Summary
Lifelong blood cell production is governed through the poorly understood integration of cell-intrinsic and -extrinsic control of hematopoietic stem cell (HSC) quiescence and activation. MicroRNAs (miRNAs) coordinately regulate multiple targets within signaling networks, making them attractive candidate HSC regulators. We report that miR-126, a miRNA expressed in HSC and early progenitors, plays a pivotal role in restraining cell-cycle progression of HSC in vitro and in vivo. miR-126 knockdown by using lentiviral sponges increased HSC proliferation without inducing exhaustion, resulting in expansion of mouse and human long-term repopulating HSC. Conversely, enforced miR-126 expression impaired cell-cycle entry, leading to progressively reduced hematopoietic contribution. In HSC/early progenitors, miR-126 regulates multiple targets within the PI3K/AKT/GSK3β pathway, attenuating signal transduction in response to extrinsic signals. These data establish that miR-126 sets a threshold for HSC activation and thus governs HSC pool size, demonstrating the importance of miRNA in the control of HSC function.
Graphical Abstract
Highlights
► miR-126 is a novel regulator of the HSC quiescence/proliferation equilibrium ► Reduction in miR-126 induces an expansion of long-term HSC without exhaustion ► Constitutive miR-126 expression promotes HSC quiescence and progenitor proliferation ► miR-126 attenuates PI3K/AKT activation in response to cytokine stimulation
miR-126 regulates multiple targets within the PI3K/AKT/GSK3β pathway to promote HSC quiescence and progenitor proliferation.
Introduction
Blood cell production is sustained for life by the continuous differentiation of multipotent hematopoietic stem cells (HSC) into at least ten distinct lineages of mature blood cells (Doulatov et al., 2010). This is achieved by balancing self-renewal and differentiation among proliferating HSC. Additionally, some HSC are maintained in a quiescent state to protect against proliferative exhaustion, yet remain poised for activation (Wilson et al., 2008). This homeostatic balance between quiescence, proliferation, and differentiation is tightly controlled by integrating intrinsic and extrinsic mechanisms that govern the HSC state. Little is known about posttranscriptional programs that aid in establishing and maintaining the quiescence-activation equilibrium in HSC.
MicroRNAs (miRNA) are now recognized as fundamental effectors of posttranscriptional gene expression control. miRNA regulate gene expression by binding to complementary sequences within multiple target messenger RNAs (mRNAs), inducing mRNA destabilization and translational inhibition (Wilson et al., 2008; Bartel, 2009).
Several studies have shown important roles for miRNA in lineage commitment at the level of restricted hematopoietic progenitors (Havelange and Garzon, 2010). Genetic ablation of Dicer or Ars2, both principal factors for miRNA biogenesis, induces bone marrow failure, suggesting that normal HSC are dependent on miRNA function (Gruber et al., 2009; Guo et al., 2010). Recently, two miRNAs have been investigated in murine HSC. Enforced expression of miR-29a induced aberrant self-renewal in downstream progenitors, resulting in a low penetrant acute myeloid leukemia (AML) disease (Han et al., 2010). Enforced expression of miR-125b induced an initial myeloproliferative disorder, leading to frank AML or an increase in lymphoid-biased HSC, depending upon the ectopic expression levels (O’Connell et al., 2010; Ooi et al., 2010). Guo et al. (2010) showed that enforced expression of miR-125a could also augment HSC activity in vitro and expand the HSC pool in vivo. Although these studies indicate that miRNAs may be important regulators of hematopoiesis, miRNA loss-of-function studies are needed to ascertain whether miR-125 is required for normal HSC function. Moreover, these studies were performed in murine models while the relevance to human HSC remains unsubstantiated.
Previously, we demonstrated that miR-126 is both highly expressed and functionally active within the murine and human HSC compartments, with progressive downregulation during early steps of hematopoietic commitment. Using only miR-126 bioactivity as a marker, we prospectively isolated human HSC that have xenograft repopulating potential (Gentner et al., 2010). These data prompted us to investigate the biological function of miR-126 in HSC. Here, by using enforced expression and knockdown strategies, we report that miR-126 has a conserved role in mouse and human HSC to help maintain quiescence by restricting cell-cycle progression in response to extrinsic stimuli that activate the PI3K/AKT/GSK3β signaling axis.
Results
miR-126 Levels Peak in Human HSC
Within the human hematopoietic hierarchy, we previously demonstrated peak miR-126 expression and bioactivity within the CD34+CD38−CD90+ primitive compartment (Gentner et al., 2010). However, the human progenitor hierarchy model was recently fractionated into more precise subpopulations (Figure 1A) (Doulatov et al., 2010; Notta et al., 2011). We sorted seven functionally characterized populations from human lineage-depleted cord blood hematopoietic stem and progenitor cells (lin− CB HSPC) to obtain a detailed expression profile of miR-126. The highest expression levels of miR-126 were restricted to the HSC-enriched CD34+CD38−CD45RA−CD49f+ fraction (Figure 1B). miR-126 levels remained high in CD34+CD38−CD45RA−CD90−CD49f− multipotent progenitors (MPPs) and became significantly downregulated in multilymphoid progenitors (MLPs) and committed CD34+CD38+ fractions. These data demonstrated a strong correlation between the HSPC hierarchy and miR-126 expression level, suggesting a role for miR-126 in regulating stem cell function.
Figure 1.
High miR-126 Expression in Hematopoietic Progenitor Cells Modulates In Vitro Expansion
(A) Sorting scheme to separate human HSC, MPP, and progenitor cell subsets from lin− CB cells.
(B) miR-126 expression levels were probed by quantitative PCR (qPCR) in seven lin− CB fractions sorted according to the scheme in (A). Data were normalized to RNU48 control. Each fraction is compared to HSC for statistical analysis. Results are shown as mean +SEM of n = 3 independent experiments of pooled human lin− CB.
(C) Lentiviral (LV) constructs for miR-126 overexpression (126/OE, upper scheme) or miR-126 knockdown (126/KD, lower scheme). 126OE-transduced cells were marked by mOrange2 (orange fluorescent protein, OFP), whereas 126KD was marked by green fluorescent protein (GFP).
(D) PIK3R2 mRNA levels were measured in a CD34+ CB sample 3 days after transduction with 126/KD (red), 126/OE (blue), or CTRL (white) LV. PIK3R2 mRNA levels (normalized to CTRL) are shown as mean +SEM of n = 3 technical replicates. ∗∗∗p < 0.001.
(E) Long-term expansion of CD34+CD38− lin− CB cells upon miR-126 modulation. Cells were transduced and expanded in suspension culture for 48 days. Fold expansion (normalized to CTRL) is shown as mean +SEM of n = 3 (126/OE) or n = 4 (126/KD) independent experiments. ∗p < 0.05.
miR-126 Attenuates Proliferation of Early Progenitors
To interrogate the function of miR-126 in primitive human blood cells, we performed miR-126 overexpression (OE) and knockdown (KD) in primary CB HSPC by using lentiviral vectors (Figure 1C). The design and validation of these vector tools is described in Supplemental Results and Figure S1 (available online). The degree of miR-126 up- or downmodulation that we obtained was functionally relevant as the expression levels of the previously validated miR-126 target PIK3R2 were specifically and significantly decreased upon OE and increased upon KD, respectively (Figure 1D). We then assessed whether modulating miR-126 could influence the proliferation and differentiation of HSPC in vitro. Cytokine-driven serum-free long-term culture of CD34+CD38− cells revealed a trend toward decreased total cell output upon 126/OE and a significantly increased expansion upon 126/KD without disturbance of myeloid differentiation into CD15+ granulocytes and CD14+ monocytes (Figure 1E and data not shown). To examine whether this response was associated with a specific CB subpopulation, we generated growth curves from CD34+ subpopulations that were transduced with the 126/KD or a scrambled control vector (Figure 2A). Interestingly, the growth of primitive CD34+CD38−CD133+ cells was significantly enhanced upon 126/KD, in contrast to the more committed CD34+CD38+CD133+ subset. Moreover, the growth advantage was most pronounced in the first 5 days of culture when the cells are the least differentiated, suggesting that the 126/KD phenotype may specifically affect stem and early progenitor cells, reflecting the physiological expression pattern of miR-126. To assess whether the cycling status of HSPC was altered upon 126/KD, cell-cycle analysis was performed 3 to 4 days after transduction. 126/KD induced a 1.7-fold increase in the proportion of cells in the S/G2/M phases of the cell cycle as compared to nontransduced or CTRL vector-transduced cells (Figure 2B). Taken together, these data indicate that 126/KD increases the proliferation of primitive HSPC.
Figure 2.
miR-126 Modulation Alters Proliferation of Primitive Hematopoietic Progenitor Cells
(A) CD34+ CB cells were fractionated according to CD133 and CD38 expression as shown in the representative plot (left) and transduced with CTRL or 126KD LV. On day 3, fractions were plated in suspension culture (TSF6), and cell numbers were monitored. Shown is the fold increase in population doublings of 126KD (red) over CTRL-transduced cells (n = 5 biological replicates, mean ±SEM).
(B) Cell-cycle analysis was performed on CTRL or 126KD LV-transduced CD34+ CB cells on days 3 to 4 of culture by Ki67/Hoechst staining. Representative FACS plots are shown for highly (GFP+) and poorly (GFP−) transduced cell populations. The graph on the right shows the fraction of cells in the S/G2/M phase of the cell cycle. Results are shown as mean ±SEM and points represent eight replicates.
(C–E) Clonogenic progenitor assays of 126KD or 126OE CB cells. (C) CD34+ or (D and E) CD34+CD38− lin− CB cells were transduced with (C and D) 126KD and CTRL or (E) 126OE and CTRL LV. After (C) 7 or (D, E) 14 days of liquid culture, equal cell numbers were plated in clonogenic assays. Counts of colony types are shown as mean +SEM of n = 4 (C) or 3 (D and E) independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
We then investigated whether 126/KD affected the proliferation and/or differentiation of functionally defined myeloid and erythroid progenitors in clonogenic assays. When CD34+ CB cells were plated 1 day after transduction, no quantitative differences in colony number or type were observed between 126/KD or control groups (Figure S2A). However, when the cells were cultured for 7 days before performing the clonogenic assay, 126/KD CD34+ CB yielded significantly more myeloid colonies than control vector-transduced cells (Figure 2C). When 126/KD CB were further enriched for primitive HSPC and expanded for 1 to 2 weeks, there was a significant increase in erythroid colonies upon 126/KD and a trend toward increased myeloid colonies, whereas the less primitive CD38lo/+ fractions only yielded more myeloid colonies as seen with total CD34+ cells (Figures 2D and S2B). Overall, these results indicate that in the CD38− compartment, 126/KD expands erythroid and myeloid clonogenic progenitors, whereas in the less primitive CD38+/lo compartment, only myeloid progenitor output is increased. The role of miR-126 in modulating primitive HSPC is reinforced by the finding that 126/OE in CD34+CD38− HSPC yielded a 1.23-fold decrease in total colony numbers (p = 0.017) involving both myeloid and erythroid colonies (Figure 2E and data not shown). Moreover, megakaryocyte/erythroid specification was altered by modulating miR-126 (Figures S2C–S2H and Supplemental Results). The regulatory function of miR-126 was conserved across species with very similar effects seen in murine HSPC subpopulations (Figures S2I–S2K).
Together, our data suggest that miR-126 constrains proliferation of primitive hematopoietic progenitors, consequently reducing the overall output of mature erythroid and myeloid cells upon in vitro culture.
miR-126 Knockdown Expands HSC In Vivo without Exhaustion
Although our in vitro results suggest a functional role for miR-126 in primitive HSPC, in vivo repopulation studies, where HSC reside in a physiological niche, remain the gold standard to prove that HSC self-renewal and differentiation are affected. To determine how reduced miR-126 expression would affect human HSC function, we xenotransplanted CD34+CD38− lin− CB HSPC transduced with 126/KD and CTRL vector to similar levels as assayed in vitro. Nine weeks after transplantation, 126/KD resulted in an increase in hematopoietic output as measured by human CD45+GFP+ engraftment of immune-deficient mice (Figure 3A). Both CD33+ myeloid and CD19+ lymphoid lineages were similarly affected, suggesting that an upstream progenitor cell is expanded by 126/KD. Phenotypic analysis of the most primitive human HSPC compartment was undertaken 9 weeks after transplantation. The frequency of CD34+CD38−CD90+CD45RA− cells within the lineage-depleted CD45+GFP+ graft was increased upon 126/KD (Figures 3B and 3C). Assessment of cycling characteristics of the CD34+CD38−CD90+ HSC-enriched fraction showed a 2.8-fold increase in the fraction of cells in S/G2/M upon 126/KD (Figure 3B), indicating that the observed expansion coincides with increased cell-cycle progression. Although increased cycling typically induces progressive exhaustion of HSCs, 126/KD HSPC still showed increased contribution to hematopoiesis at 20 weeks after transplantation (data not shown). To determine whether the observed increase in phenotypic HSC correlated with an increase in HSC function, human CD45+GFP+ CTRL and 126/KD cells from primary recipient mice were sorted and transplanted into secondary mouse recipients in a limiting dilution analysis (LDA). After 10 weeks, encompassing an overall total 30 weeks repopulation, there was a 3-fold increase in functional HSC within 126/KD mice (p = 0.01; Figure 3D)—results that strongly correlate with the phenotypic analysis.
Figure 3.
miR-126 Knockdown Expands HSC In Vivo
(A) Effect of 126/KD on CB engraftment levels. Sorted CD34+CD38− lin− CB cells were transduced with 126/KD or scrambled CTRL LV. Transduction efficiency was 92% for CTRL and 72%–75% for 126/KD. BM of xenografted mice was analyzed for human CD45+GFP+ engraftment 9 weeks after transplantation of equal cell numbers. Results are shown as mean +SEM of n = 10 CTRL and n = 10 126/KD mice per experiment.
(B and C) Phenotypic and cell-cycle analysis of the human stem cell compartment in xenografted BM. Lin− human cells were isolated from BM from mice shown in (A). The frequency of HSC-enriched CD34+CD38−CD45RA−CD90+ cells within human CD45+GFP+ cells was assessed as well as the cell-cycle status of CD34+CD38−CD90+ cells. (B) Left and middle show representative plots of n = 3 independent experiments. Right shows cell-cycle analysis of n = 1 experiment. (C) Phenotypic HSC frequency upon 126/KD normalized to CTRL; results are shown as mean +SEM of n = 3 independent experiments.
(D) Limiting dilution analysis (LDA) of CB cells with 126/KD. Sorted CD34+CD38− lin− CB cells were transduced with 126/KD or CTRL LV. CD45+GFP+ cells were sorted from the mouse BM and injected in limiting doses into secondary recipients 20 weeks after transplantation. After 10 weeks, BM of secondary mice was assessed for CD45+GFP+ engraftment. Results are summarized as the percentage of secondary mice with >0.5% engraftment.
(E) Congenic Ptprca and Ptprcb murine Lin− BM cells were transduced with the 126/KD (CD45.2) or CTRL LV (CD45.1). Equal numbers of 126/KD and CTRL cells were pooled and competitively transplanted into heterozygous CD45.1/CD45.2 recipients. The contribution of CD45.2 cells (>90% transduced with the 126/KD LV) to peripheral blood granulocytes (top) and B cells (bottom) is shown at the indicated time points posttransplant. The arrow indicates when a serial BM transplant into CD45.1/CD45.2 recipients was performed. Results are shown as mean ±SEM, n = 9 mice.
(F) Contribution of the GFP+CD45.2+ (126/KD vector-expressing) cells (red bars) to the indicated BM populations relative to the sum of all GFP+ cells (either CD45.2+ or CD45.1+). HSC were defined as Lin−Sca+Kit+ (LSK) CD150+CD48− cells. Within the Lin+ compartment (B220+ or CD3+ or GR1+ or CD11b+ or Ter119+), myeloid cells (Myelo) were defined as CD48lo/−SSChi granulocytes, whereas lymphocytes (Lympho) were CD48+SSCloFSClo. Results are shown as mean ±SEM of n = 9 mice (primary mice, top, analyzed 8 months after transplant) and n = 8 mice (secondary mice, bottom, analyzed after an additional 6 months).
(G) Cell-cycle distribution of CTRL and 126/KD cells in the LSK (left) or Lin−Sca−Kit+ (right) compartment of secondary transplant recipients. Results are shown as mean +SEM of n = 8 mice.
(H) Representative cell-cycle distribution of CTRL and 126/KD cells in the LSK (left) or LSK CD150+ HSC (right) compartment from primary transplant recipients. Top, gating scheme on Lin− BM. Bottom, Ki67/Hoechst marking within the gated subpopulations.
(I) LDA of CD45.2+ CTRL or 126/KD BM total nucleated cells injected into CD45.1 recipients is summarized, with the proportion of mice with multilineage CD45.2+ engraftment (0.1% cutoff) and estimated HSC frequency. ∗p < 0.05 and ∗∗p < 0.01.
It is possible that the expansion of phenotypic human HSC within xenografts following 126/KD could be influenced artificially due to the species mismatched physiological niche. Therefore, competitive transplantation assays were performed in the Ptprc congenic mouse model, transducing Ptprcb (CD45.2) or Ptprca (CD45.1) lin− BM HSPC, respectively, with either the 126/KD or CTRL vector. Complete blood counts (CBC) of repopulated mice were normal, even though 126/KD vector-transduced cells tended to contribute less to active hematopoiesis, particularly within the B and T lymphoid lineages (Figures 3E and S3). Steady-state bone marrow (BM) chimerism analyzed 5–8 months after transplantation showed much higher chimerism within lin−Sca+Kit+ (LSK) Cd150+Cd48− phenotypic HSC-enriched compartments of the 126/KD group (Figures 3F and S3). In contrast, 126/KD cell chimerism was reduced in Lin−Sca+Kit− lymphoid precursors (Harman et al., 2008) and lymphocyte compartments. These data suggest that 126/KD expanded the murine HSC compartment and reduced lymphopoiesis in vivo. To substantiate this hypothesis, we performed serial BM transplantation into secondary recipients. The contribution of 126/KD cells increased in myeloid and lymphoid lineages and surpassed the CTRL in the myeloid compartment (Figures 3E and S3), confirming that the relative increase in phenotypically defined 126/KD HSC detected in the primary BM translates into increased multilineage contribution to hematopoiesis in the secondary hosts. BM analysis of the secondary hosts mirrored the differences in chimerism observed in corresponding populations within the primary BM but shifted toward higher contribution from 126/KD cells (Figure 3F). Switching congenic strains between 126/KD and CTRL groups yielded similar results, confirming the role of 126/KD on HSC function (Figure S3B). We also measured the cell cycle of freshly isolated LSK cells from the BM of transplanted mice and found a significantly increased proportion of 126/KD LSK cells in S/G2/M as compared to CTRL vector-transduced cells, whereas there was no significant difference in cell-cycle distribution of the committed Lin−Sca−Kit+ progenitors (Figure 3G). Furthermore, we confirmed the increased S/G2/M fraction in 126/KD cells highly enriched for HSC (LSK CD150+; Figure 3H). These data indicate that 126/KD increased HSC proliferation within an in vivo physiological niche.
The gold standard proof of HSC expansion requires frequency analysis using transplantation at limiting doses (LDA). Ptprca mice were reconstituted with BM cells from Ptprcb mice transduced with either the 126/KD or CTRL vectors. Sixteen weeks after transplantation, mice selected on the basis of similar levels of peripheral blood chimerism (Figure S3D) showed expansion of phenotypically defined HSC in 126/KD mice (Figure S3E). We then set up an LDA from 126/KD and CTRL BM cells. Recipient mice were scored as engrafted if they displayed >0.1% Ptcrpb cells in multiple lineages. HSC frequency was 3.9-fold higher in 126/KD versus CTRL BM (Figure 3I).
Collectively, our results establish that stable knockdown of miR-126 expanded mouse and human long-term repopulating HSC, assessed by using the most stringent phenotypic and functional assays available. Although increased cycling of HSCs is frequently associated with their exhaustion, despite 1.5 years of follow up, no HSC exhaustion or malignancy was observed in our studies, pointing to the unique regulatory properties of miR-126 in HSC.
Enforced miR-126 Expression Progressively Reduces Hematopoietic Output In Vivo
Because 126/KD induced an expansion of the phenotypic HSC compartment through increased cycling, we predicted that forced expression of miR-126 may provoke the opposite response in vivo. Following xenotransplantation of lin− CD34+CD38− CB HSPC, the progeny of cells that were transduced with the 126/OE vector transiently engrafted mice but were progressively lost between 12 and 16 weeks (Figure 4A). No significant alteration of the myeloid/lymphoid ratio was detected; however, at 16 weeks, the mice injected with 126/OE-transduced cells displayed an 11-fold lower percentage of GlyA+ erythroid cells compared to the control group, which was consistent with our in vitro observations (Figure S2H). We focused on the remaining HSC compartment and were able to carry out detailed analysis by pooling BM from four to five recipient mice per time point. Evaluation of primitive cell marker expression in the human lin− compartment revealed that the loss of human 126/OE cells in transplanted mice was not due to a decrease in the CD34+CD38−CD90+ stem cell compartment size (Figure 4B). On the contrary, we observed, on average, a 2.5-fold increase in the phenotypic CD34+CD38−CD90+ stem cell compartment for 12 weeks posttransplant, after which marked primitive cells were no longer recoverable (Figure 4C). In addition, cell-cycle analysis of HSC-enriched Lin−CD34+CD38−CD90+ cells with 126/OE in the xenografted BM over time showed a consistently higher percentage of cells in G0 (Ki67−; Figures 4B and 4D). These data indicate that miR-126 negatively regulates the cell-cycle progression of HSC, which is consistent with our in vitro experiments and is contrary to the 126/KD results. Surprisingly, no change in the numbers of quiescent cells in the more heterogeneous CD34+CD38+ progenitor-enriched compartment was observed; instead, a significant increase in the fraction of cells in S/G2/M phase was noted (Figures 4B and 4D). Thus, the effects of 126/OE appeared to be context dependent and distinct between stem cell and progenitor-enriched fractions.
Figure 4.
miR-126 Overexpression Results in Progressively Reduced Hematopoietic Output In Vivo
(A) Bone marrow xenograft kinetics of lin− CB cells upon 126/OE. CD34+CD38− lin− CB cells were transduced with 126/OE or CTRL LV and were transplanted. Mice were analyzed every 4 weeks for total human CD45+OFP+ BM engraftment. Results are shown as mean +SEM of n = 4 CTRL and n = 4 126/OE mice per time point. Week 16 results are representative of two experiments.
(B and C) Gating strategy for cell-cycle analysis of 126/OE lin− CB. CTRL or 126/OE-transduced human lin− CB cells were isolated from xenografted BM 4–12 weeks after transplantation. The frequency and cell-cycle status of HSC-enriched CD34+CD38−CD90+ cells was assessed, as well as the cell-cycle status of committed CD34+CD38+ progenitors. (B) Representative plots showing the lin− CB cell phenotype and cell-cycle analysis at 4 weeks after transplantation. (C) Percentage of CD34+CD38−CD90+ cells over time is shown (left) and normalized to CTRL for statistical analysis (right).
(D) Cell-cycle analysis of lin− CB primitive cell populations upon 126/OE. Using the gating strategy depicted in (B), CD34+CD38−CD90+ (left) and CD34+CD38+ (right) human cells in the xenografted BM were analyzed for cell-cycle stage at 4, 8, and 12 weeks posttransplant. BM of four to five mice was pooled at each time point. Data were normalized to CTRL, and time points were combined for statistical analysis. Results are shown as mean of n = 2 independent experiments at 4 weeks; for 8 and 12 weeks, results represent n = 1 experiment.
(E) Congenic Ptprca and Ptprcb murine Lin− BM cells were transduced with CTRL or 126/OE LV. Equal numbers of CTRL and 126/OE cells were pooled and competitively transplanted. The contribution of OFP-expressing 126/OE cells to peripheral blood granulocytes (top) and B cells (bottom) is shown at the indicated times posttransplant. Hematopoietic stress was induced by 5-fluorouracil (5-FU) administration (Exp1) or serial BM transplantation (Exp2). Results are shown as mean ±SEM of n = 9 mice (Exp1) and n = 6 mice (Exp2).
(F) Top bar graph shows a representation of murine BM HSPC subsets 3 weeks after transplantation of 126/OE or 223OE LV-transduced cells relative to CTRL LV-transduced cells. Results are shown as mean +SEM of n = 24 mice for 126/OE, n = 13 mice for 223OE, and n = 13 mice for CTRL. Significant differences between 126/OE and both control LVs are shown, as assessed by one-way ANOVA with Bonferroni correction. Bottom bar graph shows in vivo nucleoside incorporation after two EdU pulses (24 hr and 12 hr before sacrifice) in the Lin− and Lin+ BM subsets. Results are normalized to the combined control group. Results are shown as mean +SEM of five pools of two to three mice for the 126/OE group and six pools of two to three mice for the combined control group (three pools of CTRL LV and three pools of 223OE LV).
(G) BM chimerism 100 days after competitive transplantation of CTRL and 126/OE LV-transduced cells. Blue bars represent the fraction of OFP-expressing 126/OE vector-transduced cells in relation to all other BM cells (untransduced and CTRL LV transduced). HSC were defined as LSK CD150+CD48− cells. Lin+ cells were defined as in Figure 3F. Results are shown as mean +SEM of n = 4 mice.
(H) Cell-cycle distribution of the cells shown in (G) as determined by Ki67/Hoechst staining in HSC-enriched LSK CD150+ and total LSK cells (n = 5 pools of three mice). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. ns, not significant.
To examine the effect of 126/OE on the fate of murine HSC in vivo, we performed competitive transplantation experiments. Lin− BM cells from congenic mice (Ptprca and Ptprcb, respectively) were transduced with a 126/OE or CTRL vector and injected in a 1:1 ratio into myeloablated recipients (Figure 4E). In the first month after transplantation, myeloid (Cd11b+) and B cell (Cd19+) output from 126/OE cells was increased relative to the output of control vector-transduced cells. Following this initial burst of differentiated cells, 126/OE cells declined and became progressively lost from the peripheral blood upon hematopoietic stress induced by 5-Fluorouracil administration or serial transplantation into secondary recipient mice (Figure 4E). A similar phenotype was observed when ectopically expressing miR-126 from a weaker promoter (see Supplemental Results and Figure S4).
Next, we assessed the impact of 126/OE on the early engraftment phase in a noncompetitive transplantation setting. A significant advantage of early blood reconstitution by 126/OE cells was seen for platelets and leukocytes, whereas neutropenia and thrombocytopenia were shortened (see Supplemental Results). BM analysis conducted at 3 weeks posttransplantation showed that, whereas the Lin+ compartment was reconstituted to similar levels in all groups, absolute numbers of Lin− cells were significantly increased in 126/OE BM, primarily in the Sca+Kit− and Sca−Kit− compartments (Figure 3F, top). In vivo nucleoside incorporation assays revealed increased labeling in LSK, Lin−Sca+Kit−, and Lin−Sca−Kit− cells, but not in Lin−Sca−Kit+ or Lin+ cells (Figure 3F, bottom). Analysis of the BM of engrafted mice 3 months after transplantation revealed a substantially decreased 126/OE chimerism in the LSK CD150+CD48− HSC compartment with respect to all other BM populations (Figures 4G and S4F). Moreover, although more 126/OE LSK cells were in the S/G2/M phase of the cell cycle, HSC-enriched fractions (LSK CD150+) contained significantly more 126/OE cells in G0 (Figure 4H). This is strikingly reminiscent of the situation in human cells, in which CD34+CD38+ progenitors were cycling more frequently upon 126/OE, whereas the HSC-enriched CD34+CD38−CD90+ cells displayed increased quiescence.
Overall, these results illustrate that miR-126 regulation of cell fate differs depending on cellular context. During the first weeks after transplantation of 126/OE cells, specific progenitor subsets are expanded, which is reflected in an increased multilineage output. Over time, a competitive disadvantage of 126/OE at the HSC level results in a progressive reduction of the progenitor supply, leading to the collapse of hematopoietic output. A constant reduction in cell-cycle entry of HSC may be the underlying explanation for the finding that 126/OE HSC are eventually outcompeted by nontransduced HSC with normal cycling characteristics.
PI3K/AKT Signaling Is Targeted by miR-126 in Early Hematopoietic Progenitors
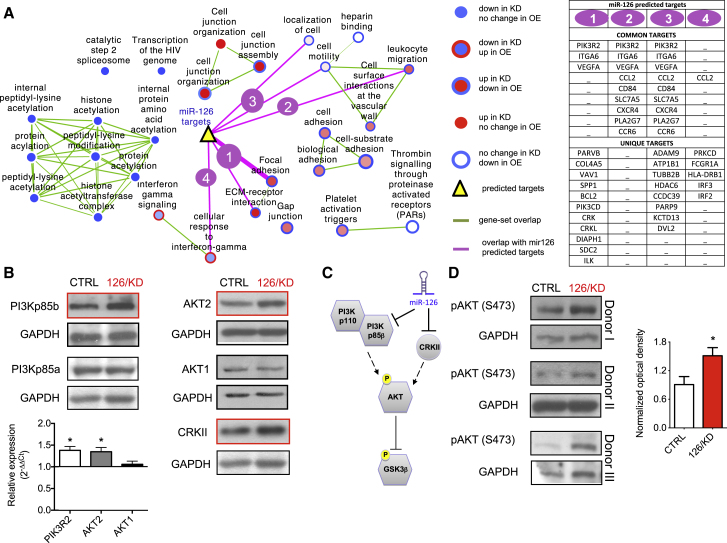
To gain insight into the mechanism whereby miR-126 was exerting its biological effects on HSPC, gene expression profiling was performed on sorted CD34+CD38− lin− CB HSPC transduced with 126/KD, 126/OE, and their respective CTRL vectors. Gene set enrichment analysis (GSEA) was applied to HSPC gene expression data to identify miR-126 modulated cellular pathways. Opposing effects between 126/KD and 126/OE groups were observed with a majority of enriched pathways (Figure 5A). Furthermore, pathways most highly correlated with miR-126 expression levels centered on cell-cell contact (cell junction and adhesion), motility, migration (all upregulated in 126/KD and downregulated in 126/OE), interferon-gamma signaling, and protein acetylation (both downregulated in 126/KD). Using four published algorithms, a list of predicted miR-126 target genes was generated and compared with the gene expression data set. Predicted targets were found in six pathways that are significantly influenced by miR-126 (Figure 5A, purple lines, p < 0.05). The focal adhesion pathway showed the most significant overlap with the list of miR-126 predicted targets (Figure 5A, list 1, p = 0.0003), indicating that this may be the primary pathway under the influence of miR-126 in primitive lin− CB cells. In this pathway, PIK3R2, ITGA6, and ILK, all predicted miR-126 targets, are upregulated upon 126/KD and downregulated upon 126/OE. As the effect of miRNA modulation of gene expression may be stronger at the protein level, and to provide another unbiased approach, we performed a quantitative analysis on the proteome of K562 cells stably transduced with 126/OE or CTRL vectors by using the stable isotope labeling of amino acids in culture (SILAC) technique (Figures S5A–S5C). Interrogation of this data set by using unsupervised Ingenuity Pathway Analysis (IPA) found that, upon 126/OE, the PI3K/AKT signaling pathway was among the most significantly affected canonical pathways (Figures S5B and S5C), validating the bioinformatic analysis of the mRNA arrays. These data suggest that PI3K/AKT signaling is a major pathway through which miR-126 exerts its effects in primary human hematopoietic cells.
Figure 5.
PI3K/AKT Signaling Is Targeted by miR-126 in Hematopoietic Progenitors
(A) Functional enrichment map for 126/KD and 126/OE fractions revealing miR-126 modulated pathways. Nodes (circles) represent gene sets (pathways), red indicates gene sets enriched in the 126/KD (node center) or in the 126/OE (node border) fractions, and blue indicates gene sets enriched in CTRL. Green line width between pathways (nodes) corresponds to the number of shared genes. miR-126 predicted targets (yellow triangle) are connected to enriched pathways by pink lines; line thickness indicates significance level (proportional to –log (p value)). Overlap of miR-126 targets was significant (p < 0.05) with six enriched gene sets. miR-126 targets were present in list 1 (KEGG:HSA04510 and KEGG:HSA04512 gene sets), list 2 (GO:0050900), list 3 (GO:0048870 and GO:0051674), and list 4 (GO:0071346). Gene targets shared between lists (common targets) and targets unique to a list (unique targets) are listed on the right.
(B) Protein (blots) and transcript (bar graph) levels of PI3K p85 isoforms, AKT isoforms, and CRKII were measured in CD34+ CB cells 3–5 days after transduction with CTRL or 126/KD LV. Top, western blots are representative of n = 5 (PI3Kp85α/β) or n = 2 (AKT1/2 and CRKII) independent experiments; GAPDH was used as a loading control. Bottom, qPCR graph shows the fold change of the indicated transcript in 126/KD over CTRL-transduced cells. Results are shown as mean +SEM of n = 5 independent experiments (PIK3R2 and AKT2) or n = 3 independent experiments (AKT1).
(C) Simplified scheme illustrating the flow of signal through the PI3K/AKT pathway.
(D) Left, pAKT (Ser473) was measured in CTRL or 126/KD LV-transduced CD34+ CB cells by western blot performed after 4 days in liquid culture. GAPDH was used as a loading control. Three independent experiments are shown. Right, bar graph showing the densitometric quantification of pAKT (Ser473) normalized to GAPDH. Mann-Whitney test was used for statistical analysis. Results are shown as mean +SEM of n = 7 independent experiments. ∗p < 0.05.
Because the PI3K/AKT pathway transduces crucial signals for HSC proliferation and differentiation (Buitenhuis and Coffer, 2009; Buitenhuis, 2011), we assessed the expression levels of individual components of this pathway in both K562 cells (Figure S5D) and CD34+ CB cells (Figure 5B) upon 126/KD. Among the investigated genes, PIK3R2/PI3Kp85β and AKT2 were significantly upregulated upon 126/KD at both the transcript and protein level, whereas their respective isoforms, PIK3R1/PI3Kp85a and AKT1, were unchanged. CRKII protein was also upregulated by 126/KD as predicted by bioinformatic analysis. Whereas PIK3R2 is well established as a direct miR-126 target (Fish et al., 2008; Guo et al., 2008), AKT2 may be regulated through indirect mechanisms because its transcripts are not predicted to contain binding sequences for the miR-126 seed sequence.
miR-126 Attenuates the PI3K/AKT/GSK3β Signaling Axis upon Extrinsic Stimuli
We next assessed whether miR-126-dependent regulation of the PI3K/AKT pathway would have an effect on signal transduction. AKT interacts with several downstream effectors implicated in stem cell regulation such as mTOR complexes and GSK3β and FOXO proteins (Buitenhuis and Coffer, 2009). AKT is activated upon phosphorylation of Ser473. We found increased phosphorylation of AKT at Ser473 (p-AKT S473) in 126/KD-transduced CD34+ CB HSPC under steady-state culture conditions (Figures 5C and 5D; see also Figure S5E for K562 cells). To investigate the acute response to cytokine stimulation at the single cell level in primary human cells, we assayed pAKT (S473) by flow cytometry in CD34+ HSPC starved in serum and cytokine-free medium prior to stimulation. Stem cell factor (SCF) containing cytokine cocktails rapidly and strongly induced pAKT (S473) in CD34+ cells (Figure S6). This response was significantly enhanced in 126/KD HSPC, and this enhancement was most evident when the analysis was focused on highly transduced cells (Figure 6A). By using the same starvation/stimulation protocol, we also found significantly increased phosphorylation of GSK3β at Ser9 in 126/KD CD34+ CB cells (Figure 6A). An opposite effect on AKT and GSK3β phosphorylation was observed upon 126/OE (Figure 6B). Overall, these data indicate enhanced transduction of the signal induced by SCF through the PI3K/AKT pathway in human HSPC upon 126/KD and decreased signal transduction upon 126/OE. Finally, we were able to abrogate the effect of 126/KD on HSPC proliferation by a single administration of the PI3K inhibitor LY294002 at the time of cytokine stimulation, reinforcing the notion that activation of PI3K signaling is required for the observed functional effects of 126/KD in HSPC (Figure 6C).
Figure 6.
Attenuation of PI3K/AKT/GSK3β Signaling upon Extrinsic Stimuli Contributes to miR-126 Control of Proliferation
(A) Cytokine-induced activation of AKT and GSK3β upon 126/KD in CD34+ CB cells after 30 min starvation and restimulation with SCF, TPO, FLT3L, and IL6 (TSF6 medium, see Experimental Procedures). Left FACS plots show GFP expression in CD34+ CB cells transduced with 126/KD-LV, and CTRL-LV (not shown) was stratified into quartiles, with brighter cells expressing more miR-126 decoy targets. Middle histograms show representative induction of pAKT (S473) (top) and pGSK3β (S9) (bottom) in the top 25% GFP-expressing 126/KD cells (red), CTRL cells (gray), or in 126/KD cells pretreated with the PI3K inhibitor Wortmannin (dashed line). Right graphs show relative mean fluorescence intensity (MFI) of phospho-AKT S473 (top) and phospho-GSK3β S9 (bottom) from untransduced (first quartile, Q) to highly transduced (top 10%) cells for CTRL (gray), 126/KD (red), and Wortmannin-treated (dashed) cells that were stimulated with TSF6. Note that the effect of 126/KD is most pronounced in highly transduced cells. Results are shown as mean +SEM of n = 6 independent experiments (AKT) and n = 3 independent experiments (GSK).
(B) Cytokine-induced activation of AKT and GSK3β upon 126/OE in CD34+ CB cells stimulated as described in (A). Histograms represent the induction of pAKT (S473) (top) and pGSK3β (S9) (middle) in the top 25% of OFP-expressing 126/OE cells (blue), CTRL (gray), or 126/OE cells pretreated with the PI3K inhibitor Wortmannin (dashed line). The bar graph on the bottom represents pAKT (S473) or pGSK3β (S9) activation in 126/OE relative to CTRL cells in the corresponding first Q and top 25% OFP-expressing fractions.
(C) Effect of the PI3K inhibitor Ly294002 (Ly) treatment on the growth of 126/KD cells. Left, representative experiment depicting the growth advantage of 126/KD CD34+ CB cells over CTRL (dotted lines) and its abrogation upon single-dose administration of 10 μM Ly (solid lines). Right, fold change in cell numbers of 126/KD normalized to CTRL cells, treated (black) or not (red) with Ly. Results are shown as min/max box and whiskers plots of n = 5 independent experiments. ∗p < 0.05 and ∗∗p < 0.01.
Collectively, these data indicate that miR-126 regulates the expression of several components of the PI3K/AKT pathway and, consequently, regulates its activation by extrinsic stimuli such as SCF.
Discussion
Our data establish that miRNAs, differentially expressed in HSPC, represent an important regulatory mechanism that governs the biological properties of mouse and human HSC. By gain- and loss-of-function studies, we demonstrate that miR-126 functionally regulates HSC and early progenitor cells by modulating signal transduction in response to extrinsic stimuli, such as SCF, and by inducing specific changes in the cell cycle of HSC and progenitors within their BM niche in vivo. miR-126 knockdown resulted in increased cell-cycle progression and expansion of the phenotypic and functional HSC compartment in vivo. Overexpression of miR-126, on the other hand, resulted in increased HSC quiescence and transiently increased proliferation of progenitor cell populations, which was followed by diminishing hematopoietic output. Our work demonstrates the conservation of miR-126 function between mouse and human HSC and establishes a conceptual framework to understand how miR-dependent adjustment of intracellular signals fine-tunes the balance between HSC quiescence and activation, enabling them to sustain the blood system throughout life.
We identify components of the PI3K/AKT signaling pathway as direct or indirect targets of miR-126 in human HSC and show that miR-126 activity attenuates the degree of activation of this pathway in response to cytokine signals—such as SCF—known to be crucial for HSC maintenance (Thorén et al., 2008). Thus, miR-126 appears to act mostly by imposing a threshold on the level of stimulation required to activate HSC. Importantly, the PI3K/AKT pathway plays a central role in regulating HSC homeostasis through downstream effectors, which include FOXO transcription factors, mTOR, GSK3β, and WNT (Buitenhuis and Coffer, 2009; Buitenhuis, 2011). Several groups have engineered mice to modify PI3K/AKT pathway activation. Recently, a detailed analysis of HSC in an AKT1/2 double-deficient mouse model revealed a deficiency in competitive long-term reconstitution (Juntilla et al., 2010). This defect was attributed to HSC persistence in G0, arguing that the impaired HSC function and cell-cycle progression that we observe upon 126/OE may indeed be due to attenuated PI3K/AKT signaling. Conversely, hyperactivation of the PI3K/AKT pathway has been studied by using conditional genetic deletion models of negative regulators such as Pten, Ship, Fbw7, Pml, and Tsc1 (Li, 2011). These studies reported a transient increase in cycling HSC, which was followed by exhaustion and/or impaired repopulation capacity. Similar phenotypes were obtained by inactivating negatively regulated downstream targets of the PI3K/AKT pathway such as GSK3β (Huang et al., 2009) or FOXO (Tothova et al., 2007). Remarkably, our 126/KD studies also hyperactivated the PI3K/AKT pathway and induced expansion of HSC without inducing their exhaustion when assayed long-term after transplant or in secondary transplanted mice. We speculate that the reason why 126/KD may expand HSC without exhaustion might be because, rather than affecting a single regulator, miR-126 likely modulates several functionally important targets in interconnected pathways that together orchestrate an HSC program. Further studies will be needed to investigate whether additional miR-126 targets in addition to those identified within the PI3K/AKT pathway contribute to the observed phenotypes. We note that the sponge vector achieves a knockdown rather than knockout of miR-126. Thus, it remains to be determined what influence a full miR-126 knockout would have on HSC. Germline miR-126 knockout results in a severe angiogenesis defect with most mice dying in utero (Kuhnert et al., 2008; Wang et al., 2008). However, the few miR-126 knockout mice that survived were reported to have no major disturbances of the hematopoietic system, except for some degree of erythrocytosis. This observation is in line with our findings that 126/KD increased erythroid and megakaryocytic differentiation of CD34+ lin− CB cells, whereas ectopic miR-126 expression inhibited erythropoiesis, as also recently shown in an embryonic stem cell (ESC) differentiation model (Huang et al., 2011).
Another important difference between 126/KD and Pten knockout mice is that the latter mode of PI3K/AKT pathway activation invariably resulted in the emergence of leukemia (Yilmaz et al., 2006), whereas 126/KD never produced hematologic malignancies in up to 1.5 years of follow-up in mice (including serial transplants). Indeed, vector integration site analysis revealed the polyclonal nature of 126/KD expansion (Figure S3C). Whereas PI3K/AKT signaling is one of the most frequently activated pathways in cancer, miR-126 is a tumor suppressor as it is lost in a number of solid cancers (Guo et al., 2008; Zhang et al., 2008; Miko et al., 2009; Feng et al., 2010). The PI3K/AKT pathway is highly branched, with many inputs, outputs, and feedbacks (Chalhoub and Baker, 2009). Follow-up studies will clarify which specific output ultimately induces the biological effects of miR-126 deregulation.
The variety of effects we observed by modulating miR-126 in hematopoietic cells adds evidence that miRNAs exhibit extensive context-dependent functions. Murine cell transplantation showed that 126/KD resulted in a cell-cycle increase only within the stem-cell-enriched compartment, which expresses the highest levels of miR-126, but not in downstream progenitors. Even more striking is the bimodal response to 126/OE, whereby the proliferation of the HSC fraction is reduced, whereas cell-cycle entry of immediate downstream progenitors is increased both in human and murine experiments. This suggests that stem and progenitor cells have distinct programs that govern which targets are available for miR-126 to influence, resulting in differences in signaling responses within each cell type. Alternative 3′ UTR sequences and/or a different pool of competing RNA targets for miR-126 could account for this apparent paradox (Mayr and Bartel, 2009; Poliseno et al., 2010). Thus, the availability of different sets of mRNA targets in distinct cell types may contribute to the context specificity of miRNAs, and further work is required to identify the whole complement of miR-126 targets in hematopoietic stem versus progenitor cells.
This report links a miRNA to the regulation of human HSC, thus opening an avenue to develop therapeutic strategies for human HSC maintenance and expansion. There is great need to understand how HSC are regulated in order to take a more rational approach to harness human HSC for clinical purposes. The discovery that miRNAs occupy a central regulatory role opens an alternative avenue to manipulate HSC quiescence and cell-cycle entry with the ultimate goal of HSC expansion for clinically important therapies. The significant expansion of long-term HSC, here induced through the manipulation of the miR-126/PI3K/AKT signaling axis, supports the plausibility of this approach. It is conceivable that manipulation of miR regulation may provide a more physiological approach to fine-tune a cellular response. Finally, given the close relationship between normal HSC and AML leukemic stem cells (Eppert et al., 2011), future studies should examine whether miR-126 deregulation contributes to leukemia stem cell properties.
Experimental Procedures
Vector Transduction of Cord Blood and In Vitro Expansion
The following transduction protocols were used with similar results in Toronto (1) and Milan (2). (1) Lin− CB cells were thawed and plated in X-VIVO (BioWhittaker) supplemented with 20% BIT 9500 (StemCell Technologies), 2 mM L-glutamine, and 100 U/ml penicillin/streptomycin (P/S). Viral particles were resuspended in the same medium and added to cells at a multiplicity of infection (MOI) of 30–50. The mixture was then supplemented with stem cell factor (SCF) 100 ng/ml, Flt3 ligand (Flt3-L) 100 ng/ml, thrombopoietin (TPO) 20 ng/ml, and interleukin 6 (IL-6) 20 ng/ml for a minimum of 16 hr. Subsequent expansion was performed in the same medium with half the concentration of these cytokines. (2) When indicated, the following conditions, referred to as TSF6, were used: CD34+ cells were cultured in StemSpan medium (Stem Cell Technologies) with the same supplements and cytokines as mentioned above except BIT. After 12–18 hr of prestimulation, cells were exposed to lentiviral vectors for 12–24 hr at an MOI of 50–100. Optionally, a second hit transduction was performed after a 12 hr resting phase in fresh medium. Subsequent expansion was performed in StemSpan medium containing half the concentration of the above-mentioned cytokines (TSF6half medium).
Mouse Xenotransplantation and Human Lin− Cell Isolation
Mouse xenografts were performed as described previously (McDermott et al., 2010). Briefly, NOD/Lt-scid/IL2Rɣnull (NSG) mice were sublethally irradiated (225 cGy) 1 day prior to injection. Transduced cells were injected with 25 μl PBS into the right femur of each recipient mouse. After euthanizing the mice, bone marrow cells were flushed with 2 ml PBS 2% FCS, and 50 μl was stained for surface markers. Remaining cells were lineage depleted and/or used for additional surface or intracellular antigen staining. For lineage depletion, BM cells flushed from the femurs and tibias of three to five mice (>3% human cells) were combined and processed with the StemSep Mouse/Human Chimera Enrichment Kit (Stem Cell Technologies) according to the manufacturer’s instructions. However, during the antibiotin incubation step, an additional 50 μL/ml human hematopoietic progenitor enrichment antibody cocktail from the StemSep Human Progenitor Cell Enrichment Kit was added to deplete human-lineage-positive cells.
Isolation and Transduction of Murine HSPC and Bone Marrow Transplants
Donor mice between 6 and 8 weeks of age were euthanized by CO2, and BM was retrieved by femurs, tibias, and humeri. Murine HSPCs were purified by lin− selection, using the Biotin Mouse Lineage Panel (BD PharMingen) and the StemSep Mouse Progenitor Enrichment Kit (Stem Cell Technologies). LVs transduction was performed as previously described (Gentner et al., 2010). For noncompetitive transplants, ≥106 transduced cells were transplanted via tail injection into 6-week-old lethally irradiated mice (10 Gy total, fractionated into two doses with an interval of 4 hr). For competitive transplants, congenic HSPC from either B6.SJL-Ptprca(CD45.1) or C57BL/6NTac-Ptprcb(CD45.2) mice transduced with either 126/KD or 126/OE and with CTRL LVs were mixed in a 1:1 ratio and transplanted (≥106 cells/mouse) into 6-week-old lethally irradiated mice. Please see Supplemental Information for details of the analysis of chimerism.
Limiting Dilution Analysis
For murine experiments, Lin− BM cells (CD45.2) were transduced with 126/KD or CTRL LV and transplanted into recipient mice (CD45.1). Mice with similar chimerism and transduction levels in both the 126/KD and CTRL group were identified 16 weeks after transplant, and a 126/KD BM pool (n = 3 mice) and CTRL BM pool (n = 3 mice) were generated. Limiting doses of total nucleated BM cells (BM TNCs) from each pool were mixed with 3 × 105 CD45.1 supporter BM TNCs and transplanted into n = 13 lethally irradiated CD45.1 recipient mice. A mouse was scored as positive if it had >0.1% engraftment in multiple lineages by serial bleeding at 8 and 14 weeks after transplantation. Limiting dilution analysis of human CB cells was performed by sorting human CD45+GFP+ cells from pooled BM of primary mice 20 weeks after transplantation and injection of different doses into secondary recipients. A secondary mouse was scored as positive if it had >0.5% BM engraftment 10 weeks after transplantation. HSC frequency was estimated by linear regression analysis and Poisson statistics using publicly available ELDA (Extreme Limiting Dilution Analysis, http://bioinf.wehi.edu.au/software/elda/) software (Hu and Smyth, 2009).
5-FU Treatment and Secondary Transplants
5-Fluorouracil (Sigma) was administered intraperitoneally at a dose of 2.5 mg. For secondary transplants of murine experiments, stably engrafted mice were euthanized (≥14 weeks after primary transplant), and 107 BM-nucleated cells from each mouse were injected into one or two secondary recipients.
Statistical Analysis
Unless otherwise indicated, mean ±SEM values are reported in the graphs. For pairwise comparisons, a Student’s t test (paired if appropriate) was used unless otherwise indicated. For three or more matched groups, a one-way analysis of variance (ANOVA) for repeated measures using a Bonferroni posttest correction was used. Chimerism (chim) values were transformed into a log-odds scale (log (%chim/(100-%chim)), given that percentages are by definition constrained between 0 and 100.
Acknowledgments
We thank G. Schira, F. Benedicenti, and L. Sergi Sergi for technical help; Arian Khandani and P.A. Penttilä and the UHN/Sickkids flow cytometry facility, A. Palini and E. Canonico from the San Raffaele flow cytometry facility, K. Moore and the obstetrics unit of Trillium Hospital (Mississauga, Ontario), and the Obstetrics and Gynecology Unit of the San Raffaele Hospital for providing CB; R. Tsien for providing the mOrange2 FP construct; E. Zonari, M. Amendola and Marie F. André for help with experiments; and A. Kreso and the entire Dick and Naldini labs for editorial and critical review of the manuscript. This work was supported by grants to L.N. from Telethon (TIGET grant), EU (FP7 GA 222878 PERSIST, ERC Advanced Grant 249845 TARGETINGGENETHERAPY), and the Italian Ministry of Health and to J.E.D. from the Canadian Institutes for Health Research, Canadian Cancer Society Research Institute, Terry Fox Foundation, Genome Canada through the Ontario Genomics Institute, Ontario Institute for Cancer Research with funds from the province of Ontario, and a Canada Research Chair. This research was funded in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC. A.G. and F.E.B. conducted this study as partial fulfilment of their PhD program in Molecular Medicine, program in Cellular and Molecular Biology and Biology and Biotherapy of Cancer, respectively (San Raffaele University, Milan, Italy).
Published online: November 8, 2012
Footnotes
Supplemental Information for this article includes six figures, Supplemental Results, and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2012.09.001.
Contributor Information
John E. Dick, Email: jdick@uhnresearch.ca.
Luigi Naldini, Email: naldini.luigi@hsr.it.
Supplemental Information
References
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitenhuis M. The role of PI3K/protein kinase B (PKB/c-akt) in migration and homing of hematopoietic stem and progenitor cells. Curr. Opin. Hematol. 2011;18:226–230. doi: 10.1097/MOH.0b013e32834760e5. [DOI] [PubMed] [Google Scholar]
- Buitenhuis M., Coffer P.J. The role of the PI3K-PKB signaling module in regulation of hematopoiesis. Cell Cycle. 2009;8:560–566. doi: 10.4161/cc.8.4.7654. [DOI] [PubMed] [Google Scholar]
- Chalhoub N., Baker S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Eppert K., Nguyen L.T., Ohashi P.S., Dick J.E. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Feng R., Chen X., Yu Y., Su L., Yu B., Li J., Cai Q., Yan M., Liu B., Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.-F., Wythe J.D., Ivey K.N., Bruneau B.G., Stainier D.Y.R., Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentner B., Visigalli I., Hiramatsu H., Lechman E., Ungari S., Giustacchini A., Schira G., Amendola M., Quattrini A., Martino S. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci. Transl. Med. 2010;2:58ra84. doi: 10.1126/scitranslmed.3001522. [DOI] [PubMed] [Google Scholar]
- Gruber J.J., Zatechka D.S., Sabin L.R., Yong J., Lum J.J., Kong M., Zong W.-X., Zhang Z., Lau C.-K., Rawlings J. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Sah J.F., Beard L., Willson J.K.V., Markowitz S.D., Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939–946. doi: 10.1002/gcc.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Lu J., Schlanger R., Zhang H., Wang J.Y., Fox M.C., Purton L.E., Fleming H.H., Cobb B., Merkenschlager M. MicroRNA miR-125a controls hematopoietic stem cell number. Proc. Natl. Acad. Sci. USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-C., Park C.Y., Bhagat G., Zhang J., Wang Y., Fan J.-B., Liu M., Zou Y., Weissman I.L., Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman B.C., Northrup D.L., Allman D. Resolution of unique Sca-1highc-Kit- lymphoid-biased progenitors in adult bone marrow. J. Immunol. 2008;181:7514–7524. doi: 10.4049/jimmunol.181.11.7514. [DOI] [PubMed] [Google Scholar]
- Havelange V., Garzon R. MicroRNAs: emerging key regulators of hematopoiesis. Am. J. Hematol. 2010;85:935–942. doi: 10.1002/ajh.21863. [DOI] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhang Y., Bersenev A., O’Brien W.T., Tong W., Emerson S.G., Klein P.S. Pivotal role for glycogen synthase kinase-3 in hematopoietic stem cell homeostasis in mice. J. Clin. Invest. 2009;119:3519–3529. doi: 10.1172/JCI40572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Gschweng E., Van Handel B., Cheng D., Mikkola H.K.A., Witte O.N. Regulated expression of microRNAs-126/126∗ inhibits erythropoiesis from human embryonic stem cells. Blood. 2011;117:2157–2165. doi: 10.1182/blood-2010-08-302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntilla M.M., Patil V.D., Calamito M., Joshi R.P., Birnbaum M.J., Koretzky G.A. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115:4030–4038. doi: 10.1182/blood-2009-09-241000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnert F., Mancuso M.R., Hampton J., Stankunas K., Asano T., Chen C.Z., Kuo C.J. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- Li J. Quiescence regulators for hematopoietic stem cell. Exp. Hematol. 2011;39:511–520. doi: 10.1016/j.exphem.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Mayr C., Bartel D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott S.P., Eppert K., Lechman E.R., Doedens M., Dick J.E. Comparison of human cord blood engraftment between immunocompromised mouse strains. Blood. 2010;116:193–200. doi: 10.1182/blood-2010-02-271841. [DOI] [PubMed] [Google Scholar]
- Miko E., Czimmerer Z., Csánky E., Boros G., Buslig J., Dezső B., Scholtz B. Differentially expressed microRNAs in small cell lung cancer. Exp. Lung Res. 2009;35:646–664. doi: 10.3109/01902140902822312. [DOI] [PubMed] [Google Scholar]
- Notta F., Doulatov S., Laurenti E., Poeppl A., Jurisica I., Dick J.E. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Chaudhuri A.A., Rao D.S., Gibson W.S.J., Balazs A.B., Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi A.G.L., Sahoo D., Adorno M., Wang Y., Weissman I.L., Park C.Y. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorén L.A., Liuba K., Bryder D., Nygren J.M., Jensen C.T., Qian H., Antonchuk J., Jacobsen S.E.W. Kit regulates maintenance of quiescent hematopoietic stem cells. J. Immunol. 2008;180:2045–2053. doi: 10.4049/jimmunol.180.4.2045. [DOI] [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Laurenti E., Oser G., van der Wath R.C., Blanco-Bose W., Jaworski M., Offner S., Dunant C.F., Eshkind L., Bockamp E. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- Yilmaz Ö.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Zhang J., Du Y.-Y., Lin Y.-F., Chen Y.-T., Yang L., Wang H.-J., Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem. Biophys. Res. Commun. 2008;377:136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.