Summary
HLA-DR molecules bind microbial peptides in an endosomal compartment and present them on the cell surface for CD4 T cell surveillance. HLA-DM plays a critical role in the endosomal peptide selection process. The structure of the HLA-DM – HLA-DR complex shows major rearrangements of the HLA-DR peptide binding groove. Flipping of a tryptophan away from the HLA-DR1 P1 pocket enables major conformational changes that position hydrophobic HLA-DR residues into the P1 pocket. These conformational changes accelerate peptide dissociation and stabilize the empty HLA-DR peptide binding groove. Initially, incoming peptides have access to only part of the HLA-DR groove and need to compete with HLA-DR residues for access to the P2 site and the hydrophobic P1 pocket. This energetic barrier creates a rapid and stringent selection process for the highest-affinity binders. Insertion of peptide residues into the P2 and P1 sites reverses the conformational changes, terminating selection through DM dissociation.
Introduction
T cells continuously patrol the body, sensing pathogen entry by T cell receptor (TCR) recognition of proteolytic pathogen fragments displayed on the cell surface bound to MHC proteins (York and Rock, 1996). Only a very small fraction of naïve T cells in the repertoire recognizes a given peptide (in mice, approximately 20–200 cells) (Moon et al., 2007). Thus, peptides acquired in intracellular compartments need to be displayed by MHC proteins on the cell surface for extended time periods for efficient recruitment of naïve T cells into an anti-microbial response. MHC class II (MHCII) molecules present peptides from exogenous (and some internal) antigens to CD4 T cells which coordinate ensuing immune responses (Cresswell, 1994). The importance of CD4 T cells is illustrated by the severity of the acquired immunodeficiency syndrome (AIDS) in patients infected by the human immunodeficiency virus (HIV) (Gandhi and Walker, 2002).
MHCII proteins aggregate in the absence of peptide (Germain and Rinker, 1993; Rabinowitz et al., 1998), and the hydrophobic peptide-binding groove is initially protected by invariant chain (Roche and Cresswell, 1990b). Upon arrival in a late endosomal compartment, invariant chain is degraded by several proteases, leaving class II-associated invariant chain peptides (CLIP) in the peptide binding groove (Riberdy et al., 1992; Roche and Cresswell, 1991). DM (HLA-DM in humans, H2-DM in mice) catalyzes CLIP dissociation, stabilizes empty MHCII proteins, and enables rapid binding of microbial peptides generated by limited proteolysis (Denzin and Cresswell, 1995; Denzin et al., 1996; Kropshofer et al., 1997; Sherman et al., 1995; Sloan et al., 1995). A substantial antigen presentation defect is observed in human cell lines and mouse strains deficient in DM expression (Fung-Leung et al., 1996; Martin et al., 1996; Mellins et al., 1990; Miyazaki et al., 1996; Morris et al., 1994).
DM can catalyze dissociation of any MHCII-bound peptide and drives selection of peptides with the highest binding affinity (Katz et al., 1996; Sloan et al., 1995; Weber et al., 1996). This selection process (also referred to as ‘editing’) has a major impact on the peptide repertoire: DM extinguishes presentation of many low-affinity peptides, while greatly enhancing presentation of high-affinity binders. For example, direct quantification of peptides from hen egg lysozyme bound to I-Ak (a murine MHCII protein) showed that the abundance of the immunodominant peptide was reduced >1,100-fold in the absence of DM (Lovitch et al., 2003). The structural mechanisms of DM-driven peptide selection are not well understood. Mutagenesis experiments identified large lateral surfaces of DM and MHCII proteins involved in the interaction, and several mutants have implicated a region in the vicinity of the peptide N-terminus (Anders et al., 2011; Doebele et al., 2000; Painter et al., 2011; Pashine et al., 2003). A central problem is that most peptides bind with similar on-rates to MHCII proteins, and many ‘low-affinity’ peptides have a half-life of several hours (Rabinowitz et al., 1998). It is unknown how DM antagonizes presentation of peptides with a t1/2 of <14 hours, while enhancing display of peptides with a t1/2 of >33 hours (Lazarski et al., 2006). The structure of a MHCII-DM complex is required to answer these questions, but this has been a significant challenge for the field since the structures of the individual proteins were reported (Brown et al., 1993; Fremont et al., 1998; Mosyak et al., 1998; Stern et al., 1994). This challenge is closely related to the function of DM, which is to stabilize an unknown MHCII conformational intermediate that binds peptides with rapid kinetics.
Results
Crystallization of the HLA-DM – HLA-DR1 complex
We recently showed that DM does not detectably bind to HLA-DR (a human MHCII protein, abbreviated as DR) when the groove is fully occupied by a covalently linked peptide. Rather, DM binding requires truncation of several N-terminal peptide residues, including the anchor for the P1 pocket (Anders et al., 2011). The hydrophobic P1 pocket is critical for stable peptide binding to DR molecules: mutation of the P1 peptide side chain to a polar residue prevents stable binding, while introduction of an aromatic residue into a polyalanine sequence (with a single lysine at a TCR contact residue) yields a high-affinity DR1 binder (Jardetzky et al., 1990; Stern et al., 1994).
We reasoned that covalent linkage of an N-terminally truncated peptide might enable crystallization of the DM-DR1 complex by trapping of a short-lived DM-sensitive DR1 conformer (Figure 1A). Soluble DR1 was first expressed with a full length low-affinity peptide (CLIPLow) and the linker between the peptide C-terminus and the N-terminus of the DRβ chain (Kozono et al., 1994) was cut with endoproteinase GluC. CLIPLow was then exchanged with an N-terminally truncated peptide, which was trapped in the groove under mildly oxidizing conditions by formation of a disulfide bond between a cysteine at peptide position P6 and DR αV65C. This peptide was similar to the influenza hemagglutinin (HA306–318) peptide previously used to crystallize DR1 (Stern et al., 1994), but lacked three N-terminal residues (P-2, P-1, P1) and thus spanned from P2 to P11. Initial co-crystallization experiments did not yield crystals. However, it was previously shown that the DM and DR transmembrane domains increase DM activity ~200 to 400-fold by properly positioning the molecules (Weber et al., 2001). Similarly, attachment of leucine zippers to both β chains yielded an active complex (Busch et al., 2002). We mimicked this positioning effect by linking DRβ and DMβ C-termini using sortase A, a transpeptidase from Staphylococcus aureus (Mao et al., 2004; Mazmanian et al., 1999). This directional approach avoided formation of homodimers of input proteins, a problem encountered during attempts to link DM and DR through a disulfide bond. Sortase A cleaves the peptide bond between threonine and glycine of a C-terminal LPATG motif and forms a covalent bond with the N-terminal amino group of an oligo-glycine. We attached the sortase tag to DRβ and linked a peptide with N-terminal glycine residues to an introduced cysteine at the C-terminus of the DMβ extracellular domain. The linked DM-DR1 complex crystallized under a variety of different conditions, and structures were determined for crystals grown at pH 5.5 and 6.5. DM has optimal activity at an acidic pH (Denzin and Cresswell, 1995), and the description therefore focuses primarily on the pH 5.5 structure.
Figure 1. Overview of the HLA-DM – HLA-DR1 complex.
(A) Generation of a covalent DM-DR1 complex with a linked peptide that lacks three N-terminal residues.
(B, E) Top-view of the DM-DR1 complex shows DM interaction site on the DRα chain and a partially empty peptide binding groove. DMα is colored red, DMβ yellow, DRα cyan and DRβ blue. Peptide and the DR αV65C covalent attachment site are colored orange.
(C, D, F) Side views of the DM-DR1 complex, with contact residues of DR1 (C) or DM (D) presented as surfaces, emphasizing interactions between DRα and DMα chains. The C-termini of DRβ and DMβ chains are indicated (C).
(F) The location of the peptide N-terminus in the DR1-HA306–318 structure is indicated with an arrow. See also Figure S1 as well as Movies S1 and S2.
Overview of the HLA-DM – HLA-DR1 complex
The structure of the complex of DM and DR1 was determined at a resolution of 2.6 Å in space group P212121 (one molecule in the asymmetric unit). The structure was refined to final R/Rfree factors of 19.7/24.1% (Table S1). An example of electron density at the interface is shown in Figure S1A. There are no crystal contacts involving the peptide or DR1 helices in relevant parts of the groove (Figure S1C, D and Table S2). No electron density for the linker is observed in either of the two structures. The distance between the C-termini of DRβ and DMβ is 19.9 Å, and no electron density is visible for 25 amino acids and a maleimide group connecting the C-termini (~extended length of ~75 Å).
The ectodomains of DM and DR1 are oriented parallel to each other, consistent with their co-localization on membranes in late endosomal compartments (Figure 1C, D). The area of the DM-DR1 interface is 1595 Å2 and the shape complementarity is 0.644. The interface is dominated by the α chains of DM and DR1 (66% of interface) (Figure 1C). DM binds to a lateral surface of the DRα1 domain, close to the peptide-binding groove, without contacting the DRβ1 domain or obstructing the open end of the groove where peptide N-termini can exit (Figure 1B, E, F). In fact, a large cavity is present at this site, explaining how long antigen fragments can bind before being trimmed to shorter peptides (Castellino et al., 1998; Nelson et al., 1997). The interface with the DRα1 domain is formed by DMα1 and to a lesser extent DMβ1 (Figure 1B, C, E and Figure S2A–D). A small interface among the β2 domains of DM and DR (19.5% of total) stabilizes the overall topology (Figure S2E). Previously reported mutations localize to the DM-DR interface (Anders et al., 2011; Doebele et al., 2000; Painter et al., 2011; Pashine et al., 2003), providing independent support for the structure (Figure 2A, B). The linked peptide spans from P2 to P11, but unambiguous density is only observed for the P5-P11 segment (Figure 1B, E). Thus, more than half of the binding groove is largely devoid of peptide.
Figure 2. Key residues involved in the DM – DR1 interaction.
(A) Molecules are shown in cartoon representation, and interacting residues are surface rendered. Mutations that affect the interaction are indicated.
(B) List of DM and DR mutants (new and previously identified), grouped by functional activity (new mutations are underlined). These residues are located at the DM-DR1 interface, except DR αP96 (mutation introduces glycan), DM βA55 (residue not surface exposed) and DM βE8 (mutation to lysine may affect DM conformation). DR αF51 and αE55 have moved into the peptide binding groove in the DM-DR1 complex, as described in Figures 4 and 5.
(C) Conservation of key contact residues. Alignments focus on the DR1 α1 domain (res. 31–61) and two segments of DMα (res. 91–101 and 121–141). Interacting residues are colored blue (DRα) and yellow (DMα). Sequences were retrieved from the www.ensembl.org website (June 2012). Key DRα - DMα contact residues are highlighted in red; DR residues that have move into the groove are colored green. See also Figure S2.
Flipping of DR αW43 from the P1 pocket towards DM
In the previously reported DR1/HA306–318 structure (PDB:1DLH) (Stern et al., 1994), DR αW43 forms a lateral wall of the P1 pocket and interacts with the P1 tyrosine of the HA peptide (Figure 3A). In addition, it stabilizes residues in the vicinity of the P1 pocket through many interactions. In the DM-DR1 complex, αW43 is rotated out of the groove and its indole ring nitrogen forms a hydrogen bond with DM αN125 (Figure 3B, D and Figure S3A–D). DM αN125 lies on the edge of a hydrophobic pocket with which the indole ring interacts (Figure 3E, Table S2). The interaction between DR αW43 and DM αN125 represents a key feature of the complex. Mutation of DM αN125 to alanine results in an almost complete loss of DM activity (Figure 3F, Figure S3E), and responsiveness to DM is greatly reduced by a conservative DR αW43F mutation (Anders et al., 2011). Also, both residues are entirely conserved among DMA and DRA homologs across all examined species, including chicken (Figure 2C). DR αW43 is even conserved in coelacanth, an ancient fish species that was thought to be extinct before its rediscovery. DR αW43 is also conserved among the polymorphic human DQA and DPA genes (except DPA1*01:10, possibly a loss of function variant).
Figure 3. HLA-DR αW43 is critical for HLA-DM binding.
(A) Structure of DR1-HA306–318 complex showing packing interactions of DR αW43 (red) with hydrophobic residues, including peptide P1 tyrosine (green).
(B) W43 stabilizes P1 pocket in DR1-HA306–318 (white) but is rotated away from the P1 pocket in the DM-DR1 complex (red). Conformational changes in DM-DR1 (cyan) compared to DR1-HA306–318 (yellow) include movement of β strands and extension of the helix to include DR α52–55.
(C) Overall conformation of DR binding groove in DM-DR1 complex compared to DR1-HA306–318.
(D) Hydrogen bonding network established by DR αW43 and αK38/αE40.
(E) Pocket on DM surface occupied by DR αW43.
(F) Effect of DM αN125 mutations on rate of peptide binding, measured using a real-time peptide binding assay based on fluorescence polarization (FP). Binding of a fluorescent peptide to a large protein reduces its tumbling rate in solution (measured in mP units). Graph shows change in rate of initial peptide binding (mP min−1). See also Figure S3.
In MHC class I (MHCI) molecules the entire peptide-binding groove is flanked by two long α helices (α1 and α2) (Bjorkman et al., 1987). In MHCII-peptide complexes, the α chain does not form a helix along the entire length of the peptide. Rather, DRα chain residues 52–55 form a short strand parallel to the bound peptide (Figure 3A) which is stabilized by conserved hydrogen bonds to the backbone of N-terminal peptide residues (P-2, P-1 and P1) (Stern et al., 1994). This entire region is substantially changed in the DM-DR1 complex. The strand and the neighboring 310 helix (DR α46–50) (Figure 3A) are merged into a helix (Figure 3B, C). Essentially, this results in formation of an extended helix from DR α46 to 77, with a break at A56-G58. This conformational change would not be possible without the DR αW43 flip which creates the required void. The conformational change is facilitated by loss of stabilizing interactions of αW43 with neighboring structural elements, including the β sheet floor of the groove, the 310 helix (DR α46–50) and the DR α52–55 strand (Figure 3A, B).
In addition, substantial conformational changes are observed on the floor of the peptide-binding groove in the vicinity of the P1 pocket. In the DR1-HA306–318 structure, strands S3 and S4 (res. α29–35 and α39–44, respectively) are in a strained conformation. In the DM-DR1 structure, both strands have moved away from the DRα1 helix and towards the main β sheet platform (Figure 3B), which positions two key residues of the S4 strand and S3/S4 loop (DR αE40 and αK38, respectively) at the interface with DM (Figure 3D and Figure S1A, B). These charged residues form an extended hydrogen-bonding network which includes DM αD183, αR98 and αH180 (Figure 3D and Figure S3F, G). The importance of this network is highlighted by previous mutagenesis data: mutation of DM αR98 greatly impairs DM function (Anders et al., 2011), while mutation of DR αE40 results in unresponsiveness to DM (Doebele et al., 2000).
Stabilization of the P1 pocket by DR αF51 and βF89
Previous mutagenesis studies showed that mutation of DR αF51 to alanine abrogates responsiveness to DM (Doebele et al., 2000; Painter et al., 2011). F51 points out of the groove in the DR1-HA306–318 structure (Figure 4A), which suggested that it represents an important DM interaction site close to the peptide N-terminus. However, the structure of the DM-DR1 complex reveals a dramatic repositioning of F51 into the P1 pocket (a 13Å movement, measured from the tip of the phenyl ring) (Figures 4B, 5B and Figure S4). This movement results from folding of the DR α52–55 strand into a helical conformation and the accompanied helix tilt, as described above (Figure 3B, C). In the DM-DR1 complex, F51 thus stabilizes the most hydrophobic site of the groove (Figure 4B, 5B). The P1 area is further stabilized by a conformational change in the DR β86–91 segment that positions βF89 near the P1 pocket where it closely interacts with DR αF51 (Figure 5C, D). This explains why empty DR molecules are stable when bound to DM (Kropshofer et al., 1997).
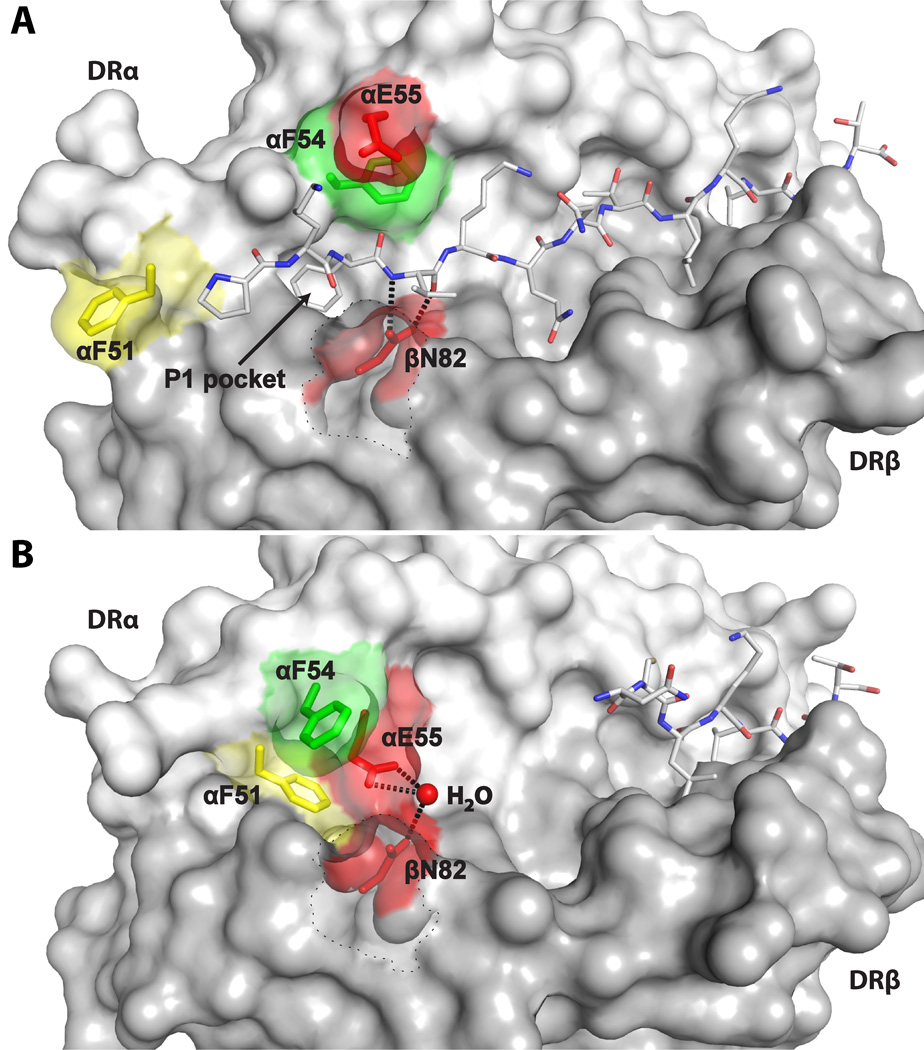
Figure 4. Large conformational changes stabilize the empty binding groove around the P1 pocket in the DM-DR1 complex.
(A) Surface representation of DR1 with fully bound HA306–318 peptide showing αF51 (yellow), αF54 (green) and αE55 and βN82 (both red). αF51 is pointing out of the groove and αE55 is located outside the binding site. DR βH81 (dotted line) is rendered transparent to improve visibility of DR βN82. DR β82 forms bidentate hydrogen bonds with the peptide backbone at P2.
(B) In the DM-DR1 complex, αF51 has moved into the P1 pocket, stabilizing this hydrophobic site. DR αE55 has moved into the groove where it forms a water-mediated hydrogen bond with DR βN82. See also Figure S4.
Figure 5. Stabilization of P1 site by DR αF51 and DR βF89.
(A) Top view of DR1-HA306–318 complex, with peptide P1 tyrosine (green) in P1 pocket.
(B) DR αF51 moved into highly hydrophobic P1 pocket in DM-DR1 complex.
(C&D) DR βF89 (red) also moved into groove in DM-DR1 complex, stabilizing αF51. Position of DR βF89 (*) in DR1-HA306–318 complex is indicated in grey (C).
(E) SPR analysis of DM binding by DR αF51 mutants: injection of DR-CLIP (stage 1, DR binding), buffer (stage 2, dissociation of empty DR), peptide (stage 3, peptide-dependent dissociation of DM-DR complex).
(F) Kinetics of peptide binding by DR αF51 mutants +/− DM. Peptide binding was examined in a real-time fluorescence polarization (FP) readout (mP units), as explained in Figure 3F. See also Figure S5.
The P1 pocket of DR1 is optimally filled by bulky aromatic residues, and we tested DR αF51 mutants for DM binding (Figure 5E) and DM-catalyzed peptide exchange (Figure 5F). The size of the hydrophobic DR α51 residue was critically important for DM binding, with a tryptophan at this position having more activity than the wild type phenylalanine, while smaller leucine and valine mutants were less responsive (some HLA-DR alleles have a smaller P1 pocket, which accommodates a phenylalanine residue but not tyrosine or tryptophan). A previously reported DRβ G86Y mutation fills the P1 pocket and would prevent the conformational changes we describe for the DM-DR1 structure, consistent with the observation that this mutation greatly reduces responsiveness to DM (Chou and Sadegh-Nasseri, 2000).
DR αE55 also moves into the groove where it forms a water-mediated hydrogen bond with DR βN82 (Figure 4B, Figure S5). The bidentate hydrogen bonds formed by DR βN82 and the P2 peptide backbone (Figure 4A) make a crucial contribution to stable peptide binding: mutation of this residue accelerates peptide dissociation from DR1 approximately 3000-fold (Zhou et al., 2009). Movement of DR αE55 into the groove may therefore contribute to peptide dissociation.
Effect of pH on DM-DR1 conformation
DM activity is regulated by pH, with an optimum in the 5.0–5.5 range of late endosomes. We determined a second structure at pH 6.5 in order to examine how pH affects the DM-DR1 interaction (Figure 6 and Tables S1, S3). SPR studies demonstrated DM binding by DR1 with a linked P2-P11 peptide at pH 6.5, with a ~3-fold lower affinity compared to pH 5.5 (Figure S6A, B). The complex crystallized in the same space group (P212121, one molecule in the asymmetric unit), and the unit cell dimensions are similar to the pH 5.5 condition. Overall, the pH 6.5 structure is very similar to the pH 5.5 structure with an overall RMSD of 0.44. Also, the overall interface with DM is similar, including the interaction of DR αW43 and DM αN125.
Figure 6. Conformational differences between pH 6.5 and pH 5.5 DM-DR1 structures.
(A & B) Peptide electron density at pH 5.5 (A) and pH 6.5 (B).
(C) Differences in position of key DR residues at pH 6.5 (colored) and pH 5.5 (gray); pH 5.5 residues (*).
(D) Space filling model of hydrophobic DR residues of P1 pocket in pH 6.5 structure, DR αF51 (yellow) located partially in P1 site and βF89 (red) outside the groove.
(E & F) Differences in the interaction of an acidic pair (DMβ D31-E47) with DR1 at pH 5.5 (E) and pH 6.5 (F). See also Figure S6.
However, several major differences are evident. Very little density is present for the P2-P4 peptide segment in the pH 5.5 structure (with some residual density in the P4 pocket) (Figure 6A). This peptide segment is presumably in rapid motion and may bind at a low occupancy. Also, the P5 residue is pointing out of the groove and even the linked P6 residue is elevated. Mass spectrometry analysis confirmed the presence of the entire P2-P11 peptide in crystals grown at pH 5.5 (data not shown). In contrast, at pH 6.5 electron density is observed for all peptide residues at full occupancy (Figure 6B), and the conformation of the peptide backbone is similar to the DR1-HA306–318 structure. These pH-dependent differences in peptide electron density were confirmed by generating simulated annealing omit maps (Fo-Fc) in which the peptide model was omitted (data not shown).
Interestingly, DR αF51 is not located in the P1 pocket in the pH 6.5 structure, but at an intermediate position compared to the pH 5.5 DM-DR1 and DR1-HA306–318 structures (Figure 6C, D). DR βF89 is located outside the groove and does not stabilize DR αF51 in the P1 pocket (Figure 6C, D). Also, DR αE55 is present in a different rotamer in the pH 6.5 structure that does not sterically hinder access of peptide P2 to DR βN82 but rather forms a hydrogen bond with P2 (Figure 6C). The pH 6.5 structure thus shows an intermediate conformation of the DM-DR complex in which the P1 pocket is not fully stabilized and DR αE55 does not sterically hinder the P2 peptide residue.
A pair of acidic DM residues, βD31 and βE47, was previously shown to contribute to the pH dependence of DM activity (Nicholson et al., 2006). Both residues interact with DR1 in the pH 5.5 structure, but DM βE47 is located outside the interface at pH 6.5 (Figure 6E, F and Figure S6E–J, Table S3). These changes affect the position of several DR residues, including DR αQ57, αE55 and αF54 (Figure 6E, F and Figure S6E–J). The relevance of these interactions is supported by a DR αQ57A mutation, which reduces DM activity in a pH-dependent manner (Figure S6D).
Model of DM-driven peptide selection
Proteolytic degradation of invariant chain leaves CLIP in the DR peptide binding groove (Figure 7, step 1). No DM binding is detectable when the DR binding groove is fully occupied by a peptide. The peptide N-terminus needs to dissociate from the groove, a step that may be closely linked with flipping of DR αW43 (highlighted in red) away from the P1 pocket (step 2). The flipped DR αW43 side chain becomes a key DM interaction residue, and three DR1 residues move into the groove (step 2 to step 3). In the DM-bound state, these DR1 residues (DR αF51, βF89 and αE55, circled) stabilize the empty groove. Incoming peptides bind with rapid kinetics, but part of the groove initially remains inaccessible, driving a rapid peptide exchange process (step 4). Only peptides that successfully compete with DR residues for access to the P2 site and the P1 pocket are stably bound. Full occupancy of the groove reverses the conformational changes associated with DM binding and results in DM dissociation (step 5). DM thus binds an otherwise labile transition state in which a peptide is partially bound in the groove. It stabilizes such conformers and thereby reduces the energetic barriers associated with peptide dissociation and binding (Weber et al., 1996).
Figure 7. Model for key steps in DM – DR interaction.
(1) CLIP is bound in DR groove and DR αW43 (red, arrow) stabilizes P1 pocket.
(2) Peptide N-terminus dissociates from DR groove and DR αW43 rotates away from the P1 pocket, becoming available for interaction with DM. Other DR residues (arrows) move into groove during transition to DM-bound state.
(3) DM stabilizes empty DR, and DR αF51 and βF89 protect the hydrophobic P1 pocket.
(4) Rapid binding of peptides to partially accessible groove; peptides that do not successfully compete with DR residues (yellow) for P2 site and P1 pocket are not stably bound.
(5) Binding of the peptide N-terminus reverses conformational changes and results in DM dissociation.
Discussion
The two structures provide mechanistic insights into three central aspects of DM function: DM-catalyzed peptide dissociation, DM-dependent stabilization of empty DR and rapid selection of high-affinity peptides by the DM-DR complex.
How does DM accelerate dissociation of peptides from DR?
A series of studies showed that DM greatly accelerates dissociation of CLIP and other peptides from DR molecules, but the structural mechanisms have remained largely unclear (Denzin and Cresswell, 1995; Sherman et al., 1995; Sloan et al., 1995). We previously reported that DM binding can only be detected when the peptide N-terminus has dissociated from the DR groove (Anders et al., 2011), and the structure provides a mechanistic explanation for this finding. Loss of hydrogen bonds with the peptide backbone (at P-2, P-1 and P1) is required for folding of the DR α52–55 segment into a helical conformation, and the peptide P1 anchor residue needs to vacate the P1 pocket for movement of the DR αF51 side chain into this hydrophobic site.
The structures suggest that flipping of DR αW43 away from the P1 pocket and dissociation of the peptide N-terminus are closely linked events. Rotation of the DR αW43 side chain out of the groove should destabilize the P1 pocket and favor dissociation of the peptide N-terminus. Alternatively, if the N-terminus dissociates first, the resulting loss of hydrophobic packing interactions with the P1 side chain should facilitate rotation of the DR αW43 side chain away from the P1 pocket. In the absence of DM, the most likely outcome is a rapid reversal of these conformational changes, depending on the DR binding affinity of the more C-terminal parts of a peptide. The structures further suggest that flipping of DR αW43 is important for subsequent conformational changes. In the structure of the DR1-HA306–318 complex, DR αW43 forms lateral packing interaction with the β sheet floor of the groove (DR αF32), the 310 helix (DR αF48) and the DR α52–55 strand (Stern et al., 1994). In the DM-bound state, substantial conformational changes are observed for all three structural elements. The most significant change is the formation of a longer tilted α-helical segment stretching from DR α46–55. DR αW43 would clash with this helical segment if its side chain would not move away from the P1 pocket, suggesting that the DR αW43 flip is an early step in this sequence. This conformational change repositions DR αF51 into the P1 pocket, a step that is critical for DM binding, as shown by extensive mutagenesis experiments.
DM and DR molecules are localized on the same endosomal membranes, pre-positioning them for the lateral interaction mode seen in the structure (Pashine et al., 2003). Indeed, the presence of DM and DR transmembrane domains enhances DM activity ~200–400-fold (Busch et al., 1998; Weber et al., 2001). It is possible that low-affinity, short-lived binding interactions can occur between the membrane-proximal Ig domains of DM and DR when both molecules are membrane-tethered. The DM α1 and β1 domains may thus be able to rapidly interact with a short-lived DR conformer in which the peptide N-terminus is outside the groove and DR αW43 is available for binding to DM αN125. Both residues are fully conserved among all examined species, and their importance in the DM-DR interaction is further underscored by mutagenesis experiments.
Comparison of the pH 5.5 and 6.5 structures identifies another feature relevant for DM-catalyzed peptide dissociation. A large body of work has shown that spontaneous peptide dissociation from MHCII molecules is greatly accelerated by a low pH, even in the absence of DM (Reay et al., 1992). Furthermore, DM activity is highly pH dependent, with an optimum at the pH of late endosomes (pH 4.5–5.5) (Sloan et al., 1995). Very little electron density is observed for the P2-P4 peptide segment in the pH 5.5 structure, but the entire covalently linked peptide (from P2 to P11) is bound at pH 6.5. Of particular interest is the pair of acidic DMβ D31-E47 residues that interact with each other at pH 5.5, but have moved apart at pH 6.5. Mutation of both residues to the corresponding carboxamide (DMβ D31N-E47Q) was previously shown to extend the pH range of DM activity (~9-fold increase of DM activity at neutral pH) (Nicholson et al., 2006). These data suggest that the DMβ D31-E47 pair may be (partially) protonated at pH 5.5, but deprotonated at a higher pH. These changes affect the interaction with DRα, in particular the conformation of the critical DR α54–57 segment: only at the lower pH, the side chain of DR αE55 forms a water-mediated hydrogen bond with DR βN82. In this position, DR αE55 sterically hinders an interaction between the peptide P2 residue and DR βN82 and prevents formation of energetically important hydrogen bonds (Zhou et al., 2009).
The following steps are thus important for peptide dissociation: Short-lived DM-sensitive DR conformers form following dissociation of the peptide N-terminus from the DR groove. Rotation of DR αW43 out of the groove creates a key DM interaction site and makes other conformational changes energetically more favorable. Rebinding of the peptide N-terminus is inhibited by movement of DR residues into the P1 pocket (DR αF51 and βF89). Peptide dissociation is further accelerated by loss of hydrogen bonds between the peptide backbone at P2 and DR βN82. This scenario is supported by the two structures and a large body of functional data. It should be noted that these steps are unlikely to always occur in a particular sequence. For example, a peptide may completely dissociate prior to DM binding if it has a very low affinity for the DR molecule. However, alternative conformations, including inactive states of the DR molecule, become more likely when a large part of the groove is devoid of peptide (Painter et al., 2008; Rupp et al., 2011). Given that DM can accelerate peptide dissociation >1,000-fold (Weber et al., 1996), it must bind to most DR molecules prior to complete peptide dissociation.
How does HLA-DM stabilize empty MHC class II molecules?
Empty DR1 rapidly transitions into an inactive state (the half-life is allele-dependent, approximately 5 minutes for DR1), yet DR molecules maintain a highly peptide-receptive conformation in the presence of DM (Grotenbreg et al., 2007; Kropshofer et al., 1997; Rabinowitz et al., 1998). DM-DR complexes have been purified from cells and were found to be largely devoid of peptide (Denzin et al., 1996; Kropshofer et al., 1997; Sanderson et al., 1996). Furthermore, SPR experiments showed that empty DR dissociates from DM with very slow kinetics (Anders et al., 2011). Movement of DR αF51 and βF89 into the P1 site protects the most hydrophobic part of the groove and prevents transition into alternative, inactive conformers with a tendency to aggregate. Stable empty MHCII – DM complexes are functionally important: DM deficiency results in an antigen presentation defect even for MHCII proteins that do not require DM for CLIP dissociation, due to a low affinity for CLIP (Koonce et al., 2003). Rapid binding to MHCII molecules protects proteolytic fragments from degradation, and proteolytic trimming can occur after binding to a MHCII molecule (Castellino et al., 1998; Nelson et al., 1997).
How does DM enable rapid selection of the highest affinity peptide ligands?
The mechanisms of DM-catalyzed peptide selection have been difficult to explain. All peptides have similar on-rates and differ primarily in their off-rates (Rabinowitz et al., 1998). The structure implies how peptide editing can occur with rapid kinetics, even though peptides have a very long half-life (many hours or days) when fully bound in the groove (Jardetzky et al., 1990; Roche and Cresswell, 1990a). In the pH 5.5 DM-DR1 structure, the two most important structural elements for stable peptide binding are inaccessible - the P1 pocket and the DR βN82 side chain. Incoming peptides thus need to compete with DR1 residues for access to these binding sites, which creates an energetic and kinetic barrier for full occupancy of the groove. This mechanism ensures that high-affinity peptides, which are present in limited quantities in the loading compartment (Germain and Hendrix, 1991), are not outcompeted by low-affinity peptides. This proposed mechanism is supported by functional experiments in which DM and DR1 were linked by leucine zippers. Peptides dissociated with very rapid kinetics from this linked complex, CLIP with a t1/2 of 10 seconds and HA306–318 with a t1/2 of 2 minutes (note that HA306–318 has a t1/2 of ~1 month in the absence of DM) (Busch et al., 2002).
Structural features along the entire peptide are relevant for DM editing (Belmares et al., 2002; Weber et al., 1996), even though DM interacts with a site close to the peptide N-terminus. Anchor residues in the middle and C-terminal part of the peptide need to properly engage available pockets (such as P4, P6 and P9 pockets) during initial stages of binding, positioning peptides to access DR βN82 and the P1 pocket. Peptides without a hydrophobic P1 anchor do not induce DR dissociation from DM and are subject to continued editing (Anders et al., 2011). This mechanism explains how DM catalyzes selection of peptides with a very long half-life for presentation on the cell surface.
Which mechanisms limit DM-catalyzed displacement of high-affinity peptides?
SPR experiments have shown that peptides induce dissociation of a complex of DM and empty DR, depending on the affinity of the peptide for the DR molecule (Anders et al., 2011). This result can now be explained based on the new structures: the peptide P1 anchor and DR αF51 compete for access to the same DR1 pocket. DM binding is abrogated by the DR αF51A mutation. This suggests that insertion of a hydrophobic peptide side chain into the P1 pocket is one of the key steps that trigger DM dissociation from the complex. High affinity peptides thus rapidly terminate editing, and re-binding of such DR-peptide complexes to DM occurs with slow kinetics (Anders et al., 2011).
Relevance for MHC class I molecules
MHC class I (MHCI) molecules bind peptides in the ER that are generated by the proteasome and transported into the ER by the TAP (transporter associated with processing) heterodimer. Empty MHCI molecules become part of the ‘peptide loading complex’: they are bound by calreticulin and the disulfide-linked tapasin-ERp57 dimer, which also recruits them to the TAP heterodimer (Chen and Bouvier, 2007; Peaper and Cresswell, 2008; Sadegh-Nasseri et al., 2008; Wearsch and Cresswell, 2007). Even though all involved proteins, the compartment and the pH are different, the essential problem for peptide loading is the same: how are empty molecules stabilized in a manner that enables rapid selection of the highest-affinity ligands? The tapasin-ERp57 heterodimer appears to stabilize the N-terminal part of the α2 helix of MHCI molecules (which flanks the C-terminal part of peptides) (Dong et al., 2009). Also, monoclonal antibodies (such as mAb 64-3-7) have been identified that bind to peptide-receptive but not peptide-loaded states of a MHCI molecule. The 64-3-7 antibody recognizes a short peptide segment of the MHCI heavy chain corresponding to a 310 helix (res. 46–52) in the vicinity of the peptide N-terminus. In the peptide-loaded state, a conserved tryptophan residue (W51) and M52 buttress invariant tyrosine residues (Y59 and Y171) at the amino-terminal end of the peptide binding groove. W51 and M52 become solvent-exposed in the peptide-receptive form, apparently by movement of the 310 helix containing W51 and M52 (Mage et al., 2012). Conformational changes in the vicinity of the peptide N-terminus may thus not only be relevant for peptide-receptive states of MHCII but also MHCI molecules.
Supplemental Information
Supplemental information includes Extended Experimental Procedures, six figures, three tables and two movies.
Experimental Procedures
Crystallization, data collection and structure determination
Crystallization trials were performed by hanging drop vapor diffusion at room temperature. Crystals appeared under a variety of different conditions. Crystals from two conditions were used for structure determination: 1) 0.2 M sodium acetate, 0.1 M sodium citrate pH 5.5, 10% PEG 4,000, 2) 0.1 M MES pH 6.5, 6% PEG 20,000. Crystals were cryoprotected using either 30% (v/v) ethylene glycol or 20% (v/v) glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at 100K at the NE-CAT beamline 24-ID-E while performing a continuous vector scan over the crystal. The data were processed with the HKL2000 program, and the structure was determined by molecular replacement using HLA-DR1 (1DLH) and HLA-DM (1HDM) as search models (for details, see Supplemental Experimental Procedures). Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank (PDB) with accession codes 4FQX (pH 5.5) and 4GBX (pH 6.5).
Biacore analysis of DM-DR binding
Mutations were introduced into HLA-DR15 (DRA*01:01, DRB1*15:01) because a large body of data are available for comparison (Anders et al., 2011). The sequence of the DRα chain is identical for DR1 and DR15. DR15-CLIP binding to DM was measured by surface plasmon resonance (SPR) using a Biacore 3000 instrument (GE Healthcare). Wild-type and mutant DM were biotinylated at a BirA site on the C-terminus of DMα and captured on a streptavidin surface (~500 response units, RU). DR15-CLIP complexes were diluted in running buffer (50 mM citrate-phosphate pH 5.35, 150 mM NaCl, 0.06% C12E9 detergent) immediately prior to injection. DR15-CLIP WT and indicated mutants were injected at 25µl/min (stage 1, DR binding to DM), followed by running buffer (stage 2, dissociation of empty DR from DM) and a peptide (myelin basic protein, MBP res. 85–99, 1µM) that binds to DR15 (stage 3, dissociation of DR-peptide from DM) (300 seconds for each stage) (Anders et al., 2011). Experiments were performed at 30°C. Measurements from the reference flow cell (immobilized DM double mutant, DMα R98A-R194A) were subtracted from experimental values.
Peptide binding assay
A real-time assay was used to assess binding of a fluorescent peptide to DR15, employing a fluorescence polarization (FP, mP units) readout (Nicholson et al., 2006). The linker connecting the CLIP peptide to the N-terminus of the DRβ chain was cut with thrombin (1 hour, 20U/mg, RT). DR15-CLIP complexes (150nM) were incubated +/− DM (typically 25nM) in the presence of a high-affinity Alexa-488 labeled MBP85–99 peptide (30nM) at 30°C. Assays were set up in a volume of 40µl in black polystyrene 384-well plates (Corning, NY, USA) in 50mM citrate, pH 5.2 at 30°C (triplicates). Initial binding rates were calculated using Graphpad 5.0.
Supplemental Experimental Procedures describe details on the formation of the DM-DR1 complex not described in the Results Section and Figure 1, as well as structure determination.
Supplementary Material
Research Highlights.
Structure of HLA-DM - HLA-DR1 complex identifies key steps in antigen presentation.
Two aromatic HLA-DR residues move in and stabilize the hydrophobic P1 pocket.
Occlusion of P1 pocket enables rapid selection of high-affinity peptides.
Only peptides that successfully compete for the P1 pocket are stably bound.
Acknowledgements
This work was supported by grants from the National Institutes of Health (RO1 NS044914 and PO1 AI045757 to K.W.W.), postdoctoral fellowships from The National Multiple Sclerosis Society (D.K.S.) and the American Diabetes Association (W.P.) as well as a Ruth L. Kirschstein National Research Service Award (to M.J.C.). We thank the staff at the UCLA-DOE crystallization core and the CSIRO Collaborative Crystallization Centre, Melbourne, Australia for setting up screens. Diffraction data were collected at the Advanced Photon Source using beamlines allocated to the Northeastern Collaborative Access Team. These beamlines are supported by grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8P41 GM103403-10) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders AK, Call MJ, Schulze MS, Fowler KD, Schubert DA, Seth NP, Sundberg EJ, Wucherpfennig KW. HLA-DM captures partially empty HLA-DR molecules for catalyzed removal of peptide. Nature immunology. 2011;12:54–61. doi: 10.1038/ni.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Busch R, Pashine A, Garcia KC, Mellins ED. Stabilization of soluble, low-affinity HLA-DM/HLA-DR1 complexes by leucine zippers. J Immunol Methods. 2002;263:111–121. doi: 10.1016/s0022-1759(02)00034-0. [DOI] [PubMed] [Google Scholar]
- Busch R, Reich Z, Zaller DM, Sloan V, Mellins ED. Secondary structure composition and pH-dependent conformational changes of soluble recombinant HLA-DM. The Journal of biological chemistry. 1998;273:27557–27564. doi: 10.1074/jbc.273.42.27557. [DOI] [PubMed] [Google Scholar]
- Castellino F, Zappacosta F, Coligan JE, Germain RN. Large protein fragments as substrates for endocytic antigen capture by MHC class II molecules. Journal of immunology. 1998;161:4048–4057. [PubMed] [Google Scholar]
- Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. The EMBO journal. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresswell P. Assembly, transport, and function of MHC class II molecules. Annual review of immunology. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II alpha beta dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLA-DR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebele CR, Busch R, Scott MH, Pashine A, Mellins DE. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13:517–527. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fremont DH, Crawford F, Marrack P, Hendrickson WA, Kappler J. Crystal structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/s1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- Fung-Leung WP, Surh CD, Liljedahl M, Pang J, Leturcq D, Peterson PA, Webb SR, Karlsson L. Antigen presentation and T cell development in H2-M-deficient mice. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- Gandhi RT, Walker BD. Immunologic control of HIV-1. Annu Rev Med. 2002;53:149–172. doi: 10.1146/annurev.med.53.082901.104011. [DOI] [PubMed] [Google Scholar]
- Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by post-endoplasmic reticulum antigen binding. Nature. 1991;353:134–139. doi: 10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363:725–728. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- Grotenbreg GM, Nicholson MJ, Fowler KD, Wilbuer K, Octavio L, Yang M, Chakraborty AK, Ploegh HL, Wucherpfennig KW. Empty class II major histocompatibility complex created by peptide photolysis establishes the role of DM in peptide association. J Biol Chem. 2007;282:21425–21436. doi: 10.1074/jbc.M702844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardetzky TS, Gorga JC, Busch R, Rothbard J, Strominger JL, Wiley DC. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990;9:1797–1803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JF, Stebbins C, Appella E, Sant AJ. Invariant chain and DM edit self-peptide presentation by major histocompatibility complex (MHC) class II molecules. J Exp Med. 1996;184:1747–1753. doi: 10.1084/jem.184.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce CH, Wutz G, Robertson EJ, Vogt AB, Kropshofer H, Bikoff EK. DM loss in k haplotype mice reveals isotype-specific chaperone requirements. Journal of immunology. 2003;170:3751–3761. doi: 10.4049/jimmunol.170.7.3751. [DOI] [PubMed] [Google Scholar]
- Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- Mage MG, Dolan MA, Wang R, Boyd LF, Revilleza MJ, Robinson H, Natarajan K, Myers NB, Hansen TH, Margulies DH. The peptide-receptive transition state of MHC class I molecules: insight from structure and molecular dynamics. Journal of immunology. 2012;189:1391–1399. doi: 10.4049/jimmunol.1200831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H, Hart SA, Schink A, Pollok BA. Sortase-mediated protein ligation: a new method for protein engineering. J Am Chem Soc. 2004;126:2670–2671. doi: 10.1021/ja039915e. [DOI] [PubMed] [Google Scholar]
- Martin WD, Hicks GG, Mendiratta SK, Leva HI, Ruley HE, Van Kaer L. H2-M mutant mice are defective in the peptide loading of class II molecules, antigen presentation, and T cell repertoire selection. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mellins E, Smith L, Arp B, Cotner T, Celis E, Pious D. Defective processing and presentation of exogenous antigens in mutants with normal HLA class II genes. Nature. 1990;343:71–74. doi: 10.1038/343071a0. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris P, Shaman J, Attaya M, Amaya M, Goodman S, Bergman C, Monaco JJ, Mellins E. An essential role for HLA-DM in antigen presentation by class II major histocompatibility molecules. Nature. 1994;368:551–554. doi: 10.1038/368551a0. [DOI] [PubMed] [Google Scholar]
- Mosyak L, Zaller DM, Wiley DC. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- Nelson CA, Vidavsky I, Viner NJ, Gross ML, Unanue ER. Amino-terminal trimming of peptides for presentation on major histocompatibility complex class II molecules. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:628–633. doi: 10.1073/pnas.94.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, Wucherpfennig KW. Small molecules that enhance the catalytic efficiency of HLA-DM. J Immunol. 2006;176:4208–4220. doi: 10.4049/jimmunol.176.7.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter CA, Cruz A, Lopez GE, Stern LJ, Zavala-Ruiz Z. Model for the peptide-free conformation of class II MHC proteins. PLoS One. 2008;3:e2403. doi: 10.1371/journal.pone.0002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter CA, Negroni MP, Kellersberger KA, Zavala-Ruiz Z, Evans JE, Stern LJ. Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:19329–19334. doi: 10.1073/pnas.1108074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, Nolan GP, Mellins ED. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- Peaper DR, Cresswell P. Regulation of MHC class I assembly and peptide binding. Annu Rev Cell Dev Biol. 2008;24:343–368. doi: 10.1146/annurev.cellbio.24.110707.175347. [DOI] [PubMed] [Google Scholar]
- Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, Davis MM, McConnell HM. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- Reay PA, Wettstein DA, Davis MM. pH dependence and exchange of high and low responder peptides binding to a class II MHC molecule. EMBO J. 1992;11:2829–2839. doi: 10.1002/j.1460-2075.1992.tb05350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riberdy JM, Newcomb JR, Surman MJ, Barbosa JA, Cresswell P. HLA-DR molecules from an antigen-processing mutant cell line are associated with invariant chain peptides. Nature. 1992;360:474–477. doi: 10.1038/360474a0. [DOI] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J Immunol. 1990a;144:1849–1856. [PubMed] [Google Scholar]
- Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990b;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- Roche PA, Cresswell P. Proteolysis of the class II-associated invariant chain generates a peptide binding site in intracellular HLA-DR molecules. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3150–3154. doi: 10.1073/pnas.88.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp B, Gunther S, Makhmoor T, Schlundt A, Dickhaut K, Gupta S, Choudhary I, Wiesmuller KH, Jung G, Freund C, et al. Characterization of structural features controlling the receptiveness of empty class II MHC molecules. PLoS One. 2011;6:e18662. doi: 10.1371/journal.pone.0018662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadegh-Nasseri S, Chen M, Narayan K, Bouvier M. The convergent roles of tapasin and HLA-DM in antigen presentation. Trends Immunol. 2008;29:141–147. doi: 10.1016/j.it.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson F, Thomas C, Neefjes J, Trowsdale J. Association between HLA-DM and HLA-DR in vivo. Immunity. 1996;4:87–96. doi: 10.1016/s1074-7613(00)80301-5. [DOI] [PubMed] [Google Scholar]
- Sherman MA, Weber DA, Jensen PE. DM enhances peptide binding to class II MHC by release of invariant chain-derived peptide. Immunity. 1995;3:197–205. doi: 10.1016/1074-7613(95)90089-6. [DOI] [PubMed] [Google Scholar]
- Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nature immunology. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- Weber DA, Dao CT, Jun J, Wigal JL, Jensen PE. Transmembrane domain-mediated colocalization of HLA-DM and HLA-DR is required for optimal HLA-DM catalytic activity. J Immunol. 2001;167:5167–5174. doi: 10.4049/jimmunol.167.9.5167. [DOI] [PubMed] [Google Scholar]
- Weber DA, Evavold BD, Jensen PE. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- York IA, Rock KL. Antigen processing and presentation by the class I major histocompatibility complex. Annual review of immunology. 1996;14:369–396. doi: 10.1146/annurev.immunol.14.1.369. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.