or flow cytometry (FACS scans at the right). Fluorescent protein expression from the transgenes fell upon removal of G418 selection (compare +G418 with –G418 FACS panels). (b) Immunostaining of H7.S6 OCT4-mCherry and NANOG-mCherry reporter lines (63×, zoom) for endogenous OCT4 or NANOG expression showed that most cells expressed the endogenous proteins but many did not express the fluorescent reporter (arrowheads), indicating that the mosaic expression was due to silencing of the transgene and not differentiation of the cells. (c) Double stable reporter H7.S6 OCT4-mCherry/OCT4-GFP and NANOG-mCherry/NANOG-GFP lines were generated after transfecting the single OCT4 and NANOG reporters above. Both OCT4 and NANOG double mCherry/GFP reporters showed partially overlapping mosaicism, indicating random silencing of the reporter. Arrows and asterisk show cells that exhibited only GFP or mCherry fluorescence, respectively.
Generation of hESC BAC reporters using PiggyBac transposition
To circumvent silencing, we decided to use BAC transgenes. BACs containing OCT4 and NANOG genes were modified with the GFP-IRES-neo-pA reporter cassettes and stable hESC clones were established using nucleofection (20). However, once again we observed mosaic expression (data not shown). Because small transposons have been shown to mediate efficient gene transfer in mESC, hESC and human cell lines (25,29,34–36), we decided to evaluate whether transposition could be applied to BAC transgenesis. Among several transposons, we first chose PiggyBac based on its reported efficiencies in mammalian cells (26,45). The OCT4 and NANOG BACs were further recombineered to insert a cassette into the BAC backbone that contained the PiggyBac ITRs (5′-313 bp, 3′-235 bp (46)); flanking an ampicillin resistance gene. For this purpose, we built a recombineering cassette in an R6K plasmid so that PacI or PacI/AscI restriction digestion releases a fragment that will recombine with most common human and mouse BAC vectors; pBACe3.6, pBeloBAC11, pTARBAC1, pTARBAC1.3, pTARBAC2, pTARBAC2.1 and pTARBAC6 (Figure 2a and Supplementary Figure S2). Then, the OCT4-GFP and NANOG-GFP BACs were transfected into H7.S6 hESCs using either nucleofection or lipofection, with or without co-transfected codon optimized PiggyBac transposase expression plasmid, mPBase (47). Without co-transfected mPBase, both transfection methods produced a similar number of resistant colonies (22 and 16 clones for OCT4-GFP and 5 and 4 clones for NANOG-GFP per 107 transfected cells; Table 1 and Supplementary Table S1). With nucleofection, co-transfection of mPBase did not increase the colony number (29 resistant clones for OCT4-GFP and 3 clones for NANOG-GFP). However, co-lipofection of mPBase produced more colonies for both BACs (413 for OCT4-GFP and 14 for NANOG-GFP).
Figure 2.
BAC transgenesis using PiggyBac transposition. (a) Human BACs were modified by recombineering with GFP reporter cassettes that were inserted directly after the start codon (ATG) and contained a selectable marker expressed either by the gene promoter (for genes expressed in hESCs), or by PGK promoter (for genes that are not expressed in hESCs). The figure shows the hNANOG example, which is expressed in hESCs. A second recombineering step inserted a standardized cassette containing PiggyBac ITRs (PB5 and PB3) flanking the ampicillin resistance gene (Amp) into the BAC backbone. The PiggyBac ITR/ampicillin cassette was cloned into an R6K vector so that PacI/AscI digestion releases a restriction fragment that is flanked by homology regions that will recombine with most BAC vectors. The modified BACs were co-transfected with a PiggyBac transposase (mPBase) expression plasmid into hESCs. (b) Transposition of the BAC by PiggyBac will be full-length, flanked by the ITRs and ampR will be omitted. Hence, PCR assays for the presence of PB5 and PB3 with simultaneous loss of Amp indicates transposition. The copy number of the BAC was determined by quantitative, allele counting PCR (qPCR) on the genomic DNA using 5–6 primer pairs at about 30–40 kb intervals along the BAC (a–e).
Table 1.
Summary of BAC transgenesis and transpositions. H7.S6 and H9 hES clones were screened for the PiggyBac transpositional signature by PCR after nucleofection or lipofection with or without a PBase expression construct (either mPBase or hyPBase) as indicated
| Transposition events (PB3+ Amp− PB5+) | N analysed | N total | |||
|---|---|---|---|---|---|
| Nucleofection | H7.S6 OCT4 (IRES-neo) 149 kb | BAC | 0 | 10 | 22 |
| BAC+mPBase | 0 | 18 | 29 | ||
| H7.S6 NANOG (IRES-neo) 164 kb | BAC | 0 | 1 | 5 | |
| BAC+mPBase | 0 | 2 | 3 | ||
| Lipofection | H7.S6 OCT4 (IRES-neo) 149 kb | BAC | 0 | 12 | 16 |
| BAC+mPBase | 9 (18.8%) | 48 | 413 | ||
| H7.S6 NANOG (IRES-neo) 164 kb | BAC | 0 | 2 | 4 | |
| BAC+mPBase | 6 (75%) | 8 | 14 | ||
| H7.S6 GATA4 (PGK-neo) 196 kb | BAC | 0 | 20 | 130 | |
| BAC+mPBase | 16 (36.4%) | 44 | 222 | ||
| H7.S6 PAX6 (PGK-neo) 150 kb | BAC | 0 | 24 | 120 | |
| BAC+mPBase | 17 (37.8%) | 45 | 200 | ||
| BAC+hyPBase | 21 (61.7%) | 34 | 454 | ||
| H7.S6 PAX6 (PGK-neo) ITR-backbone 150 kb | BAC | 0 | 8 | 8 | |
| BAC+mPBase | 3 (11.5%) | 26 | 35 | ||
| BAC+hyPBase | 28 (50.0%) | 56 | 83 | ||
| H7.S6 PAX6 (UbiC-BSD) ITR-backbone 150 kb | BAC+hyPBase | 41 (43.2%) | 95 | 240 | |
| H9 PAX6 (PGK-neo) 150 kb | BAC+hyPBase | 55 (57.3%) | 96 | 201 |
All BACs contained a GFP reporter integrated at the initiating ATG codon and the antibiotic resistance gene for selection either under an IRES (for OCT4 and NANOG) or expressed from the PGK or UbiC promoter (for GATA4 and PAX6). The BAC sizes are indicated. The PiggyBac inverted repeats either flanked the AmpR gene (1 kb apart) or the whole BAC backbone (9.5 kb apart—indicated as ‘ITR backbone’). The data in the transpositional events column show the number and percentage of clones positive for the transpositional signature. ‘N analysed’ presents the number of clones that were screened for the transpositional signature and ‘N total’ presents the yield of clones per 107 transfected cells in that experiment.
To check whether the BACs had been integrated by transposition, we established a PCR-based strategy for colony screening based on primers directed to the PiggyBac ITRs (PB5 and PB3) and the ampicillin (Amp) gene (Figure 2b). Transposition should integrate the ITRs yet exclude the ampicillin gene, whereas random integration with or without fragmentation could give any combination of PCR signals. Most of the colonies established by nucleofection, with or without mPBase, contained either the whole PiggyBac cassette (PB3+ Amp+ PB5+) or none of it (PB3− Amp− PB5−). The same was observed for the lipofection of the BACs alone (Table 1 and Supplementary Table S1). Notably, only lipofection with mPBase resulted in clones that had the signature of transposition (PB3+ Amp− PB5+; 18.8% of OCT4-GFP and 75% of NANOG-GFP clones). Characterization of clones obtained from nucleofection with or without mPBase indicated that all contained only limited pieces of the BAC and none of them was due to transposition, suggesting that nucleofection breaks BACs.
Given these encouraging results, we generated BAC transposon GFP reporters for the lineage-specific genes PAX6 and GATA4 and lipofected them into H7.S6 cells with or without mPBase. Because these genes are not expressed in hESCs, the IRES could not be used to drive neomycin resistance so we used the PGK promoter, which consequently provided similar efficiencies for both transfections (Table 1 and Supplementary Table S1). Application of the mPBase resulted in 1.7-fold increase of the colony number for both GATA4 and PAX6 BACs, and about one-third of those clones gave the transpositional signature. The use of the hyperactive version of PBase, hyPBase (48), further increased the total number of colonies and the proportion of transpositional events (61.7%). These lipofection results were essentially reproduced using another hESC line, H9 (Table 1 and Supplementary Table S1), which is more difficult to transfect (our unpublished data). The optimum ratio of BAC transgene to PBase expression vector was evaluated (Supplementary Table S2). In most cases, 10–30 µg BAC with 3–10 µg PBase per 10 cm dish (∼3 × 106 cells), which corresponds to molar ratios of BAC to PBase from 1:9 to 1:30, gave the most colonies.
Furthermore, we generated a PAX6-GFP reporter BAC that contained PiggyBac ITRs flanking the vector backbone (ca. 9.5 kb apart from each other). This version also contained the Blasticidin resistance gene driven by the ubiquitin C promoter at one end of the genomic sequence next to the PiggyBac 3′-ITR (Supplementary Figure S2). Transposition by PBase (mPBase or hyPBase) resulted in clones that contained integrated BAC without the vector backbone, albeit with apparent lower efficiency. Thus, PiggyBac transposition can be used to exclude integration of the prokaryotic vector sequences into the genome.
Copy number of BAC integrations
To further characterize the BAC integrations, we analysed OCT4-GFP, GATA4-GFP and PAX6-GFP hESC clones for transgene copy number using qPCR on genomic DNA. Primer pairs were selected across the full-length BAC at a distance of 30–40 kb from each other (Figures 2b and 3). The Ct-values were normalized to wild-type DNA to calculate the number of additional copies arising from the integrated transgenes.
Figure 3.
BAC transgene copy number and integrity in H7.S6 hESC clones containing OCT4-, GATA4- and PAX6-GFP reporters. Copy number was evaluated by qPCR assays similar to that illustrated in Figure 2b. (a) Results from 17 clones that were positive for the transpositional signature (PB5+ Amp− PB3+) are depicted. (b) Results from clones that did not present the transpositional signature, including nine from experiments with co-lipofected transposase expression plasmid (+mPBase) and 10 from experiments without co-transfected mPBase (−mPBase). The bars show the additional copy number for each primer pair in the clones and the transpositional signature results are shown below.
We analysed 17 clones positive for the transpositional signature (PB5+ Amp− PB3+; Figure 3). Most clones showed single-copy signals for all primer pairs indicative of a single transpositional event (12/17; 70.5%). A further three clones showed full-length, multi-copy integrations (three, two and four copies) suggesting multiple transpositional events. The remaining two clones showed signs of a partial integration of a second copy in addition to a single full-length copy, suggesting a combination of a single transpositional event and a partial random event. In addition, we characterized 19 clones that did not present the transpositional signature (either PB5+ Amp+ PB3+ or PB5− Amp− PB3−; Figure 3b). Most of these clones had missing or inconsistent signals for at least one primer pair indicating random integration of fragments.
Analysis of integration sites of the BACs
The integration sites were examined by splinkerette PCR and sequencing (41). We analysed 21 H7.S6 clones (including OCT4-GFP, OCT4-mCherry, NANOG-GFP, PAX6-GFP, SOX1-GFP and GATA4-GFP) and 4 H9 clones that were positive for the transpositional signature. In all cases, the integration locus was continuous on the 5′- and 3′-sides of a duplicated TTAA sequence (Table 2), which is characteristic of PiggyBac insertion sites (49). As controls we analysed the junctions of PiggyBac ITRs in several PB3+ Amp+ PB5+ clones that were established by transfection of the BAC without mPBase. As expected, the PiggyBac ITRs were followed by the ampicillin gene sequence and not genomic DNA, consistent with random integration of the BAC (data not shown).
Table 2.
Analysis of integration sites of the BACs in stable transfected clones (H7.S6 or H9 when indicated)
| Clone | Chrom | Locus genomic contig GRCh37.p2 | 5′-Junction | 3′-Junction | |
|---|---|---|---|---|---|
| PAX6-GFP | T3 | 3 | NT 022517.18 (pos. 8137952) intergenic | CGCTGAGACTTTAAccctagaaag | ctttctagggTTAACAAGATGGCA |
| T4 | X | NT 011651.17 (pos. 30782387) collagen a6 (IV) | CAACAGTTGCTTAAccctagaaag | ctttctagggTTAATGGTACAGAA | |
| T6 | 3 | NT 005612.16 (pos. 74499330) intergenic | CCCCGTATCCTTAAccctagaaag | ctttctagggTTAAAAGAGGGGAG | |
| T5 | 10 | NT 030772.10 (pos. 544627) intergenic | TATCAGTATCTTAAccctagaaag | ctttctagggTTAAGAGCTTGTTC | |
| T9 (H9) | X | NT 167197.1 (pos. 31005589) dystrophin | GGAATGCAAATTAAccctagaaag | ctttctagggTTAAAACCACAATG | |
| T54 (H9) | 20 | NT 011387.8 (pos. 12184592) intergenic | TCAGTTCTTATTAAccctagaaag | ctttctagggTTAATTGAGGGACC | |
| T93 (H9) | 22 | NT 011520.12 (pos. 16525887) intergenic | CCAGTCTTTTTTAAccctagaaag | ctttctagggTTAATAAGGACAAT | |
| SOX1-GFP | T5 | 6 | NT 007592.15 (pos. 46428206) intergenic | ATAATTTTCTTTAAccctagaaag | ctttctagggTTAAAAGAATGACA |
| T6 | 1 | NT 167186.1 (pos. 5485465) LPGAT1 | TGAAACTGCTTTAAccctagaaag | ctttctagggTTAATATCTTCTCC | |
| T11 | 10 | NT 008818.16 (pos. 5518212) intergenic | GGCCCTTTCGTTAAccctagaaag | ctttctagggTTAAGCACCACTCA | |
| T13 | Xq22.3-24 | NT 011651.17 (pos. 34049525) intergenic | TCAACAAATCTTAAccctagaaag | ctttctagggTTAATGCTGTTGCT | |
| T12 | 14 | NT 026437.12 (pos. 66717241) intergenic | AATGCTGCAGTTAAccctagaaag | ctttctagggTTAACAGCCCAACA | |
| GATA4-GFP | T2 | 21q21.1-q21.2 | NT 011512.11 (pos. 4443106) intergenic | GAGCATTTCCTTAAccctagaaag | ctttctagggTTAACGATCATTCA |
| T11 | Xq25-26.3 | NT 011786.16 (pos. 15279393) intergenic | ATGCATCCTTTTAAccctagaaag | ctttctagggTTAAATTCCCATCA | |
| NANOG-GFP | T2 | 11 | NT 033899.8 (pos. 38119006) intergenic | CCTTACTATCTTAAccctagaaag | ctttctagggTTAACAGAGATTTT |
| T3 | 21q22.2 | NT 011512.11 (pos. 22449081) intergenic | TTTGTTTTTGTTAAccctagaaag | ctttctagggTTAAACATGACCGA | |
| T4 | 2 | NT 022171.15 (pos. 1305535) ankyrin repeat prot | TAAATTTTTTTTAAccctagaaag | ctttctagggTTAAAAAAGAAAAA | |
| OCT4-Cherry | T1 | 5 | NT 006713.15 (pos. 5958679) intergenic | GTGCCCAGCCTTAAccctagaaag | ctttctagggTTAAAGTACTTTTT |
| T2 | 1 | NT 032977.9 (pos. 62226542) Tgfb receptor III a | AATTCTTTTTTTAAccctagaaag | ctttctagggTTAAAGCCAAGGGC | |
| T9 | 1 | NT 032977.9 (pos. 18736795) SPATA 6 | GCCTCCCCTTTTAAccctagaaag | ctttctagggTTAAAATGTCCCTC | |
| OCT4-GFP | T2 | 12 | NT 029419.12 (pos. 14861721) keratin 83 (KRT83) | TTGTGCCTTCTTAAccctagaaag | ctttctagggTTAATGGATTTTTG |
| T4 | 5 | NT 006713.15 (pos. 28880477) ARSB | TATAATATGCTTAAccctagaaag | ctttctagggTTAATTTCCACATA | |
| T11 | 3 | NT 005612.16 (pos. 95506050) tumour protein p63-regulated gene 1 protein | GTTGCCAACTTTAAccctagaaag | ctttctagggTTAAAGAAAAGAAA | |
| T5 | 6 | HLA | ATTCTTTTCTTTAAccctagaaag | ctttctagggTTAAGAATGTTGAA | |
| T1 (H9) | 5 | NT 023133.13 (pos. 15037738) GABA receptor, pi (intron 7) | TCCTGACTTTTTAAccctagaaag | ctttctagggTTAATGATTGCCAT | |
The junctions of PiggyBac terminal repeats with genomic DNA were amplified using splinkerette PCR and sequenced. In all cases, the site of integration was a single genomic TTAA duplicated at each end of the BAC transgene. Genomic sequences are shown in upper cases, transposon ITRs in lower cases and duplicated TTAA sequences are highlighted in bold.
Previous studies showed that the PiggyBac transposon can integrate into any chromosome with a preference for AT-rich sequences within 10 kb from transcription units (25,29,34). We analysed the sites of integrations of the BACs and they were either located in intergenic (n = 14) or intronic regions (n = 11) (Table 2) without any obvious chromosomal preference or DNA consensus around the TTAA site of the integrations (Supplementary Figure S3).
Functionality of the reporter clones
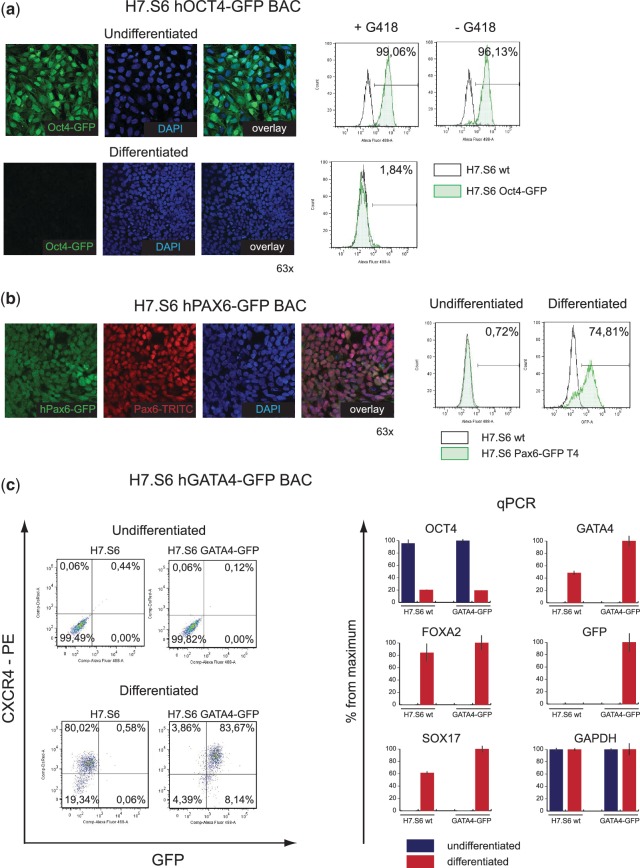
The integrity of the reporter BAC transgenes after transposition into hESCs can be functionally evaluated. Various clones were examined for the stability and pattern of expression before and after differentiation. Using single-copy OCT4-GFP clones integrated by mPBase (n = 7), we found that all clones uniformly expressed GFP (99%) in the undifferentiated state in the presence of G418 and notably after >1 month of culture without G418 selection pressure (96.13%; Figure 4a). The clones were induced to differentiate by bFGF removal (50,51) leading to the loss of GFP expression after 8 days. Hence, the OCT4-GFP BAC transgenes integrated by transposition are functional, reliable and not prone to position effects.
Figure 4.
Validation of H7.S6 BAC transposon reporter lines. H7.S6 clones containing verified PiggyBac BAC transpositions for OCT4-GFP, PAX6-GFP and GATA4-GFP reporters were analyzed. (a) An H7.S6 OCT4-GFP reporter clone showed homogeneous expression of the transgene as evaluated by immunofluorescence (panels at left) and flow cytometry (+G418 FACS panel). GFP expression was without selection pressure in undifferentiated cells (−G418 FACS panel). After differentiation for 8 days, GFP expression was uniformly down-regulated (bottom right FACS panel). (b) An H7.S6 PAX6-GFP reporter clone did not express any fluorescence before differentiation (FACS panel—undifferentiated). After 10 days of differentiation to neural epithelial cells, most cells in rosette-like structures (74.81%; FACS panel—differentiated) expressed GFP, which co-localized with endogenous Pax6 by immunofluorescence. (c) An H7.S6 GATA4-GFP reporter clone and the parental H7S6 line were differentiated to definitive endoderm cells for 7 days and gene expression was compared with undifferentiated cells for selected genes. CXCR4 expression was evaluated by FACS (left hand panels). OCT4, GATA4, FOXA2, GFP, SOX17 and GAPDH levels were compared by qRT–PCR (right hand panels).
To validate lineage reporters for the genes that are not expressed in hESCs, we used PAX6-GFP and GATA4-GFP BAC transfected clones, created by transposition. H7.S6 PAX6-GFP cells were differentiated into neural epithelium cells in N2B27 through EBs in the presence of TGFβ receptor inhibitor (42,43). The attached EBs formed neural rosettes, which is the characteristic phenotype for neural progenitor cells. 74.81% of the cells were GFP positive and co-expressed GFP with endogenous PAX6 as shown by flow cytometry and immunostaining (Figure 4b).
H7.S6 GATA4-GFP BAC reporter cells were used to generate definitive endoderm by treatment with a high concentration of activin A (44). Quantitative reverse transcription–PCR (qRT–PCR) confirmed up-regulation of the endodermal markers GATA4, SOX17 and FOXA2 after 7 days of differentiation (Figure 4c). Most (83.67%) cells were GFP positive and co-expressed the marker of definitive endoderm CXCR4 shown by flow cytometry. Interestingly, the dynamic of the reporter expression closely reproduced recent observations on definitive endoderm differentiation, which showed that GATA4 expression preceeds CXCR4 (52) (data not shown). These data support the conclusion that the BAC transposon reporters are functional and faithfully reproduce endogenous patterns of gene expression during differentiation.
Sleeping Beauty BAC transposition
To determine whether the ability to transpose a very large cargo was a feature of PiggyBac, we undertook BAC transposon experiments with SB (Supplementary Figure S4). SB ITRs, separated by a spectinomycin resistance gene, were inserted into the PAX6-GFP BAC vector and co-lipofected with SB100xco transposase (a human codon optimized version provided by Zsuzsanna Izsvak). The combination of BAC and transposase expression plasmid resulted in >20-fold increase in the colony number compared with the control co-transfection of BAC with catalytic mutant of transposase (D3 construct). PCR screening identified the clones with transpositional signature (23/32; 72%) and most of them contained a single copy of the BAC. We mapped the transgene positions in three clones and confirmed a TA integration site that is characteristic for SB transposition (Supplementary Figure S4).
DISCUSSION
All genomes from bacterial to mammalian have been bombarded by transposons. Consequently, transposons have been widely successful as transgenic vehicles and genetic tools in many cell types and organisms (7,8,24). However, applications have been almost entirely limited to transposons with small cargos due to the facts that the majority of natural transposons are <2 kb long and larger cargos appear to suppress transposition. Indeed, many studies have described an inverse length dependence of transpositional efficiency in the range of 30–50% per each extra kilobase of transposon size (37,53–55). In apparent contradiction to this literature, transpositions of ∼60 kb transgenes were reported using ‘MycoMar’ transposon in prokaryotic hosts by our group (38) and Tol2 in zebrafish and mice by others (39). Furthermore, recent model experiments in mESCs reported BAC transpositions up to 100 kb using PiggyBac (56). The question therefore arises: if transposition with very large cargos is possible, how can this apparent contradiction be resolved? The answer may lie with the suggestion that transposon length dependence is determined by the shortest distance between the ITRs, regardless of the orientation inside or outside the transposon (57). In this case, the transposons reported here are efficient because the ITRs are only 1 kb apart and the transposase is largely indifferent to the 100 kb+ size of the cargo (Supplementary Figure S5). Consistent with this proposition, we observed similar efficiencies of transposition for the BACs that contained the same cassette for the selection of resistant clones (PGK-neo-pA), regardless of the cargo size difference (PAX6, 150 kb and GATA4, 196 kb; Figure 2 and Table 1). Furthermore, experiments using the BACs with increased distance between the ITRs (when placed at either end of the BAC vector to achieve transgene integration without the inclusion of prokaryotic vector sequences) indeed indicate a reduced efficiency (about 2–3-fold reduced when the ITRs were spaced about 9.5 kb apart; Table 1 and Supplementary Table S1). This explanation also accounts for the observation that very different types of transposases can mediate BAC transposition [PiggyBac, SB/MycoMar and Tol2 come from three distinct classes (23,25,58,59)]. It also suggests that there is no inherent limitation for transposon cargo size, which is an unanticipated conclusion.
The transposition mechanism implies that BAC transposons must be covalently closed circular molecules when transfected. It therefore follows that the quality of the BAC DNA preparation will affect the ratio of transposon to random integration events. Breaks in the BAC will tend to promote random transgenesis whereas covalently closed circles will favour transposition. In line with this point, we only achieved BAC transposition by lipofection and failed when nucleofection was used, apparently because nucleofection promotes BAC breakage. Whether electroporation also promotes breakage is unclear. We note that Li et al. (56) used electroporation to achieve BAC transpositions. It is also notable that Li et al. achieved transpositions of BACs with the ITRs separated by large distances. Because they use more elaborate protocols, involving transfection of the PiggyBac expression plasmid 3 days before electroporation of the BAC, followed by positive and negative selection for transposition, it is not possible to deduce efficiencies that could be compared with those reported here. We show that a much simpler protocol based on co-lipofection and positive selection only is sufficient to achieve satisfactory frequencies of BAC transposition when the ITRs are within 1 or a few kb of each other.
BAC transgenesis by transposition brings three major advantages over the widely used methods for BAC transgenesis by random integration (60,61). For random integration, the BAC can be transfected either after linearization by restriction digestion or as uncut circles. Linearization is useful because it determines how the BAC integrates into the genome. In contrast the uncut BAC needs to break, which can occur anywhere, before integration. However, linearized BACs are more difficult to handle because they are prone to shearing. BAC transposition avoids this conundrum because the uncut BAC is used and the site of integration on the BAC is determined by the positions of the ITRs. Hence, the continuity of the integrated BAC can be assured. Furthermore, as a transposon, the BAC will be integrated as a single copy, which ensures physiological expression and avoidance of tandem repeat-associated silencing. (Note, as shown here, it is possible to obtain cells that have more than one integrated transposon; however these will almost always be independent events resulting in single copies integrated at different genomic sites.) Furthermore, advantages over random integration include the fact that transposition increases the frequency of transgenesis, which is particularly important for difficult systems such as hESCs and that the genomic integration site can be identified by a standard splinkerette assay based on the ITRs. Because the splinkerette assay is laborious, for ease of detection we developed a PCR assay for the transpositional signature (PB5+ Amp− PB3+), which can be applied for fast screening of the clones in a large scale. All clones positive for the signature that we also examined by splinkerette sequencing were bona fide transpositions. Hence, we suggest that BAC transpositions can be evaluated using this convenient PCR test with high confidence. Given these many advantages, and because we show that BAC transposition requires no additional transfection steps when compared with random transgenesis, we strongly recommend that all applications of BAC transgenesis now use transposition.
To facilitate this recommendation, we made PiggyBac and SB ITR cassettes flanked by homologies to the BAC vector in R6K plasmids for ease of recombineering. R6K vectors do not replicate in common Escherichia coli hosts; hence, the most common source of recombineering background is eliminated (62). These standardized ITR cassettes are released by restriction digestion with PacI and AscI, which permits asymmetric dephosphorylation so that beta recombination can be applied to enhance recombineering efficiency (63) (Supplementary Figure S2). Whereas this is not necessary for normal applications, it is relevant for high-throughput processing in recombineering pipelines (64–66), which can now be applied to existing BAC resources to rapidly convert them into transposons.
We generated a panel of fluorescent BAC reporter hESCs, including SOX1 (data not shown), OCT4, NANOG, GATA4 and PAX6. In almost all cases, the clones showed the correct expression properties quantitatively and stably. For example, all seven OCT4 BAC transposon hESC clones tested showed stable GFP expression in the undifferentiated state after prolonged passaging with or without selection pressure. This underlines the reliability of BAC transgenes when compared with, for example, the lines generated here using quite large constructs (19–25 kb). Generation of reporter lines for genes that are not expressed in hESCs is a particularly difficult task due to the silencing of the transgenes and so far this has been achieved only in few cases by gene targeting (67–71). Herein, we show with PAX6 and GATA4 hESC reporters that BAC transposition is a reliable way to establish hESC lines even for genes that are not expressed. To our knowledge, this is the first transgenic method that can be reliably applied to access the power of hESCs for developmental and disease-modelling studies.
Our work brings a further advance in BAC transgenesis in different organisms and cell types, especially those that are recalcitrant to genetic modifications such as hESCs. This technology includes the standardized insertion of transposon ITR cassettes into the BAC backbone using recombineering, co-delivery of the BAC with the transposase and fast PCR screening for transpositions. These steps are straightforward and achieve single-copy, full-length, BAC integrations in genomic loci that can be readily mapped, all of which we believe sets a new ideal for transgenesis. Our work also unravelled a fundamental property of transposition regarding the impact of cargo size that has been underestimated in the past.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–5, Supplementary Methods and Supplementary References [72–74].
FUNDING
European Union’s 7th Framework Program EUCOMMTOOLS [261492 to A.F.S.]; ESTOOLS [to K.A., A.J.H.S.]; Deutsche Forschungsgemeinschaft Research Center and Cluster of Excellence Center for Regenerative Therapies Dresden [to K.A.]; UK Medical Research Council Centre Grant Award [MRC/G0700711 to A.J.H.S.]. Funding for open access charge: EU 7th Framework Integrated project, EUCOMMTOOLS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alan Bradley (Sanger) for providing us with the mPB and hyPB expression and CAG-OSKM-puDtk vectors and Zsuzsanna Izsvak (Max Delbrück Center for Molecular Medicine, Berlin, Germany) for pT2BH vector and Sleeping Beauty SB100xco expression plasmid. H7.S6 hESCs were kindly provided by Peter Andrews (University of Sheffield, UK). We thank Anna Falk (University of Cambridge, UK) and Karolina Lundin (University of Helsinki, Finland) for the help with the protocols for hESC differentiation. PAX6 antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA.
REFERENCES
- 1.Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 4.Ciotta G, Hofemeister H, Maresca M, Fu J, Sarov M, Anastassiadis K, Stewart AF. Recombineering BAC transgenes for protein tagging. Methods. 2011;53:113–119. doi: 10.1016/j.ymeth.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 6.Johansson T, Broll I, Frenz T, Hemmers S, Becher B, Zeilhofer HU, Buch T. Building a zoo of mice for genetic analyses: a comprehensive protocol for the rapid generation of BAC transgenic mice. Genesis. 2010;48:264–280. doi: 10.1002/dvg.20612. [DOI] [PubMed] [Google Scholar]
- 7.Venken KJ, Bellen HJ. Transgenesis upgrades for Drosophila melanogaster. Development. 2007;134:3571–3584. doi: 10.1242/dev.005686. [DOI] [PubMed] [Google Scholar]
- 8.Kawakami K. Transposon tools and methods in zebrafish. Dev. Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 9.Montigny WJ, Phelps SF, Illenye S, Heintz NH. Parameters influencing high-efficiency transfection of bacterial artificial chromosomes into cultured mammalian cells. Biotechniques. 2003;35:796–807. doi: 10.2144/03354rr02. [DOI] [PubMed] [Google Scholar]
- 10.Heintz N. Analysis of mammalian central nervous system gene expression and function using bacterial artificial chromosome-mediated transgenesis. Hum. Mol. Genet. 2000;9:937–943. doi: 10.1093/hmg/9.6.937. [DOI] [PubMed] [Google Scholar]
- 11.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 13.Sarov M, Stewart AF. The best control for the specificity of RNAi. Trends Biotechnol. 2005;23:446–448. doi: 10.1016/j.tibtech.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 15.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 16.Braam SR, Denning C, van den Brink S, Kats P, Hochstenbach R, Passier R, Mummery CL. Improved genetic manipulation of human embryonic stem cells. Nat. Methods. 2008;5:389–392. doi: 10.1038/nmeth.1200. [DOI] [PubMed] [Google Scholar]
- 17.Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521–1528. doi: 10.1634/stemcells.2006-0634. [DOI] [PubMed] [Google Scholar]
- 20.Placantonakis DG, Tomishima MJ, Lafaille F, Desbordes SC, Jia F, Socci ND, Viale A, Lee H, Harrison N, Tabar V, et al. BAC transgenesis in human embryonic stem cells as a novel tool to define the human neural lineage. Stem Cells. 2009;27:521–532. doi: 10.1634/stemcells.2008-0884. [DOI] [PubMed] [Google Scholar]
- 21.Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H. Transposable element in fish. Nature. 1996;383:30. doi: 10.1038/383030a0. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 23.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 24.Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, Campos LS, Yusa K, Banerjee R, Li MA, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 29.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 30.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat. Rev. Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 31.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 32.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 33.Xue X, Huang X, Nodland SE, Mates L, Ma L, Izsvak Z, Ivics Z, LeBien TW, McIvor RS, Wagner JE, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- 34.Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 35.Wilber A, Linehan JL, Tian X, Woll PS, Morris JK, Belur LR, McIvor RS, Kaufman DS. Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells. 2007;25:2919–2927. doi: 10.1634/stemcells.2007-0026. [DOI] [PubMed] [Google Scholar]
- 36.Chen YT, Furushima K, Hou PS, Ku AT, Deng JM, Jang CW, Fang H, Adams HP, Kuo ML, Ho HN, et al. PiggyBac transposon-mediated, reversible gene transfer in human embryonic stem cells. Stem Cells Dev. 2010;19:763–771. doi: 10.1089/scd.2009.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 38.Fu J, Wenzel SC, Perlova O, Wang J, Gross F, Tang Z, Yin Y, Stewart AF, Muller R, Zhang Y. Efficient transfer of two large secondary metabolite pathway gene clusters into heterologous hosts by transposition. Nucleic Acids Res. 2008;36:e113. doi: 10.1093/nar/gkn499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Muguruma K, Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- 41.Horn C, Hansen J, Schnutgen F, Seisenberger C, Floss T, Irgang M, De-Zolt S, Wurst W, von Melchner H, Noppinger PR. Splinkerette PCR for more efficient characterization of gene trap events. Nat. Genet. 2007;39:933–934. doi: 10.1038/ng0807-933. [DOI] [PubMed] [Google Scholar]
- 42.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 44.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 45.Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Harrell RA, Handler AM, Beam T, Hennessy K, Fraser MJ., Jr piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol. Biol. 2005;14:17–30. doi: 10.1111/j.1365-2583.2004.00525.x. [DOI] [PubMed] [Google Scholar]
- 47.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl Acad. Sci. USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Lin G, Martins-Taylor K, Zeng H, Xu RH. Inhibition of caspase-mediated anoikis is critical for basic fibroblast growth factor-sustained culture of human pluripotent stem cells. J. Biol. Chem. 2009;284:34054–34064. doi: 10.1074/jbc.M109.052290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu RH, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan Y, Ouyang Z, Wong WH, Baker JC. A new FACS approach isolates hESC derived endoderm using transcription factors. PLoS One. 2011;6:e17536. doi: 10.1371/journal.pone.0017536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandler M, Clerget M, Galas DJ. The transposition frequency of IS1-flanked transposons is a function of their size. J. Mol. Biol. 1982;154:229–243. doi: 10.1016/0022-2836(82)90062-6. [DOI] [PubMed] [Google Scholar]
- 55.Morisato D, Way JC, Kim HJ, Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983;32:799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- 56.Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, Rad L, Fitzgerald TW, Craig NL, Bradley A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Way JC, Kleckner N. Transposition of plasmid-borne Tn10 elements does not exhibit simple length-dependence. Genetics. 1985;111:705–713. doi: 10.1093/genetics/111.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097–1109. doi: 10.1038/emboj.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brillet B, Bigot Y, Auge-Gouillou C. Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains. Genetica. 2007;130:105–120. doi: 10.1007/s10709-006-0025-2. [DOI] [PubMed] [Google Scholar]
- 60.Vintersten K, Testa G, Stewart AF. Microinjection of BAC DNA into the pronuclei of fertilized mouse oocytes. Methods Mol. Biol. 2004;256:141–158. doi: 10.1385/1-59259-753-X:141. [DOI] [PubMed] [Google Scholar]
- 61.Hofemeister H, Ciotta G, Fu J, Seibert PM, Schulz A, Maresca M, Sarov M, Anastassiadis K, Stewart AF. Recombineering, transfection, Western, IP and ChIP methods for protein tagging via gene targeting or BAC transgenesis. Methods. 2011;53:437–452. doi: 10.1016/j.ymeth.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Sarov M, Rientjes J, Fu J, Hollak H, Kranz H, Xie W, Stewart AF, Zhang Y. An improved recombineering approach by adding RecA to lambda Red recombination. Mol. Biotechnol. 2006;32:43–53. doi: 10.1385/mb:32:1:043. [DOI] [PubMed] [Google Scholar]
- 63.Maresca M, Erler A, Fu J, Friedrich A, Zhang Y, Stewart AF. Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol. 2010;11:54. doi: 10.1186/1471-2199-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y, Hyman AA, Stewart AF. A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat. Methods. 2006;3:839–844. doi: 10.1038/nmeth933. [DOI] [PubMed] [Google Scholar]
- 65.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- 68.Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, et al. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang P, Rodriguez RT, Wang J, Ghodasara A, Kim SK. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goulburn AL, Alden D, Davis RP, Micallef SJ, Ng ES, Yu QC, Lim SM, Soh CL, Elliott DA, Hatzistavrou T, et al. A targeted NKX2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells. 2011;29:462–473. doi: 10.1002/stem.587. [DOI] [PubMed] [Google Scholar]
- 71.Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- 72.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ringrose L, Chabanis S, Angrand PO, Woodroofe C, Stewart AF. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 1999;18:6630–6641. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat. Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.