Abstract
MET receptor tyrosine kinase (MET) has been proposed as a candidate risk gene for autism spectrum disorder (ASD) based on associations between MET polymorphisms and ASD diagnosis, as well as evidence from animal studies that MET protein may regulate early development of cortical regions implicated in the neurobiology of ASD. The relevance of differences in MET signaling for human cortical development remains unexamined, however. We sought to address this issue by relating genotype at a functional single nucleotide polymorphism within the MET promoter (rs1858830, G→C), to in vivo measures of cortical thickness development derived from 222 healthy children and adolescents with 514 longitudinally acquired structural MRI brain scans between ages 9 and 22 years. We identified a statistically significant, developmentally fixed, and stepwise CT reduction with increasing C allele dose in superior and middle temporal gyri, ventral pre- and post-central gyri, and anterior cingulate bilaterally, and in the right fronto-polar cortex. We were also able to demonstrate that mean CT within these cortical regions showed a statistically significant reduction with increasing scores on a continuous measure of autistic traits (the Social Responsiveness Scale). The cortical regions highlighted by our analyses are not only established areas of MET expression during prenatal life, but are also key components of the “social brain” which have frequently shown structural and functional abnormalities in autism. Our results suggest that genetic differences in the MET gene may influence the development of cortical systems implicated in the neurobiology of ASD
Keywords: MET receptor tyrosine kinase, cortex, autism, development, MRI
Introduction
Autism spectrum disorder (ASD) is a phenotypically heterogeneous group of neurodevelopmental disorders characterized by impairments in communication and social interaction, as well as the presence of repetitive behaviors/restricted interests (American Psychiatric Association, 2000). Despite clear evidence that genetic factors play a prominent role in the etiology of ASD (Folstein & Rutter, 1977; State & Levitt, 2011), progress in identifying risk genes for ASD that carry a high population attributable risk, has been slow– likely reflecting the complex genetic architecture underlying ASD (Gillis & Rouleau, 2011), and limitations in the use of behaviorally-defined diagnostic categories as a way of parsing this complexity (Meyer-Lindenberg, 2010). Despite these challenges, certain genes have shown associations with ASD diagnosis in multiple independent studies, and one of the best such examples is MET receptor tyrosine kinase (MET) (Campbell, Li, Sutcliffe, Persico, & Levitt, 2008; Campbell et al., 2006; Jackson et al., 2009; Sousa et al., 2009; Thanseem et al., 2010). In addition to these data from genetic association studies, evidence from molecular biology also strengthens the candidacy of MET as an ASD risk gene.
MET receptor tyrosine kinase, with its ligand, hepatocyte growth factor (HGF), is now recognized to regulate several key aspects of early brain development, alongside its earlier established contributions to immune function and gastrointestinal repair (Campbell, 2009; Campbell et al., 2007). In the rat brain, Hgf and Met are expressed widely in the cerebral cortex, as well as in the hippocampus, pineal gland, choroid plexus, ependymal cells, and granule layer of the cerebellum (Hgf), and in septum and pons, (Met) (Gutierrez, Dolcet, Tolcos, & Davies, 2004). Evidence from both in vitro (Birchmeier & Gherardi, 1998; Gutierrez et al., 2004) and in vivo (Birchmeier & Gherardi, 1998; Judson, Eagleson, Wang, & Levitt, 2010; Powell, Mars, & Levitt, 2001) studies in mice suggest that Met signaling regulated processes are central to cortical developmental processes including dendritogenesis, arborization, and synaptogenesis. For example, knock-out mouse models with disrupted Met signaling during early postnatal development show cell-nonautonomous changes in dendritic morphology: proximal to the soma of anterior cingulate pyramidal neurons, knock-out mice show an increase in basal arbor length and branching complexity, while distally, they show a reduction in apical arbor length and complexity (Judson et al., 2010). Furthermore, knock-out mouse models show abnormal interneuron migration from the ganglionic eminence, with a reduction of interneurons in frontal and parietal cortex (Powell et al., 2001), and increased intracortical excitatory connections (Qiu, Anderson, Levitt, & Shepherd, 2011). These studies suggest that the MET pathway may be important for normal cortical development in humans.
The capacity of MET to regulate so many core aspects of early cortical development becomes especially relevant for models of ASD neurobiology given recent evidence for focal up-regulation of MET expression during prenatal synaptogenesis within regions of the primate brain that are crucial for social cognition (limbic structures such as the amygdala, hippocampus, and cingulate cortex) and sensory perception (extrastriate visual and auditory cortices of the temporal, inferior parietal, and occipital lobes) (Judson, Eagleson, & Levitt, 2011). Abnormal development of these structures is thought to play an important role in social aspects of the ASD behavioral phenotype– which have in turn been shown to be especially sensitive to genetic variation within MET (Campbell, Warren, Sutcliffe, Lee, & Levitt, 2010). Furthermore, a recent study charting patterns of differential gene expression in post-mortem cortical tissue found that normative regionalization of MET expression across social regions was strikingly attenuated in ASD individuals compared to controls (Voineagu et al., 2011). Taken together then, these data suggest that functional variation within MET may impact focal cortical development in humans within regions relevant for social aspects of the ASD behavioral phenotype.
The best-described source of genetic variation in MET signaling within humans is a common functional polymorphism within the MET promoter region [G→C single nucleotide polymorphism (SNP), rs1858830]. Presence of an ASD diagnosis has now been associated with carriage of a C allele at this SNP in five independent samples (Campbell et al., 2008; Campbell et al., 2006; Jackson et al., 2009). Furthermore, carriage of this variant appears to impact expression of MET within cortical regions where those with ASD show decreased MET expression as compared to healthy controls (i.e. temporal cortex) (Campbell et al., 2007). Given the aforementioned role of MET signaling in early cortical development, we hypothesized that C allele load at rs1858830 would impact structural maturation of the cortex in humans, and were particularly interested in known “hot-spots” of MET expression such as anterior cingulate, temporal, and inferior parietal cortices. We tested this hypothesis using an “imaging-genetics” (Hariri, Drabant, & Weinberger, 2006) paradigm in which we related C allele load at rs1858830 to measures of cortical thickness (CT) as derived from longitudinally acquired, structural magnetic resonance imaging (sMRI) scans in typically developing children and adolescents.
We selected CT as our morphometric index of interest because it is a highly heritable aspect of cortical anatomy (Lenroot et al., 2009), shows abnormalities in ASD (Hadjikhani, Joseph, Snyder, & Tager-Flusberg, 2006; Raznahan, Toro et al., 2010; Wallace, Dankner, Kenworthy, Giedd, & Martin, 2011), is sensitive to genotypic variation at other functional genetic polymorphisms of relevance to ASD (Raznahan et al., 2011) and can be reliably measured at several thousand vertices on each hemisphere using high-throughput automated procedures (Kabani, Le, MacDonald, & Evans, 2001). Given that genetic influences on CT variation change with age (Lenroot et al., 2009), and CT alterations in ASD may also change with age (Raznahan, Toro et al., 2010), we first tested if C allele load modified the rate of CT change in our sample, before moving on to consider any fixed and developmentally invariant associations between C allele load and CT. Finally, based on evidence that MET genotype modulates social and communication behavior in those with autism (Campbell et al., 2010), we further examined whether any observed genotype-dependent CT differences related to autism traits, using the Social Responsiveness Scale (SRS) (Constantino et al., 2003).
Methods
Participants
See Table 1 for full demographic information and genotype composition. We studied 222 healthy Caucasian individuals (124 male, 98 female) between the ages of 9 and 22 years on whom a total of 514 sMRI brain scans were collected longitudinally. Participants were recruited through local advertisement. Screening for neurological or psychiatric illness was conducted using the Child Behavior Checklist and a structured diagnostic interview administered by a child psychiatrist [as previously described, (Giedd et al., 1996) ]All participants had a full-scale intelligence quotient (IQ) of >80 (estimated using age-appropriate Wechsler intelligence scales). The institutional review board of the National Institutes of Health approved this study’s research protocol, and informed consent and assent to participate were obtained from parents and children respectively. Genotype groups did not differ significantly by age, sex, or IQ.
Table 1.
Demographics by MET Genotype Group
| GG | GC | CC | Total | |
|---|---|---|---|---|
| Participants | 41 | 95 | 86 | 222 |
| Sex | ||||
| M | 27 | 48 | 49 | 124 |
| F | 14 | 47 | 37 | 98 |
| IQ, mean (SD) | 114.02 (11.42) | 114.47 (12.59) | 111.60 (12.73) | |
| Subject no. scans | ||||
| ≥1 scan | 41 | 95 | 86 | 222 |
| ≥2 scans | 38 | 88 | 66 | 192 |
| ≥3 scans | 18 | 52 | 49 | 119 |
| Age, mean (SD) | ||||
| 1st scan | 12.74 (2.42) | 12.49 (2.79) | 12.41 (3.12) | |
| 2nd scan | 14.59 (2.72) | 15.11 (2.85) | 14.50 (2.42) | |
| 3rd scan | 17.19 (1.82) | 16.94 (2.68) | 17.16 (2.92) | |
| Age distribution | ||||
| Mean (SD) | 14.15 (3.61) | 14.67 (3.45) | 14.41 (3.56) | |
| Range | 9.00 – 22.55 | 9.00 – 22.78 | 9.00 – 22.78 | |
Genotyping
For each participant, DNA was extracted from previously prepared lymphoblastoid cell lines using standard methods (Qiagen, MD, USA). It has been established that conversion of cells into lymphoblastoid lines does not cause errors into SNP genotyping (Herbeck et al., 2009). Genotyping of rs1858830 was performed by Prevention Genetics (Marshfield, WI, USA), using sub-microliter allele-specific polymerase-chain-reactions (Hawkins et al., 2002) [Common primer (GCTCGCGCCCTCCACTCG), allele-specific primers (GGCGCTGGGCTCAGCCC / GGCGCTGGGCTCAGCCG)], with the following conditions: 94°C for 15 minutes, and 25–35 cycles of 94°C for 20 seconds, 57°C for 40 seconds and 72°C for 40 seconds. DNA sequencing of positive controls was conducted to ensure correct assignment of genotypes. Genotype frequencies did not deviate from Hardy-Weinberg equilibrium (p=0.1).
Neuroimaging
Each participant had at least one sMRI scan (mean age=12.3 years, SD=2.9), 155 had at least two scans (mean age=14.8 years, SD=2.7), and 90 had three or more scans (mean age at third scan=17.1 years, SD=2.7). Mean time between scans was 2.76 years (SD=1.04). All scans were T-1 weighted images obtained on the same 1.5-T General Electric (Milwaukee, WI) Sigma scanner, with contiguous 1.5 mm axial slices and 2.0 mm coronal slices. A 3D spoiled gradient recalled echo sequence was used, with the following parameters: 5 ms echo time, 24 ms repetition time, 45° flip angle, 256×192 acquisition matrix, 1 excitation, and 24 cm field of view. Head placement was standardized as described previously.
Cortical thickness (CT) was measured at approximately 80,000 points across the entire cortical sheet from each scan using the CIVET (version 1.1.8, http://wiki.bic.mni.mcgill.ca/index.php/CIVET) set of automated algorithms [as previously detailed, (Raznahan et al., 2011)]. A 30 mm bandwidth blurring kernel was applied as to maximize statistical power and minimize false positives (Lerch & Evans, 2005).
Statistical Analysis
Linear mixed effects models were used to model the main effects of age and C allele dose at each vertex. Genotype was modeled in an ordinal fashion based on prior behavioral genetic (Campbell et al., 2006) and molecular (Campbell et al., 2007) data. Age was modeled linearly, based on (i) previously published descriptions of age-related CT change within the age-range 9 –22 years (Shaw et al., 2008), and (i) lack of evidence from preliminary analyses within our current sample for significant non-linear age effects. Preliminary analyses found no significant interactions between age and genotype for CT, or sensitivity of our findings to the inclusion of sex, handedness, and IQ as covariates. Thus CT was modeled at each vertex for ith individuals, jth families, kth timepoint as:
CTijk = intercept + di + dij + β1 (age) + β2 (C-allele load) + eijk
where intercept and β terms are fixed effects, eijk is the residual error, and d terms represent within-family and within-person dependence of observations. Reported results were significant after applying a q ≤ 0.05 False Discovery Rate threshold across p-values, and were visualized by projecting the t-statistics onto a standard brain template. This threshold provided a “mask” that defined cortical regions where CT variation was significantly related to C-allele load, which could then be used to derive a single estimate of mean CT across all such regions for each available sMRI scan. The relationship between SRS score and mean within-mask CT for ith individuals, jth families, kth timepoint was then modeled as:
CTijk = intercept + di + dij + β1 (age) + β2 (SRS score) + eijk
where intercept and β terms are fixed effects, eijk is the residual error, and d terms represent within-family and within-person dependence of observations.
Behavioral Autism Traits Measure
Linear mixed effects models were used to test for a relationship between mean CT within cortical regions sensitive to C allele load, and inter-individual differences in a well-established continuous measure of autistic traits: the Social Responsiveness Scale (SRS) (Constantino et al., 2003). SRS scores show high sensitivity and specificity for ASD compared to controls and other mental disorders, and they correlate strongly with gold standard ASD assessments including Autism Diagnostic Interview and Autism Diagnostic Observation Schedule subscales (Bolte, Westerwald, Holtmann, Freitag, & Poustka, 2011). Participants’ parents completed this 65-item questionnaire, and the total score was used for analysis. This analysis was limited to the 131 participants within our sample on whom SRS data were available.
Results and Discussion
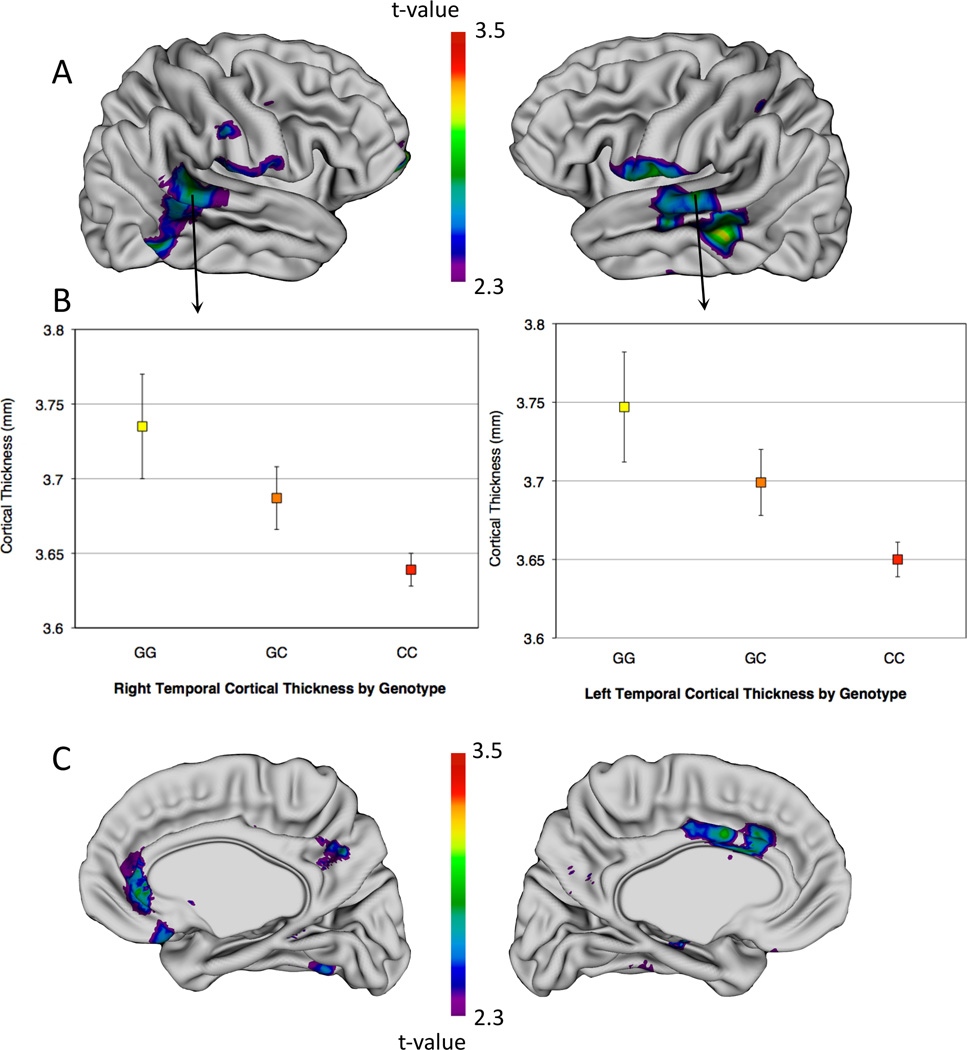
We identified developmentally fixed, stepwise CT reductions with increasing C allele dose within superior and middle temporal gyri, ventral pre- and post-central gyri, and anterior cingulate bilaterally, and in fronto-polar cortex in the right hemisphere (see Figure 1). There were no cortical regions where increasing C-dose was associated with a stepwise increase in CT. Furthermore, in the subset of participants with SRS scores, mean CT within these regions showed a statistically significant (p=0.002) reduction with greater SRS score (i.e., higher autistic-trait ratings). Genotype did not modify this relationship (p=0.7) or show a significant direct relationship with SRS score (p=0.7).
Figure 1.
Cortical thickness differences by genotype group. (A and C) T-statistics are projected onto a brain template (A, right and left lateral view; C, right and left medial view), showing regions where there was a developmentally fixed, stepwise CT reduction with increased C-allele dose (false discovery rate-adjusted p < 0.05, t > 2.3). (B) Mean CT (± s.e.) by genotype at representative right superior temporal gyrus (left) and left middle temporal gyrus (right) loci, illustrating MET dosage effects.
The links we find between MET genotype and cortical anatomy are highly convergent with existing maps of cortical MET expression (Judson et al., 2011), the step-wise manner in which rs1858830 is thought to modify MET expression levels (Campbell et al., 2007), and evidence from animal studies that modified MET signaling impacts cortical development (Gutierrez et al., 2004; Powell et al., 2001; Qiu et al., 2011). The age-invariant quality of genetic effects within our sample suggests that genetic variations in MET signaling may be more impactful during earlier neurogenesis and synaptogenesis (Judson et al., 2010; Powell et al., 2001)when cortical MET expression is thought to peak (Judson et al., 2011). Hypotheses regarding the cellular processes that might underlie our human neuroimaging findings are necessarily speculative however.
Our novel finding that anatomy within cortical regions sensitive to MET genotype also tracks a measure of autistic traits in healthy controls supplements established links between (i) brain structure and function within these same regions and socio-communicative impairments in ASD (Amaral, Schumann, & Nordahl, 2008), (ii) decreased CT in these regions and greater autistic traits amongst typically developing children and adolescents (Wallace et al., 2012), and (iii) greater rs1858830 C allele dose and more impaired social functioning within ASD (Campbell et al., 2010). It is notable that we do not find a statistically significant relationship between MET genotype and SRS score, despite there being significant relationships between genotype and cortical anatomy, as well as between cortical anatomy and SRS score. This may be because we explicitly examined behavior within the normal range (SRS total score range = 0–59, mean = 22.09, SD = 14.26), and therefore the full variability of SRS scores was not represented within our sample. More fundamentally, the expectation that genetic effects will be more easily detected for biological than behavioral phenotypes is one of the prime motivators of an imaging genetics approach (Meyer-Lindenberg, 2010). Under this paradigm, the capacity for MET genotype to shape structural development of social brain regions may still be relevant for disease models of ASD without penetrance of genetic effects for SRS score.
While our data identify a candidate biological mechanism for modification of ASD risk by MET, there are a number of limitations, and further studies will be required to establish replicability of our findings. While CT change has been found to be largely linear within our age range (Shaw et al., 2008), a larger sample spanning a greater age range could use a more complex model to examine non-linear changes over time. Thickness is only one aspect of cortical anatomy, and we did not measure folding or surface area - which can show dissociable genetic effects from CT (Raznahan, Cutter et al., 2010). Future studies could also include complementary white matter measurements. Using only healthy controls in this study is a strength in that gene effects are studied in the absence of disease-related confounds, however, further work is required in ASD populations to understand how MET regulation of CT is related to ASD risk and symptom profiles. Finally, future studies of the larger MET signaling pathway are needed to better understand how MET and its many signaling partners interact to shape histological and neuroimaging markers of brain development.
Acknowledgments
This research was supported by the National Institutes of Health, National Institute of Mental Health Intramural Research and a UK MRC Clinical Research Training Grant awarded to AR (G0701370). We thank the participants and families who took part in this study.
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
References
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends in Neurosciences. 2008;31(3):137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends in Cell Biology. 1998;8(10):404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- Bolte S, Westerwald E, Holtmann M, Freitag C, Poustka F. Autistic traits and autism spectrum disorders: the clinical validity of two measures presuming a continuum of social communication skills. Journal of Autism and Developmental Disorders. 2011;41(1):66–72. doi: 10.1007/s10803-010-1024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB. When linkage signal for autism MET candidate gene. European Journal of Human Genetics. 2009;17(6):699–700. doi: 10.1038/ejhg.2008.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, D'Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Annals of Neurology. 2007;62(3):243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Li C, Sutcliffe JS, Persico AM, Levitt P. Genetic evidence implicating multiple genes in the MET receptor tyrosine kinase pathway in autism spectrum disorder. Autism Res. 2008;1(3):159–168. doi: 10.1002/aur.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, et al. A genetic variant that disrupts MET transcription is associated with autism.[see comment] Proceedings of the National Academy of Sciences of the United States of America. 2006;103(45):16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, Warren D, Sutcliffe JS, Lee EB, Levitt P. Association of MET with social and communication phenotypes in individuals with autism spectrum disorder. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):438–446. doi: 10.1002/ajmg.b.30998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Davis SA, Todd RD, Schindler MK, Gross MM, Brophy SL, et al. Validation of a Brief Quantitative Measure of Autistic Traits: Comparison of the Social Responsiveness Scale with the Autism Diagnostic Interview-Revised. [References] Journal of Autism & Developmental Disorders. 2003;33(4):427–433. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. Journal of Child Psychology & Psychiatry & Allied Disciplines. 1977;18(4):297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Giedd JN, Snell JW, Lange N, Rajapakse JC, Casey BJ, Kozuch PL, et al. Quantitative magnetic resonance imaging of human brain development: ages 4–18. Cerebral Cortex. 1996;6(4):551–560. doi: 10.1093/cercor/6.4.551. [DOI] [PubMed] [Google Scholar]
- Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131(15):3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- Hadjikhani N, Joseph RM, Snyder J, Tager-Flusberg H. Anatomical differences in the mirror neuron system and social cognition network in autism. Cerebral Cortex. 2006;16(9):1276–1282. doi: 10.1093/cercor/bhj069. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Drabant EM, Weinberger DR. Imaging Genetics: Perspectives from Studies of Genetically Driven Variation in Serotonin Function and Corticolimbic Affective Processing. Biological Psychiatry. 2006;59(10):888–897. doi: 10.1016/j.biopsych.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Herbeck JT, Gottlieb GS, Wong K, Detels R, Phair JP, Rinaldo CR, et al. Fidelity of SNP array genotyping using Epstein Barr virus-transformed B-lymphocyte cell lines: implications for genome-wide association studies. PLoS One. 2009;4(9):e6915. doi: 10.1371/journal.pone.0006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, et al. Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2009;2(4):232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- Judson MC, Eagleson KL, Levitt P. A new synaptic player leading to autism risk: Met receptor tyrosine kinase. J Neurodev Disord. 2011 doi: 10.1007/s11689-011-9081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson MC, Eagleson KL, Wang L, Levitt P. Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain. Journal of Comparative Neurology. 2010;518(21):4463–4478. doi: 10.1002/cne.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani N, Le GG, MacDonald D, Evans AC. Measurement of cortical thickness using an automated 3-D algorithm: a validation study. Neuroimage. 2001;13(2):375–380. doi: 10.1006/nimg.2000.0652. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, et al. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Human Brain Mapping. 2009;30(1):163–174. doi: 10.1002/hbm.20494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005;24(1):163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A. Intermediate or brainless phenotypes for psychiatric research? Psychological Medicine. 2010;40(7):1057–1062. doi: 10.1017/s0033291709991929. [DOI] [PubMed] [Google Scholar]
- Powell EM, Mars WM, Levitt P. Hepatocyte Growth Factor/Scatter Factor Is a Motogen for Interneurons Migrating from the Ventral to Dorsal Telencephalon. Neuron. 2001;30(1):79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Qiu S, Anderson CT, Levitt P, Shepherd GM. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase. Journal of Neuroscience. 2011;31(15):5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Cutter W, Lalonde F, Robertson D, Daly E, Conway GS, et al. Cortical anatomy in human X monosomy. Neuroimage. 2010;49(4):2915–2923. doi: 10.1016/j.neuroimage.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lee Y, Long R, Greenstein D, Clasen L, Addington A, et al. Common functional polymorphisms of DISC1 and cortical maturation in typically developing children and adolescents. Molecular Psychiatry. 2011;16(9):917–926. doi: 10.1038/mp.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Toro R, Daly E, Robertson D, Murphy C, Deeley Q, et al. Cortical anatomy in autism spectrum disorder: an in vivo MRI study on the effect of age. Cerebral Cortex. 2010;20(6):1332–1340. doi: 10.1093/cercor/bhp198. [DOI] [PubMed] [Google Scholar]
- Shaw P, Kabani NJ, Lerch JP, Eckstrand K, Lenroot R, Gogtay N, et al. Neurodevelopmental trajectories of the human cerebral cortex. J Neurosci. 2008;28(14):3586–3594. doi: 10.1523/JNEUROSCI.5309-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa I, Clark TG, Toma C, Kobayashi K, Choma M, Holt R, et al. MET and autism susceptibility: family and case-control studies. European Journal of Human Genetics. 2009;17(6):749–758. doi: 10.1038/ejhg.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nature Neuroscience. 2011;14(12):1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, Nakamura K, Miyachi T, Toyota T, Yamada S, Tsujii M, et al. Further evidence for the role of MET in autism susceptibility. Neuroscience Research. 2010;68(2):137–141. doi: 10.1016/j.neures.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GL, Dankner N, Kenworthy L, Giedd JN, Martin A. Age-related temporal and parietal cortical thinning in autism spectrum disorders. Brain. 2011 doi: 10.1093/brain/awq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace GL, Shaw P, Lee NR, Clasen LS, Raznahan A, Lenroot RK, et al. Distinct cortical correlates of autistic versus antisocial traits in a longitudinal sample of typically developing youth. Journal of Neuroscience. 2012;32(14):4856–4860. doi: 10.1523/JNEUROSCI.6214-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]