Abstract
The therapeutic potential of autophagy for the treatment cancer and other diseases is beset by paradoxes stemming from the complexity of the interactions between the apoptotic and autophagic machinery. The simplest question of how autophagy acts as both a protector and executioner of cell death remains the subject of substantial controversy. Elucidating the molecular interactions between the processes will help us understand how autophagy can modulate cell death, whether autophagy is truly a cell death mechanism and how these functions are regulated. We suggest that despite many connections between autophagy and apoptosis, a strong causal relationship wherein one process controls the other, has not been adequately demonstrated. Knowing when and how to modulate autophagy therapeutically depends on understanding these connections.
Introduction
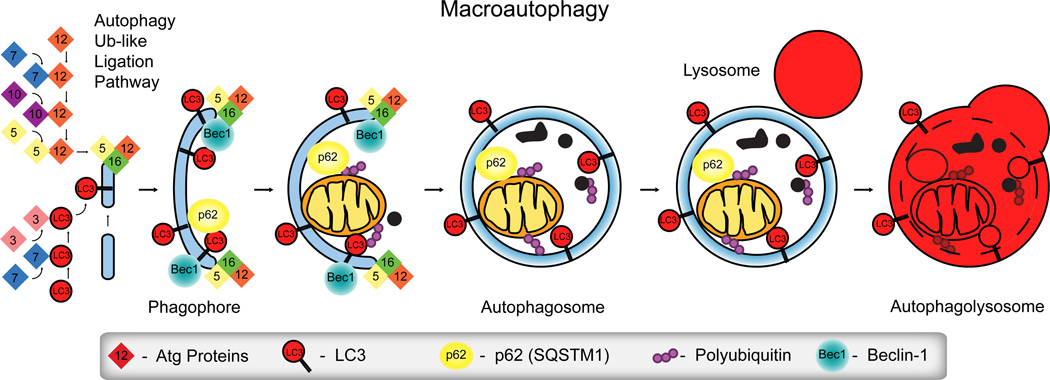
Macroautophagy (hereafter autophagy) is an evolutionarily conserved catabolic process involving the formation of vesicles (autophagosomes) that engulf cellular macromolecules and organelles, leading to their breakdown following fusion with lysosomes (Figure 1). While autophagy was described over 50 years ago, only within the last 5–7 years have strong cell biological data elucidated the form and function of this ubiquitous process [1–4]. Autophagy is active in all cells and can be upregulated in response to stress or nutrient deprivation. Induction of autophagy not only facilitates the degradation of damaged cellular components but also provides the cell with molecular building blocks and energy. Autophagy is now known to play a role in many diverse disease processes including cancer, neurodegeneration, aging, autoimmune diseases like Crohn’s disease and rheumatoid arthritis, heart disease and infection. At the level of both the organism and the cell, autophagy can, paradoxically, have pro-death or pro-survival functions depending on the context [5–7].
Figure 1. Macroautophagy.
The autophagic process degrades cellular macromolecules and organelles releasing metabolites to provide the cell with energy and anabolic building blocks to aid in survival and repair during nutrient deprivation or cellular damage. The Ubiquitin-like conjugation cascade leads to the lipidation of LC3 and conjugation of Atg5-Atg12, two key elements in the formation of the phagophore. The autophagic process begins (autophagy induction) with formation of the phagophore, its elongation and closure to form the autophagosome that then fuses with lysosomes resulting in degradation of the contents (autophagic flux).
At the cellular level, the pro-survival functions of autophagy are the most well-defined wherein autophagy aids the cell in dealing with stress by clearing damaged proteins, organelles, pathogens or aggregates, or by providing the cell with energy and anabolic building blocks during starvation. Conversely, several recent studies have indicated that autophagy itself may be a mechanism of caspase and apoptosis-independent cell death [8, 9]. Figuring out exactly when and where these disparate functions of autophagy apply are key goals in the field. The role played by autophagy in cell death is of great interest to us and others because autophagy’s potential ability to modulate cell death makes it a therapeutic target (through either up or down-regulation) in several diseases including cancer and neurodegeneration [10–13].
Autophagy and Apoptosis
The connection between autophagy and apoptosis or other forms of cell death is a burgeoning area of research. The number of publications about autophagy is growing rapidly and a great many of these papers are concerned with cell death in one context or another. The molecular connections between autophagy and cell death are multifaceted, complex and still poorly understood. Determining which interactions are important in autophagy’s regulation of cell death is critical as we try to either protect cells we don’t want to die (as in neurodegenerative diseases) or cause diseased cells to die (as in cancer treatment). While it is generally accepted that autophagy functions as a mechanism to survive cellular stresses like nutrient deprivation, the molecular mechanisms underlying the regulation and specificity of autophagic degradation are just beginning to be understood. Moreover, although there are a multitude of published connections between the processes of autophagy and cell death (both apoptosis and necrosis), there are gaping holes in our understanding of even the most basic questions regarding the interactions between autophagy, cell death and disease. For example, despite many publications, our opinion is that it is not yet firmly established whether cancer cells can use autophagy to evade being killed by chemotherapy or radiation and, even for the same treatments, different publications sometimes come to completely opposite conclusions [14–16]. Here, we try to identify some key areas of confusion regarding the connections between autophagy and apoptosis/cell death and come up with ideas to solve these problems.
Linking autophagy and apoptosis
Broadly speaking, there are two ways that autophagy and apoptosis could be directly linked (Figures 2 & 3). First, the process of autophagy could control apoptosis – either by making it more or less likely. Second, the process of apoptosis (i.e. activation of caspases etc.) could control autophagy (either increasing or decreasing it). The general view is that autophagy is usually pro-survival; it allows cells to survive prolonged starvation and other stresses, infectious agents and, as noted above it is often suggested that autophagy can protect cells against treatment with agents like anti-cancer drugs that are intended to kill them. However, we would argue that just because autophagy can protect cells, it doesn’t necessarily follow that this was because the autophagy machinery and apoptosis machinery are connected. For example, one of the primary functions of autophagy is to recycle damaged proteins, organelles and aggregates thereby cleaning up the cell and providing it with new building blocks to replace damaged or depleted cellular components [3, 4, 17]. This mechanism can also provide a protective function during nutrient deprivation that does not necessarily require a direct connection between autophagy and the death machinery. If a cell is in the process of dying because it doesn’t have enough amino acids, the fact that autophagy stops the death doesn’t necessarily mean that autophagy controls the apoptosis or necrosis machinery, but could just mean that autophagy provided the needed amino acids. However, as discussed below, a rapidly expanding number of direct molecular connections between autophagy and apoptosis open the possibility of a real causal link between the two processes (Table 1 & Figure 3).
Figure 2. Apoptosis.
Key proteins involved in the intrinsic and extrinsic apoptotic pathways are depicted focusing on the components with known links to autophagy mentioned in the text. The extrinsic pathway results from the binding of extracellular death ligands which transduce a signal resulting in the formation of the death inducing signaling complex and activation of caspase-8. This activation is potentiated by p62-mediated aggregation leading to efficient activation of caspase-8. Caspase-8 then activates mitochondrial outer membrane permeabilization through the formation of Bax/Bad/Bak pores leading to cytoplasmic Cytochrome-C, that activates the apoptosome and effector caspases (Caspase-3), and the release of SMAC (Diablo), a protein that disables Inhibitor of Apoptosis Proteins (IAPs). Activated effector (executioner) caspases then cleave multiple targets resulting in cell death.
Figure 3. Linking Autophagy and Apoptosis.
Connections between the autophagic and apoptotic processes. a. Atg5 activates DISC via an interaction with FADD. b. FLIP inhibits Atg3-LC3 association and therefore induction of autophagy. c. Atg12-Atg3 conjugation inhibits mitochondrial fission and apoptosis, independent of autophagy. d. p62 promotes aggregation and activation of Caspase-8 which, paradoxically, is degraded by autophagy, likely through its interaction with p62. e. Dap kinase phosphorylation of Beclin-1 promotes autophagy. f. Bcl-2 interaction with Beclin-1 inhibits autophagy. g. p53 can both promote and inhibit autophagy depending on the context. h. Autophagic cell death. Well established links are depicted with solid lines and less established links are shown with dashes. More information about each interaction is contained in Text Box 1 and the references therein, or in the text.
Table 1.
Proteins with dual roles in autophagy and apoptosis.
| Protein | Function | References |
|---|---|---|
| DAPK | Phosphorylates Beclin1; activates DISC | [44] |
| RIP | Activates Cell death independent of caspases; may activate autophagic cell death | [9,45] |
| NF-κB | Regulates survival pathways – inhibits apoptosis activates autophagy | [46] |
| JNK | Positively Regulates both apoptosis and autophagy | [47] |
| p62 | Critical for activation of caspase-8; regulates selective autophagy of many substrates | [23] |
| Beclin1 | Primary cellular activator of autophagy; regulated by Bcl-2 | [48] |
| Bcl-2 | Inhibits both autophagy and apoptosis by binding Beclin-1 and Bax/Bad/Bak | [36] |
| Caspase-8 | Activates apoptosis via extrinsic pathway; cleaves p62 during apoptosis | [27] |
| Caspase-3 | May cleave Beclin-1 to inhibit autophagy during terminal stages of apoptosis | [49] |
| p53 | Induces MOMP in response to stress; positively and negatively regulates autophagy | [50–52] |
| Atg5 | Critical autophagy gene; activates apoptosis via FADD and MOMP upon calpain cleavage | [53–55] |
| FLIP | c-FLIP inhibits autophagy through inhibition of Atg3-LC3 conjugation | [56] |
| Atg12-Atg3 | Novel regulator of mitochondria and apoptosis with no known function in autophagy | [40] |
There is also a substantial body of literature suggesting that under certain circumstances autophagy can promote death and recent evidence indicates that autophagy itself may be a mechanism of cell death [5, 8, 9]. Autophagy may be able to kill a cell either by actively degrading necessary cellular components (e.g. catalase, mitochondria) or by non-selectively degrading cellular components to the point that the cell can no longer survive [9, 18]. However, once again, it does not follow that these mechanisms mean that the process of autophagy is directly acting on the cell death machinery – autophagy might degrade cellular components so that the cell eventually activates the apoptosis machinery but this doesn’t mean that the process of autophagy is directly responsible for activating apoptosis; the effect might be indirect.
Adding further confusion, most studies look for relationships between autophagy and apoptosis by asking if the overall amount of death changes- i.e. does autophagy prevent death or cause death. However the connections between autophagy and apoptosis might be more along the lines of changing the way that the cells die as opposed to changing whether or not they die. For example, it was found that a targeted toxin kills glioma cells via a mechanism that does not involve caspase activation and, when autophagy is inhibited this modestly increases the amount of death, but dramatically changed the mode of death by allowing the toxin to activate caspases [19]. These and other data show that autophagy can alter the way cells die not just whether they die or not [20, 21]. One complication of this is that if the assay used to assess death was specific for a certain type of death (e.g. apoptosis) one can easily obtain a mistaken impression that there was a big effect on cell death when in fact the total number of dead cells is not greatly affected by autophagy manipulation. How then might the autophagy and apoptosis machinery talk to each other, and do they do so?
Mechanistic connections between apoptosis and autophagy
Multiple direct and indirect interactions have been described suggesting mechanistic overlap and interaction between the apoptosis machinery and autophagy proteins [6, 13, 22]. The majority of these interactions have been apoptosis altering autophagy; less is known at the mechanistic level about how autophagy controls apoptosis. We concentrate here on two proteins that have multiple connections between autophagy and cell death but there are other molecules with connections between the two processes that may also be critical links.
Two autophagy proteins at the crux of autophagy-apoptosis interactions are p62, a protein that is important for Ras-induced tumorigenesis, and the tumor suppressor Beclin-1. p62 is a key player in the selective autophagic degradation of many proteins (and mitochondria) and is known to interact directly with several apoptotic and survival pathway proteins including Caspase-8, TRAF6 (which modulates NF-kB survival pathways) and ERK [23–26]. The interaction between caspase-8 and p62 is particularly intriguing because p62 is critical for efficient activation of caspase-8 but caspase-8 also cleaves p62 in response to death receptor activation [27, 28]. Furthermore, caspase-8 has recently been shown to be degraded by autophagy (presumably via p62) [29]. This creates a paradigm where autophagy and apoptosis might be involved in a complicated balancing act wherein autophagy alters the extent and kinetics of apoptosis and apoptosis alters the autophagic degradation of p62 and p62-dependent autophagic cargos, including caspase-8.
Beclin-1 is a critical regulator of autophagy that directly interacts with anti-apoptotic Bcl-2 [30–33]. When Bcl-2 and Beclin-1 are bound, Beclin-1 is incapable of activating autophagy. Autophagy is induced by release of Beclin-1 from Bcl-2 by pro-apoptotic BH3 proteins, Beclin-1 phosphorylation by DAP kinase (DAPK) or Bcl-2 phosphorylation by JNK [34, 35]. Conversely, overexpression of Bcl-2 or Bcl-XL can inhibit autophagy [30, 36, 37]. Another Beclin-1-dependent mechanism by which apoptosis can inhibit autophagy is through caspase-3 cleavage of Beclin-1 to produce a truncated protein that is unable to promote autophagy thus leading to overall autophagy inhibition [38]. Thus, Beclin-1 regulation by components of the apoptosis machinery can either promote or inhibit autophagy perhaps depending on the relative activities of BH3 proteins and initiator and/or executioner caspases. These examples show that there can be mutual regulation of apoptosis and autophagy, so that when apoptosis is promoted autophagy is reduced to provide a mechanism to ensure that autophagy is switched off when a cell “decides” to go through with apoptosis.
Other potential avenues by which autophagy may regulate apoptosis are through the active degradation of apoptotic proteins. It should be noted however that although autophagy is capable of specifically degrading components of the apoptotic machinery (e.g. Caspase-8, mitochondria), the significance of these events in terms of actually changing the amount of cell death in a physiologic or disease setting is unclear. For example, although sequestration and degradation of mitochondria in autophagosomes occurs, this happens to only some of the mitochondria in a cell [39]. Thus an unanswered question is: even if autophagy is increased, why would an apoptotic stimulus not still cause cytochrome c release from the other mitochondria in the cell to induce apoptosome formation and apoptosis? Perhaps by reducing the number of mitochondria, autophagy might alter the kinetics of cell death or reduce the threshold of pro-apoptotic activity necessary to induce apoptosis. Therefore, autophagy might control an apoptosis “threshold”, i.e. some kind of regulator that ultimately decides whether apoptosis should proceed or not. This, in turn, may provide a way to ensure the rapid and complete demise of the cell to thus control the degree of apoptosis in the population of cells. One prediction (that has been untested so far) of this idea is that in a population of cells where some die and some don’t die, the molecular mechanisms outlined above like Beclin-1 or p62 cleavage, will only take place in those cells that actually die and these will be the same cells that have lower levels of autophagy. Thus, although we have evidence of connections between the two processes and it makes intuitive sense that such connections might exist, a clear mechanistic explanation of how autophagy can inhibit apoptosis for any defined apoptotic pathway is still lacking.
Adding confusion to an already complicated situation, just because a protein regulates autophagy and it also affects how efficiently a cell can undergo apoptosis, this does not necessarily mean that it was the process of autophagy that affected apoptosis. A provocative recent example shows that a previously unknown conjugation of Atg12 to Atg3, which are both essential components of the autophagy ubiquitin-like conjugation machinery that is required for autophagosome formation, has an autophagy-independent function that can regulate apoptosis [40]. Atg3 mutants that cannot be conjugated to Atg12 inhibit apoptosis through the mitochondrial pathway but this occurs in a manner independent of autophagy itself – inhibiting Atg12-Atg3 conjugation has no effect on starvation-induced autophagy (Atg12-Atg5 conjugation is required) and the effect on cell death is instead associated with altered fragmentation of mitochondria. This intriguing finding shows that although components of the autophagy machinery can control the apoptosis pathway, they may do so independent of autophagy and also leaves open the possibility of other unknown functions for proteins of the autophagy ligation process.
So, although we are starting to develop an understanding of how components of the apoptosis and autophagy machineries can intersect, there is much that we do not know. It is important to continue studying these connections because of the potential clinical impact of manipulating these complicated pathways.
Clinical application
In addition to the importance of understanding the biology of autophagy, there is great interest in trying to manipulate autophagy in a clinical setting. For example, there are currently almost 3 dozen clinical trials in the U.S. attempting to block the cellular survival functions of autophagy in cancer treatment by combining chemotherapy with the autophagy inhibitor hydroxychloroquine. The basis for these clinical trials is the simple premise that since autophagy protects damaged cells from dying, autophagy inhibition will make it more likely that damaged cancer cells die and the tumor will be eradicated [6, 13, 24, 41]. However, if autophagy’s function in cancer cells is not to protect them from damage, but instead to promote cell death, then these clinical trials could be pointless or even harmful unless we can find a way to predict in which patients pro-death or pro-survival functions of autophagy apply. Conversely, in other diseases can we take advantage of an autophagy-dependent survival function? For example, in neurodegenerative disease, mTOR inhibition leading to increased autophagy enhances the ability of cells to clear damaged or aggregated proteins or mitochondria improving the health and functioning of neurons and, in animal models of neurodegeneration, increasing autophagy can delay or prevent the onset of symptoms of neurological disease [11, 12, 42, 43]. This has the potential to prevent, delay or cure several types of neurodegenerative disease including Parkinson’s, Huntington’s and ALS.
Conclusion
Better understanding of how autophagy and apoptosis are related (whether it be directly or indirectly) is likely to be an important area of research in the cell death field in coming years. The autophagy proteins p62 and Beclin1 are likely critical molecular players, regulating -- and being regulated by -- a number of pro and anti-apoptotic molecules. Understanding the way that these proteins, and other components of the autophagy and apoptosis pathways, can tip the balance between survival and death is crucial to determining the utility of targeting autophagy in cancer treatment and other diseases. However, we need to answer some very basic questions before we can hope to apply this information in a useful manner (Text Box 1). It is likely that the next few years will see these fundamental questions answered as we understand at a molecular level the complex direct and indirect interactions between autophagy and apoptosis. But until we do so, we would argue that the jury should still be out regarding key questions about the relationship between autophagy and apoptosis – including perhaps the most fundamental one of all– whether or not the process of autophagy directly controls the process of apoptosis or merely share some common machinery. We believe that by continuing to probe these processes and applying more specific drugs and genetic inhibitors as well as newer, more accurate and less subjective assays to measure autophagy, we should be able to more definitively answer this question. And, if the answer is yes, we think this will help determine the full spectrum of molecular mechanisms that underlie the connections to provide a framework for more precise manipulation of these processes in a clinical setting.
Text Box 1. Outstanding questions.
Under what circumstances does autophagy control apoptosis? Are all apoptotic stimuli susceptible or only some? What about other death mechanisms such as necrosis?
If autophagy controls apoptosis what steps in the apoptosis pathway are affected? Does autophagy control the release of mitochondrial proteins? Does autophagy control enzymatic activation of caspases? Are multiple steps in the apoptosis pathway affected?
Does autophagy regulate apoptosis primarily by indirect means? For example, by merely controlling the level of macromolecular precursors rather than directly controlling the apoptosis machinery?
Are the apparent connections between autophagy and apoptosis due to the same proteins being used for different functions as with the case of Atg12-Atg3?
Does variation in the extent of autophagy within a population determine which cells in the population die or survive an apoptotic stimulus? Is this how cells determine whether they have breached the “apoptotic threshold”?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1(2):66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberg-Lerner A, et al. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16(7):966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 7.Dalby KN, et al. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6(3):322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu L, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103(13):4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington's disease models. Nat Chem Biol. 2007;3(6):331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15(17):5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaravadi RK, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117(2):326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crazzolara R, et al. Potentiating effects of RAD001 (Everolimus) on vincristine therapy in childhood acute lymphoblastic leukemia. Blood. 2009;113(14):3297–3306. doi: 10.1182/blood-2008-02-137752. [DOI] [PubMed] [Google Scholar]
- 16.Katayama M, et al. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14(3):548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- 17.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 18.Lartigue L, et al. Caspase-independent mitochondrial cell death results from loss of respiration, not cytotoxic protein release. Mol Biol Cell. 2009;20(23):4871–4884. doi: 10.1091/mbc.E09-07-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thorburn J, et al. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009;16:175–183. doi: 10.1038/cdd.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang C, et al. Autophagy is associated with apoptosis in cisplatin injury to renal tubular epithelial cells. Am J Physiol Renal Physiol. 2008;294(4):F777–F787. doi: 10.1152/ajprenal.00590.2007. [DOI] [PubMed] [Google Scholar]
- 21.Abedin MJ, et al. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14(3):500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- 22.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2007;13(1):1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70(9):3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 25.Kirkin V, et al. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Mathew R, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman JM, Cohen GM, Bampton ETW. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6(8):1–15. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137(4):721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Hou W, et al. Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6(7) doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chipuk JE, et al. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29(12):1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 32.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27(Suppl 1):S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20(6):355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei Y, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pattingre S, et al. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Erlich S, et al. Differential Interactions Between Beclin 1 and Bcl-2 Family Members. Autophagy. 2007;3(6):561–568. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 37.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4(5):600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010;17(2):268–277. doi: 10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2010;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142(4):590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11(2):127–137. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravikumar B, et al. Rapamycin pre-treatment protects against apoptosis. Hum Mol Genet. 2006;15(7):1209–1216. doi: 10.1093/hmg/ddl036. [DOI] [PubMed] [Google Scholar]
- 43.Sarkar S, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282(8):5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 44.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X(L) and induction of autophagy. EMBO Rep. 2009 doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonapace L, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance. J Clin Invest. 2010 doi: 10.1172/JCI39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr Opin Genet Dev. 2008;18(1):19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorin S, et al. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy. 2009;6(1):153–154. doi: 10.4161/auto.6.1.10537. [DOI] [PubMed] [Google Scholar]
- 48.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohn TT, et al. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer's disease brain. Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crighton D, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 51.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10(6):637–639. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tasdemir E, et al. A dual role of p53 in the control of autophagy. Autophagy. 2008;4(6):810–814. doi: 10.4161/auto.6486. [DOI] [PubMed] [Google Scholar]
- 53.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8(10):1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 54.Bell BD, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008;105(43):16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pyo JO, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280(21):20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 56.Lee JS, et al. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11(11):1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]