Abstract
Human norovirus (HuNoV) is the major cause of acute nonbacterial gastroenteritis worldwide but has no clear animal reservoir. HuNoV can persist after the resolution of symptoms, and this persistence may be essential for viral maintenance within the population. Many strains of the related murine norovirus (MNV) also persist, providing a tractable animal model for studying norovirus (NoV) persistence. We have used recombinant cDNA clones of representative persistent (CR6) and nonpersistent (CW3) strains to identify a domain within the nonstructural gene NS1/2 that is necessary and sufficient for persistence. Furthermore, we found that a single change of aspartic acid to glutamic acid in CW3 NS1/2 was sufficient for persistence. This same conservative change also caused increased growth of CW3 in the proximal colon, which we found to be a major tissue reservoir of MNV persistence, suggesting that NS1/2 determines viral tropism that is necessary for persistence. These findings represent the first identified function for NoV NS1/2 during infection and establish a novel model system for the study of enteric viral persistence.
INTRODUCTION
Human noroviruses (HuNoVs) are the leading cause of nonbacterial gastroenteritis and are a significant public health problem worldwide, causing both outbreaks and sporadic cases of acute gastrointestinal disease (reviewed in references 1, 2, and 3). There is no evidence for an animal reservoir of HuNoV, suggesting that asymptomatic shedding within the human population initiates symptomatic infection. Indeed, PCR-based detection has shown that HuNoVs are present in asymptomatic individuals within a variety of communities and ranges in prevalence from 3 to 13% (4–7). Furthermore, experimental infection of volunteers revealed that some individuals continue to shed virus for up to weeks after the resolution of symptoms (8), and there are examples where outbreaks likely originated with asymptomatic HuNoV shedders (9–11). Taken together, these studies demonstrate that asymptomatic and persistent infections are an important aspect of HuNoV biology. However, the molecular mechanisms of NoV persistence remain incompletely understood.
Noroviruses (NoVs) are positive-sense, single-stranded RNA viruses in the calicivirus family. They are classified into five genetic groups (genogroups), with HuNoVs in genogroups I, II, and IV (GI, GII, and GIV, respectively), bovine noroviruses in GIII, and murine noroviruses (MNV) in GV (12). The NoV genome is ∼7.5 kb in length and encodes three open reading frames (ORFs) (13), with an additional ORF4 in MNV (14, 15). ORF1 encodes the NoV nonstructural polyprotein which is cleaved into six mature peptides (NS1/2 and NS3 through NS7) by the ORF1-encoded viral protease NS6 (16–18). ORF2 encodes the major structural protein (VP1), which self assembles into the icosahedral viral capsid (19), and ORF3 encodes a minor structural protein (VP2) (20). MNV ORF4 is an alternate reading frame within ORF2 and encodes virulence factor 1 (VF1) (14).
NS1/2 is the most N-terminal viral protein encoded by ORF1. The cleavage site between NS1/2 and NS3 has been confirmed by mutagenesis studies, and the position of this cleavage is conserved among all NoVs. All genera of caliciviruses other than NoVs have an additional viral protease cleavage site within the region homologous to NS1/2, resulting in two mature viral proteins derived from this region (21–23). It is possible that some NoVs have lost this cleavage event, but there is one report of additional cleavage of the human NS1/2 protein in cells transfected with HuNoV ORF1 (24), and MNV NS1/2 can be cleaved by caspase 3 (18). NS1/2 localizes to organelles in the secretory pathway (25, 26), and HuNoV NS1/2 disrupts protein secretion in transfected cells (25, 27). However, very little is known about the function of NS1/2 in NoV replication or pathogenesis.
Similar to HuNoV, MNV establishes persistent asymptomatic infection (15, 28) and therefore represents an ideal model system in which to study NoV persistence. While most strains of MNV persist, the strain MNV1 is cleared. MNV1 was identified as a cause of lethality in RAG/STAT1 knockout mice (29), and it was passaged intracranially in these mice until a culture system was established (30). While MNV1 rapidly kills STAT1 knockout mice, it persists avirulently in RAG knockout mice and is cleared from immune-sufficient mice (29, 31). Subsequently, many closely related strains of MNV isolated from stool samples were shown to persist in immune-sufficient mice (15, 28). One of these persistent stool isolates, CR6, has been shown to be avirulent in STAT1 knockout mice, demonstrating a second phenotypic distinction from MNV1 (31). A reverse genetics system for MNV exists (32), making phenotypic analyses of MNV1 and persistent strains possible (14, 31, 33, 34). The increased virulence of MNV1 has been linked to the protruding domain of the capsid gene (31, 33), but the gene responsible for MNV persistence has not been identified.
Here, we used recombinant cDNA clones of the nonpersistent strain MNV1.CW3 (subsequently referred to as CW3) and persistent strain CR6 to identify a genetic determinant of persistence. In contrast to virulence in STAT1 knockout mice (31), reciprocal replacement of the capsid gene did not alter persistence. Instead, the NS1/2 gene of CR6 enabled persistence of CW3. Within NS1/2, we further narrowed the determinant of persistence to the 5′ domain and to a single amino acid within this domain. Furthermore, this same determinant increased growth of CW3 in the proximal colon, a major reservoir of CR6 persistence. This is the first demonstrated function for NS1/2 during infection and the first report of a protein other than the MNV capsid to alter viral tropism. This is also the first study identifying a genetic determinant for enteric viral persistence.
MATERIALS AND METHODS
Generation of recombinant MNV stocks.
Construction of molecular clones of CW3 (EF014462.1) and CR6 (JQ237823) and the strategy for cloning of viral gene substitutions have been previously described (31). Primers used for the substitutions of NS1/2 5′ and 3′ domains were NS1/2 forward (5′-GTGAAATGAGGATGGCAACGCCATCTTC-3′), 5′ NS1/2 reverse (5′-GCGTCCTCCGCATGGGAGGGCGC-3′), 3′ NS1/2 forward (5′-GCGCCCTCCCATGCGGAGGAC-3′), and CR6 3′ NS1/2 reverse (5′-AGGTCGAAGGGCCCTTCGGCCTGCCA-3′).
Generation of viral stocks from plasmids carrying MNV was performed as described previously (31) with minor modifications. Briefly, plasmids were transfected into 293T cells with TransIT-LT1 (Mirus Bio, Madison, WI), and virus was recovered 48 h posttransfection by freezing and thawing. Clarified 293T supernatants were passaged one to two times on RAW264.7 cells at a multiplicity of infection (MOI) of ≤0.05, and virus was recovered 48 h postinfection. Virus-containing RAW264.7 cell supernatants were cleared of debris by centrifugation for 2 min at ≥18,000 × g. All viral stocks were derived from cDNA clones rather than serially passaged MNV isolates.
Mice and infections.
C57BL/6 mice (stock number 000664) were purchased from Jackson Laboratories (Bar Harbor, ME) and housed at Washington University School of Medicine under specific-pathogen-free conditions (35) according to university guidelines. Cages of male or female mice were inoculated with virus at 6 to 8 weeks of age by the oral route in a volume of 25 to 35 μl. A dose of 3 × 104 PFU was used for all experiments, with one exception (1 × 106 PFU; see Fig. 5E).
Fig 5.
NS1/2 and amino acid 94 determine growth in the proximal colon, a major reservoir of persistent MNV. (A to E) Tissues were harvested from MNV-infected mice, and the genome copy number was measured by quantitative PCR. (A) Detection of CR6 in regions of the gastrointestinal tract 35 days postinfection. Data are combined from two experiments for a total of five mice. (B) A kinetic comparison of titers in spleen, proximal colon, and MLN following infection with either CW3 or CR6. Data are combined from three experiments for a total of nine mice per data point. (C and D) Detection of the indicated viruses in tissues at day seven postinfection. Data are combined from at least two experiments for a total of at least eight mice for each data point. CW3 and CR6 data from day seven in panel B are shown in panel C for reference. (E) Detection of CR6-VP1 in the indicated tissues at 3 or 35 days postinfection with 1 × 106 PFU. Data are combined from two experiments for a total of six mice per data point.
Stool and tissues were harvested into 2-ml tubes (Sarstedt, Germany) with 1-mM diameter zirconia/silica beads (Biospec, Bartlesville, OK). Tissues were flash frozen in a bath of ethanol and dry ice and either processed on the same day or stored at −80°C.
Plaque assay.
Culture of RAW264.7 cells and the plaque assay for determining the titer of virus stocks was performed as described previously (31). Growth curve plaque assays utilized suspension-adapted RAW264.7 cells maintained in minimal essential medium (MEM) supplemented with 10% bovine serum, 1% nonessential amino acids, 100 μg/ml penicillin-streptomycin, 2 mM l-glutamine, 10 mM HEPES, and 0.225% sodium bicarbonate. RAW264.7 cells were plated on 6-well plates at a density of 2 × 106 cells per well. Sixteen to 24 h later, medium was removed and 10-fold serial dilutions of virus inoculum were added at 500 μl per well. Plates were incubated for 1 h at room temperature on a rocking platform before removing the inoculum and overlaying the cells with 2 ml per well of 1% methylcellulose in MEM supplemented with 10% fetal calf serum, 100 μg/ml penicillin-streptomycin, 2 mM l-glutamine, 10 mM HEPES. Plates were incubated at 37°C for 3 to 4 days, and plaques were visualized by staining with 0.2% crystal violet and 20% ethanol.
Quantitative reverse transcription-PCR.
RNA from stool was isolated using either an RNeasy Miniprep (Qiagen, Valencia, CA) or Quick-RNA Miniprep (Zymoresearch, Irvine, CA) kit. RNA from tissues was isolated using TRIzol (Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. Five μl of RNA from stool or 1 μg of RNA from tissue was used as a template for cDNA synthesis with the ImPromII reverse transcriptase system (Promega, Madison, WI). When evident in the melting curve analysis, DNA contamination was removed using the DNAfree kit (Life Technologies).
MNV TaqMan was performed as described previously (36). MNV genome quantities from tissue samples were normalized to the housekeeping gene of ribosomal protein S29 (RPS29). SYBR green quantitative PCR for RPS29 was performed with the forward primer 5′-AGCAGCTCTACTGGAGTCACC-3′ and reverse primer 5′-AGGTCGCTTAGTCCAACTTAATG-3′ at a concentration of 0.2 μM in 1× Power SYBR green master mix (Life Technologies). Cycling parameters were identical to those for MNV TaqMan with the exception of an additional melting curve analysis.
Statistical analysis.
Data were analyzed with Prism 5 software (GraphPad Software, San Diego, CA). In all graphs, three asterisks indicate P < 0.001, two asterisks indicate P < 0.01, one asterisk indicates P < 0.05, and n.s. indicates not significant (P > 0.05) as determined by one-way analysis of variance (ANOVA) with Tukey's posttest (single-time-point experiments) or two-way ANOVA with Bonferroni's posttest (kinetic experiments). All error bars depict standard errors of the means.
RESULTS
CW3 and CR6 differ in quantity and persistence of shedding in stool.
Molecular clones of the representative nonpersistent and persistent strains CW3 and CR6 were used for a kinetic comparison of fecal shedding. CR6 was shed by six out of eight mice on day three and reached a peak level of shedding by day seven postinfection. The day seven shedding of CW3 was significantly lower than that of CR6, and CW3 was not detected in more than 50% of mice at any time point. Average CW3 shedding decreased below the limit of detection at day 14 and remained undetectable at days 21 and 35 (Fig. 1). In contrast, CR6 was consistently shed between days 21 and 70 with no observed clearance in any animals (Fig. 1). Therefore, CR6 infection leads to long-lived persistent shedding as well as significantly greater shedding than CW3 during acute infection.
Fig 1.
CW3 and CR6 differ in quantity and persistence of viral shedding in stool. Stool was collected at the indicated days postinfection, and the genome copy number was determined by quantitative PCR. Data show the mean shedding of at least four mice per time point.
MNV capsid does not determine persistence.
VP1 was a likely candidate for determining MNV persistence due to its role in immunity, lethality, and tropism (31, 33, 34, 37, 38). Therefore, we assessed the shedding of viruses with reciprocal substitutions in the VP1 gene (including the overlapping VF1 gene, as depicted in Fig. 2A). The number of genome copies in the stool at 35 days postinfection was not significantly different between CW3 and CW3 carrying the VP1 gene from CR6 (CW3-VP1) or between CR6 and CR6 carrying the VP1 gene from CW3 (CR6-VP1) (Fig. 2B). These data indicate that a gene or genes other than VP1 are required for persistent infection by MNV.
Fig 2.
5′ Domain of NS1/2 is necessary and sufficient for persistence. (A) Diagram of the MNV genome and the genomes of MNV chimeras used in panels B to E to map the persistence trait. (B to E) Mice were inoculated with the indicated MNV strains. Stool was collected 35 days postinfection, and MNV genomes were detected by quantitative PCR. (B) Comparison of VP1-substituted strains. (C) A screen of additional single-gene substitutions. (D) Confirmation of CW3-NS1/2 persistence with three independent viral stocks. (E) Comparison of NS1/2 domain-substituted viruses. Data in panels C and D are from single experiments; those in panels B and E are combined from at least two experiments for each virus. 1/Ct indicates the inverse of the threshold cycle.
5′ Region of the NS1/2 gene is necessary and sufficient for persistence.
To determine whether a CR6 gene other than VP1 was sufficient to enable CW3 persistence, we tested additional CW3 viruses with single CR6-derived gene replacements (Fig. 2A) (31). A comparison of threshold cycle (CT) values revealed that shedding of CW3-NS1/2 was significantly greater than that of all other single-gene substitutions (Fig. 2C). To confirm and quantify the result of this screen, we generated two additional viral stocks of CW3-NS1/2 and tested all three stocks for persistent shedding. All three biological replicates of CW3-NS1/2 were detected in stool at 35 days postinfection, and the quantity of MNV genomes was not significantly different from that of CR6 (Fig. 2D). The reciprocal insertion of NS1/2 from CW3 into CR6 did not generate virus and could not be tested for persistence. These findings indicate that the NS1/2 gene of CR6 is sufficient to enable CW3 persistence.
The NS1/2 gene encodes a 3′ domain conserved among NoVs (39) and a 5′ domain that is more divergent. To narrow the persistence determinant to a single domain, we generated CW3 with the 5′ domain of CR6 NS1/2 (CW3-5′ NS1/2) and CW3 with the 3′ domain of CR6 NS1/2 (CW3-3′ NS1/2) as depicted in Fig. 2A. While persistent CW3-3′ NS1/2 shedding was not detected, CW3-5′ NS1/2 shedding was indistinguishable from that of CR6 (Fig. 2E) To test whether the 5′ NS1/2 domain was also necessary for persistence, we generated CR6 with a CW3 substitution in the 5′ domain of the NS1/2 gene (CR6-5′ NS1/2). Persistent shedding of CR6-5′ NS1/2 was significantly lower than that of CR6 and not significantly different from that of CW3 (Fig. 2E). Taken together, these data demonstrate that the 5′ domain of the NS1/2 gene is necessary and sufficient for MNV persistence.
A change of aspartic acid to glutamic acid at amino acid 94 of NS1/2 is sufficient for persistence.
CR6 is one of seven persistent strains identified to date (15, 28). To test whether CW3 has unique polymorphisms that correlate with its inability to persist, we analyzed the 5′ domain of NS1/2 from CW3 and the seven persistent MNV strains (Fig. 3A). Amino acid alignment showed that only a single residue was both unique to CW3 and conserved among the persistent strains of MNV. This difference was a conservative change at amino acid 94, with glutamic acid present in persistent strains and aspartic acid present in CW3 (Fig. 3A). To assess whether this difference was involved in determining persistence, we generated a mutant of CW3 that encoded the conserved glutamic acid present in CR6 (CW3D94E). Additionally, we made the reciprocal mutation in CR6, changing the conserved glutamic acid to an aspartic acid (CR6E94D). CR6 persistent shedding was not significantly changed by the E94D mutation (Fig. 3B), and sequencing demonstrated no reversion to E94 in vivo (data not shown). In contrast, CW3 shedding was significantly increased by the D94E mutation to levels that were not significantly different from those of CR6 (Fig. 3B). Furthermore, a D94E mutation in the nonpersistent CR6-5′NS1/2 chimera rescued its ability to persist. This striking result demonstrates that glutamic acid at amino acid 94 of NS1/2 is sufficient, though not necessary, for persistence.
Fig 3.
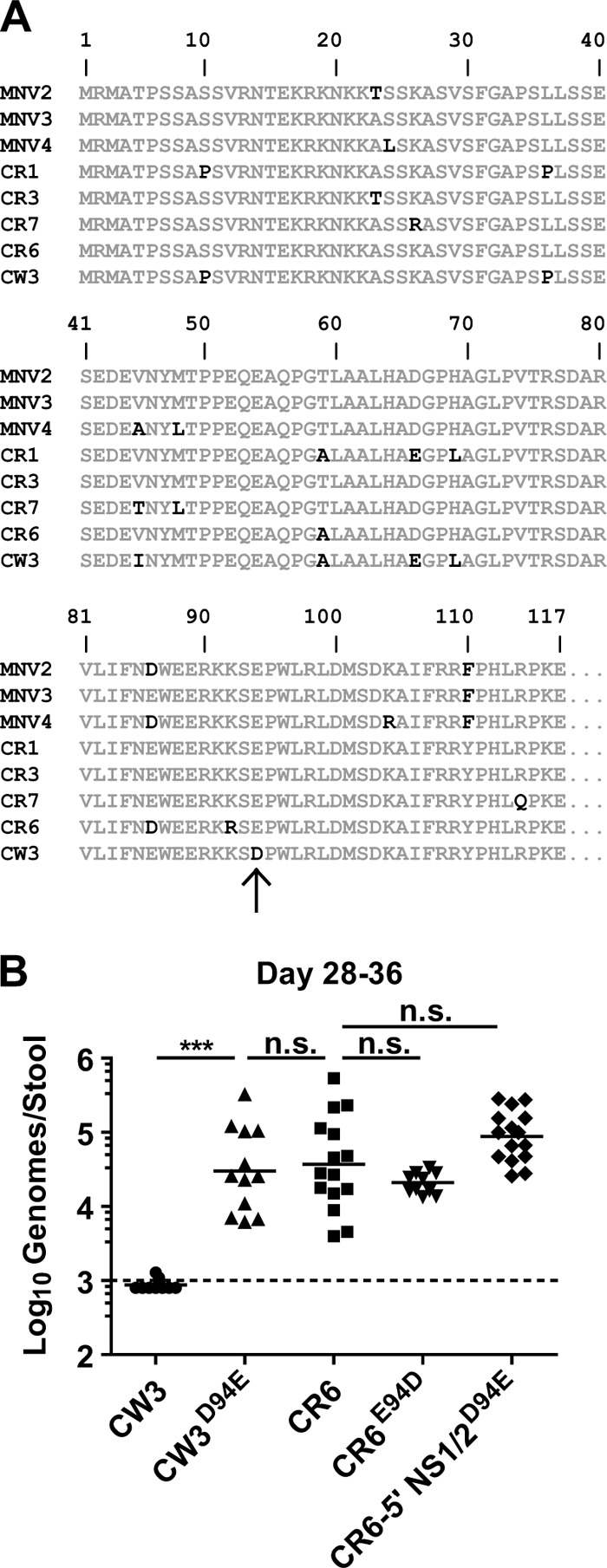
Change of aspartic acid to glutamic acid at position 94 of NS1/2 is sufficient for persistence. (A) Alignment of the first 117 amino acids of CW3 with CR6 and six additional persistent strains of MNV (accession numbers DQ223041, DQ223042, DQ223043, EU004672, EU004673, and EU004677 [15, 28]). Conserved amino acid residues are gray, and nonconserved residues are black. The arrow indicates a unique aspartic acid residue at position 94 of CW3. (B) Mice were inoculated with the indicated MNV strains. Stool was collected between 28 and 36 days postinfection, and MNV genomes were detected by quantitative PCR. Data are combined from at least two experiments for each virus.
Persistent and nonpersistent viruses have similar in vitro growth.
The association of NS1/2 with cellular membranes and the viral replication complex (40) suggested that it has a role in viral replication that is altered by the persistence determinant. To test this hypothesis, we compared in vitro growth of the persistent viruses CW3D94E and CR6 to that of the nonpersistent viruses CW3 and CR6-5′ NS1/2. Infection of RAW264.7 cells at an MOI of 5 revealed no significant overall difference in growth at any time postinfection between any pair of persistent and nonpersistent viruses (Fig. 4). At an MOI of 0.05 there were also no significant differences other than at 4 h postinfection between CR6 and CR6-5′ NS1/2 (P < 0.05) (Fig. 4). However, there were no differences between CW3 and CW3D94E at any time that would indicate a role for this early growth difference in determining the persistence phenotype. Together, these data show that the persistence determinant has little to no effect on in vitro growth in RAW264.7 cells.
Fig 4.
Viral mutants with altered persistence phenotype have similar in vitro growth. RAW264.7 cells were infected at an MOI of either 0.05 or 5, and viral growth was measured by plaque assay at the indicated times postinfection. Data are combined from three experiments.
CR6 grows better than CW3 in the proximal colon.
While no growth difference was seen in RAW264.7 cells, the significant difference in fecal shedding between CW3 and CR6 on day seven (Fig. 1) suggested that there was a difference in enteric growth during the early stages of infection in vivo. To identify the preferential enteric site(s) of CR6 replication, relative levels of persistent CR6 were measured in anatomically distinct regions along the gastrointestinal (GI) tract. CR6 was detected in the proximal colon at levels significantly greater than those of all other GI regions with the exception of the cecum (Fig. 5A). This observation was supported by recent publications which also predominantly detected persistent MNV strains in the cecum and/or proximal colon (34, 41).
Therefore, the proximal colon was included as the representative enteric tissue in a comparison between CR6 and CW3 growth during acute infection. Spleen and mesenteric lymph node (MLN) were also included in the comparison because MNV grows in macrophages and dendritic cells (DCs) and can spread to these tissues. The peak titer of both strains in all tissues was reached by day seven (Fig. 5B). In the proximal colon, CR6 grew significantly better than CW3 from day 3 through 14, and CW3 was only minimally detected at any time point (Fig. 5B, left). In the spleen, CW3 grew significantly better than CR6 between days 3 and 7 but was cleared by day 14 (Fig. 5B, middle). This difference in the spleen was in accordance with recent publications demonstrating the ability of CW3 and MNV1 to spread systemically (31, 42) and indicates that the failure of CW3 to persist in the intestine is not due to a failure to establish infection. Both strains were detected in the MLN with no significant difference before day 14 when CW3 was cleared (Fig. 5B, right). Taken together, these data demonstrate significant differences in tissue tropism between CW3 and CR6 in the spleen and proximal colon, the latter being a major site of persistence.
NS1/2 amino acid 94, rather than capsid, determines growth in the proximal colon.
Other studies have mapped MNV1 tropism changes to VP1 (31, 34), so we used CW3-VP1 and CR6-VP1 to test the role of VP1 in determining the tropism differences revealed in Fig. 5B. Growth of CW3-VP1 in the proximal colon 7 days after infection was not significantly higher than that of CW3, and growth of CR6-VP1 was not significantly lower than that of CR6 (Fig. 5C, left). In the spleen, growth of CW3-VP1 was significantly lower than that of CW3 and not significantly different from that of CR6; CR6-VP1 growth was significantly higher than that of CR6 and not significantly different from that of CW3 (Fig. 5C, middle). Therefore, VP1 determines CW3 tropism for the spleen but not CR6 tropism for the proximal colon.
In parallel, we tested day seven growth of NS1/2 mutants to address the effect of the persistence determinant on tropism during acute infection. The persistent viruses CW3-NS1/2 and CW3D94E grew in both the spleen and proximal colon at levels significantly higher than the nonpersistent CR6-5′ NS1/2, which was not detected in any tissue (Fig. 5D). Strikingly, introduction of the D94E mutation into CR6-5′ NS1/2 resulted in significantly increased growth in the colon without a significant change in the spleen (Fig. 5D, CR6-5′ NS1/2D94E). Therefore, D94E is sufficient for growth in the proximal colon, but not in the spleen, during acute infection.
MNV persistence is restricted to the intestine.
The correlation between persistent fecal shedding and growth in the proximal colon during acute infection (Fig. 2E, 3B, and 5D) raised the possibility that MNV persistence is an intestine-specific phenotype. CR6 did not persist outside the intestine and associated lymphoid tissue, but its replication during acute infection was also restricted to these same sites (Fig. 5B and data not shown). Therefore, we took advantage of the expanded tropism of CR6-VP1 during acute infection to test this hypothesis. On day three postinfection with 1 × 106 PFU, CR6-VP1 had grown in both enteric tissues (Fig. 5E, proximal colon and MLN) as well as systemic tissues (Fig. 5E, spleen and liver). However, while CR6-VP1 was still present in the intestine on day 35, it was reduced to the limit of detection in the spleen and liver (Fig. 5E). This result shows that MNV persistence is specific to the intestine and provides a possible explanation for the correlation between enteric growth during acute infection and persistence.
DISCUSSION
The findings presented here demonstrate that the 5′ domain of NS1/2 is necessary and sufficient for MNV persistence. Furthermore, a single conservative change from aspartic acid to glutamic acid at amino acid 94 within this domain is sufficient for CW3 to persist. This D94E change also determines tropism for the proximal colon but not the spleen during acute infection. Notably, the proximal colon is a major site of MNV persistence, while the spleen does not support persistent replication.
MNV as a model of persistent enteric viral infection.
There is no clear animal reservoir for HuNoV, suggesting that it has other adaptations for viral maintenance between outbreaks. Asymptomatic and persistent shedding is likely to be one way that HuNoVs are maintained (4–7, 11), yet many questions remain about the mechanism of NoV persistence. Furthermore, while asymptomatic shedding has been demonstrated for many enteric viruses (43–45), a tractable model has not been established in which to study enteric persistence. We have characterized a novel model of persistence and identified the first genetic determinant of enteric persistence. Future study using the MNV model system has the exciting potential to reveal general principles of enteric persistence and host-pathogen interactions in the intestine.
Relationship between enteric growth and persistence.
We have shown a correlation of persistent shedding with growth in the colon, but not the spleen, during acute infection (Fig. 2E, 3B, and 5D). This correlation suggests that efficient colonic replication is required for persistence. Support for this idea comes from the fact that CR6-VP1, which has broad early tissue tropism, persists in the colon and MLN but not systemic sites (Fig. 5E). Therefore, efficient early replication in colon may correlate with persistence, because MNV persistence is restricted to enteric tissues. The importance of enteric growth may reflect a requirement for fecal shedding and reexposure, replication in enteric cell types, or induction of enteric immune responses.
CR6 infection results in increased fecal shedding compared to CW3 (Fig. 1). Consequently, there are likely an increased number of virions in the environment and more frequent reexposure of the intestinal epithelium. Antigenic drift within a cage may increase exposure to viral mutations that outpace immune elimination and result in persistence. Sequencing of CW3D94E from persistently infected mice reveals that mutations in the protruding domain of the capsid are in fact present (T. J. Nice, data not shown). Furthermore, there is evidence that this type of selection takes place in human populations and is important for persistence of HuNoV (46). It is notable that infection with 3 × 107 PFU of CW3, rather than the 3 × 104 PFU used here, results in appreciable shedding in the stool during acute infection and yet is ultimately cleared (35). Therefore, while it is still possible that CR6 and other persistent strains have a unique ability to promote antigenic drift, reexposure to shed MNV per se is not likely to be sufficient to explain the capacity to persist.
A separate, but not mutually exclusive, hypothesis to explain the importance of enteric growth is that characteristics of the immune system unique to this anatomical location are critical for persistence. The mammalian mucosal immune system has evolved in proximity to abundant commensal microbes and has a unique requirement for regulatory mechanisms that prevent excessive inflammatory responses to these microbes (47–51). These mechanisms include both intestine-specific differentiation programs for innate leukocytes (52, 53) and the related ability to generate regulatory T cell responses (54). An analogous regulatory immune response may be required for tolerance of persistent MNV in the intestine. Alternatively, because MNV is known to replicate in macrophages and dendritic cells (30), the unique differentiation states of these cell types in the intestine may provide cell-intrinsic growth requirements for persistent replication. Clarification of the anatomical requirements for MNV persistence will provide complementary insights into molecular studies of NS1/2 in establishing the mechanism of persistence in this novel model system.
Potential mechanisms of NS1/2 in persistence.
Studies implicating NS1/2 in membrane reorganization and formation of the replication complex (27, 40) raise the possibility that NS1/2 determines persistence through changes in MNV replication. We found no evidence for an in vitro replication function of the NS1/2 persistence determinant in RAW264.7 cells (Fig. 4). However, its role in determining growth in the proximal colon during acute infection (Fig. 5D) means that NS1/2 may have functions specific to a colonic cell type. Therefore, a role for NS1/2 in cell type-specific viral replication remains a possibility, and identification of the colonic cell types in which MNV persists will be necessary to address this possibility experimentally.
While there are multiple potential mechanisms for persistence, all persistent viruses share the capacity to prevent clearance by the immune system. Strategies for preventing immune elimination include inhibition of antigen presentation or innate immune signaling, evasion of adaptive immunity through mutation of immune-dominant epitopes, and infection of an immune-privileged site (55–57). Therefore, potential immune-modulatory functions of NS1/2 must also be considered in addition to basic replicative functions. Humoral immunity, cellular immunity, and innate immunity cooperate to clear infection with MNV1 (29, 37, 38), so NS1/2-mediated evasion of any of these immune components could account for its role in persistence. Identification of NS1/2 structures and interacting partners will be required to generate more detailed hypotheses about a role for NS1/2 in immune modulation.
Domains of NS1/2.
The NS1/2 caspase cleavage sites (18) have been used by us and others (58) to define NS1/2 domains. The region 5′ of the caspase cleavage site, which contains the amino acid 94 persistence determinant, is poorly conserved among NoVs and is also poorly conserved within genogroups. The region 3′ of the caspase cleavage site, however, is conserved among NoVs and is bioinformatically predicted to fold into a permutated papain-like fold (39). Characteristic of this fold, there are conserved cysteine, histidine, and histidine/asparagine residues predicted to form a catalytic triad. Supportive evidence for a catalytic function comes from a study of a picornaviral orthologue, where mutation of the conserved cysteine was found to block positive-strand RNA synthesis in an in vitro replication system (59). Similarly, mutation of the conserved cysteine or histidine in MNV prevents recovery of plaquable virus from the reverse genetics system (Nice, unpublished). Whether the persistence determinant in the 5′ domain is related to a hypothetical catalytic function of the 3′ domain requires further biochemical and functional studies.
We find that the 5′ NS1/2 domain determines persistence and colonic growth of both CW3 and CR6 (Fig. 2E and 5D), but amino acid 94 determines persistence of CW3 only (Fig. 3B). This suggests that amino acid 94 interacts with CW3-specific sequences. The fact that the D94E mutation can increase colonic growth of both CW3 and CR6-5′ NS1/2 (CW3D94E and CR6-5′ NS1/2D94E in Fig. 5D) suggests that the CW3-specific interaction of amino acid 94 is within the 5′ domain. While these data do not preclude an additional role for strain-independent intergenic interactions of the entire 5′ NS1/2 domain, they support the hypothesis that discrete domains exist within NS1/2. The separation of these domains by caspase 3 cleavage and the demonstrated induction of apoptosis during MNV infection (18, 60) raises the intriguing possibility that the MNV NS1/2 5′ domain acts independently of the 3′ domain to determine persistence and colonic tropism.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grants to H.W.V. (AI0544483 and AI084887). T.J.N. was supported by an NIH training grant (5T32A100716334) and postdoctoral fellowships from the Cancer Research Institute and American Cancer Society. Washington University and H.W.V. receive income based on licenses for MNV technology.
We thank members of the Virgin laboratory for their comments on the manuscript, P. Vachharajani for technical support, and D. Kreamalmeyer for managing mouse colonies.
Footnotes
Published ahead of print 17 October 2012
REFERENCES
- 1. Atmar RL, Estes MK. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35:275–290 [DOI] [PubMed] [Google Scholar]
- 2. Estes MK, Prasad BV, Atmar RL. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467–474 [DOI] [PubMed] [Google Scholar]
- 3. Radford AD, Gaskell RM, Hart CA. 2004. Human norovirus infection and the lessons from animal caliciviruses. Curr. Opin. Infect. Dis. 17:471–478 [DOI] [PubMed] [Google Scholar]
- 4. Ayukekbong J, Lindh M, Nenonen N, Tah F, Nkuo-Akenji T, Bergström T. 2011. Enteric viruses in healthy children in Cameroon: viral load and genotyping of norovirus strains. J. Med. Virol. 83:2135–2142 [DOI] [PubMed] [Google Scholar]
- 5. Barreira DMPG, Ferreira MSR, Fumian TM, Checon R, de Sadovsky ADI, Leite JPG, Miagostovich MP, Spano LC. 2010. Viral load and genotypes of noroviruses in symptomatic and asymptomatic children in southeastern Brazil. J. Clin. Virol. 47:60–64 [DOI] [PubMed] [Google Scholar]
- 6. Cheon D-S, Jeong HS, Jeong A, Lee K-B, Lee MH, Tahk H, Choi C. 2010. Seasonal prevalence of asymptomatic norovirus infection in Korean children. Foodborne Pathog. Dis. 7:1427–1430 [DOI] [PubMed] [Google Scholar]
- 7. Phillips G, Tam CC, Rodrigues LC, Lopman B. 2010. Prevalence and characteristics of asymptomatic norovirus infection in the community in England. Epidemiol. Infect. 138:1454–1458 [DOI] [PubMed] [Google Scholar]
- 8. Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. 2008. Norwalk virus shedding after experimental human infection. Emerging Infect. Dis. 14:1553–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barrabeig I, Rovira A, Buesa J, Bartolomé R, Pintó R, Prellezo H, Domínguez A. 2010. Foodborne norovirus outbreak: the role of an asymptomatic food handler. BMC Infect. Dis. 10:269 doi:10.1186/1471-2334-10-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmid D, Kuo H-W, Hell M, Kasper S, Lederer I, Mikula C, Springer B, Allerberger F. 2011. Foodborne gastroenteritis outbreak in an Austrian healthcare facility caused by asymptomatic, norovirus-excreting kitchen staff. J. Hosp. Infect. 77:237–241 [DOI] [PubMed] [Google Scholar]
- 11. Sukhrie FHA, Siebenga JJ, Beersma MFC, Koopmans M. 2010. Chronic shedders as reservoir for nosocomial transmission of norovirus. J. Clin. Microbiol. 48:4303–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng D-P, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312–323 [DOI] [PubMed] [Google Scholar]
- 13. Jiang X, Wang M, Wang K, Estes MK. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51–61 [DOI] [PubMed] [Google Scholar]
- 14. McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. 2011. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 7:e1002413 doi:10.1371/journal.ppat.1002413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, Kelley ST, Virgin HW. 2007. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 81:10460–10473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Belliot G, Sosnovtsev SV, Mitra T, Hammer C, Garfield M, Green KY. 2003. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J. Virol. 77:10957–10974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu B, Clarke IN, Lambden PR. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sosnovtsev SV, Belliot G, Chang K-O, Prikhodko VG, Thackray LB, Wobus CE, Karst SM, Virgin HW, Green KY. 2006. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J. Virol. 80:7816–7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang X, Wang M, Graham DY, Estes MK. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527–6532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glass PJ, White LJ, Ball JM, Leparc-Goffart I, Hardy ME, Estes MK. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meyers G, Wirblich C, Thiel HJ, Thumfart JO. 2000. Rabbit hemorrhagic disease virus: genome organization and polyprotein processing of a calicivirus studied after transient expression of cDNA constructs. Virology 276:349–363 [DOI] [PubMed] [Google Scholar]
- 22. Oka T, Katayama K, Ogawa S, Hansman GS, Kageyama T, Ushijima H, Miyamura T, Takeda N. 2005. Proteolytic processing of sapovirus ORF1 polyprotein. J. Virol. 79:7283–7290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sosnovtsev SV, Garfield M, Green KY. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seah EL, Marshall JA, Wright PJ. 2003. Trans activity of the norovirus Camberwell proteinase and cleavage of the N-terminal protein encoded by ORF1. J. Virol. 77:7150–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fernandez-Vega V, Sosnovtsev SV, Belliot G, King AD, Mitra T, Gorbalenya A, Green KY. 2004. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 78:4827–4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hyde JL, Mackenzie JM. 2010. Subcellular localization of the MNV-1 ORF1 proteins and their potential roles in the formation of the MNV-1 replication complex. Virology 406:138–148 [DOI] [PubMed] [Google Scholar]
- 27. Ettayebi K, Hardy ME. 2003. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 77:11790–11797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsu CC, Riley LK, Wills HM, Livingston RS. 2006. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 56:247–251 [PubMed] [Google Scholar]
- 29. Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575–1578 [DOI] [PubMed] [Google Scholar]
- 30. Wobus CE, Karst SM, Thackray LB, Chang K-O, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, Virgin HW. 2004. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432 doi:10.1371/journal.pbio.0020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strong DW, Thackray LB, Smith TJ, Virgin HW. 2012. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol. 86:2950–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin HW, Lambden PR. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. U. S. A. 104:11050–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey D, Thackray LB, Goodfellow IG. 2008. A single amino acid substitution in the murine norovirus capsid protein is sufficient for attenuation in vivo. J. Virol. 82:7725–7728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taube S, Perry JW, McGreevy E, Yetming K, Perkins C, Henderson K, Wobus CE. 2012. Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J. Virol. 86:5584–5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cadwell K, Patel KK, Maloney NS, Liu T-C, Ng ACY, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baert L, Wobus CE, Van Coillie E, Thackray LB, Debevere J, Uyttendaele M. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 74:543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. 2008. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 4:e1000236 doi:10.1371/journal.ppat.1000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chachu KA, Strong DW, LoBue AD, Wobus CE, Baric RS, Virgin HW. 2008. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 82:6610–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11 doi:10.1186/gb-2003-4-2-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hyde JL, Sosnovtsev SV, Green KY, Wobus C, Virgin HW, Mackenzie JM. 2009. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 83:9709–9719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arias A, Bailey D, Chaudhry Y, Goodfellow IG. 2012. Development of a reverse genetics system for murine norovirus 3; long-term persistence occurs in the caecum and colon. J. Gen. Virol. 93:1432–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kahan SM, Liu G, Reinhard MK, Hsu CC, Livingston RS, Karst SM. 2011. Comparative murine norovirus studies reveal a lack of correlation between intestinal virus titers and enteric pathology. Virology 421:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cinek O, Witsø E, Jeansson S, Rasmussen T, Drevinek P, Wetlesen T, Vavrinec J, Grinde B, Rønningen KS. 2006. Longitudinal observation of enterovirus and adenovirus in stool samples from Norwegian infants with the highest genetic risk of type 1 diabetes. J. Clin. Virol. 35:33–40 [DOI] [PubMed] [Google Scholar]
- 44. Maldonado Y, Cantwell M, Old M, Hill D, Sanchez ML, Logan L, Millan-Velasco F, Valdespino JL, Sepulveda J, Matsui S. 1998. Population-based prevalence of symptomatic and asymptomatic astrovirus infection in rural Mayan infants. J. Infect. Dis. 178:334–339 [DOI] [PubMed] [Google Scholar]
- 45. Phillips G, Lopman B, Rodrigues LC, Tam CC. 2010. Asymptomatic rotavirus infections in England: prevalence, characteristics, and risk factors. Am. J. Epidemiol. 171:1023–1030 [DOI] [PubMed] [Google Scholar]
- 46. Lindesmith LC, Donaldson EF, LoBue AD, Cannon JL, Zheng D-P, Vinjé J, Baric RS. 2008. Mechanisms of GII. 4 norovirus persistence in human populations. PLoS Med. 5:e31 doi:10.1371/journal.pmed.0050031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hill DA, Artis D. 2010. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 28:623–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jarchum I, Pamer EG. 2011. Regulation of innate and adaptive immunity by the commensal microbiota. Curr. Opin. Immunol. 23:353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee YK, Mazmanian SK. 2010. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330:1768–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nava GM, Stappenbeck TS. 2011. Diversity of the autochthonous colonic microbiota. Gut Microbes 2:99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Virgin HW, Todd JA. 2011. Metagenomics and personalized medicine. Cell 147:44–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mowat AM, Bain CC. 2011. Mucosal macrophages in intestinal homeostasis and inflammation. J. Innate Immun. 3:550–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scott CL, Aumeunier AM, Mowat AM. 2011. Intestinal CD103+ dendritic cells: master regulators of tolerance? Trends Immunol. 32:412–419 [DOI] [PubMed] [Google Scholar]
- 54. Nutsch KM, Hsieh C-S. 2012. T cell tolerance and immunity to commensal bacteria. Curr. Opin. Immunol. 24:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kane M, Golovkina T. 2010. Common threads in persistent viral infections. J. Virol. 84:4116–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oldstone MBA. 2009. Anatomy of viral persistence. PLoS Pathog. 5:e1000523 doi:10.1371/journal.ppat.1000523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Virgin HW, Wherry EJ, Ahmed R. 2009. Redefining chronic viral infection. Cell 138:30–50 [DOI] [PubMed] [Google Scholar]
- 58. Baker ES, Luckner SR, Krause KL, Lambden PR, Clarke IN, Ward VK. 2012. Inherent structural disorder and dimerisation of murine norovirus NS1-2 protein. PLoS One 7:e30534 doi:10.1371/journal.pone.0030534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sasaki J, Taniguchi K. 2008. Aichi virus 2A protein is involved in viral RNA replication. J. Virol. 82:9765–9769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bok K, Prikhodko VG, Green KY, Sosnovtsev SV. 2009. Apoptosis in murine norovirus-infected RAW264.7 cells is associated with downregulation of survivin. J. Virol. 83:3647. [DOI] [PMC free article] [PubMed] [Google Scholar]






