Abstract
How tumor cells process damaged or unwanted DNA is a matter of much interest. Recently, Rello-Varona et al. (Cell Cycle 2012; 11:170–76) reported the involvement of macroautophagy (hereon autophagy) in the elimination of micronuclei (MN) from osteosarcoma cells. Prior to that, diminution of whole nuclei from multinucleated TP53-mutant tumor cells was described. Here, we discuss these two kinds of chromatin autophagy evoked after genotoxic stress in the context of the various biological processes involved: (1) endopolyploidy and the ploidy cycle; (2) the timing of DNA synthesis; (3) DNA repair; (4) chromatin:nuclear envelope interactions; and (5) cytoplasmic autophagy. We suggest that whereas some MN can be reunited with the main nucleus (through interactions with envelope-limited chromatin sheets) and participate in DNA repair, failure of repair serves as a signal for the chromatin autophagy of MN. In turn, autophagy of whole sub-nuclei in multi-nucleated cells appears to favor de-polyploidization, mitigation of aneuploidy with its adverse effects, thereby promoting the survival fitness of descendents and treatment resistance. Thus, both kinds of chromatin autophagy provide tumor cells with the opportunity to repair DNA, sort and resort chromatin, reduce DNA content, and enhance survival.
Keywords: chromatin autophagy, genotoxic damage, reversible polyploidy, micronuclei, aneuploidy
Niels Bohr once said “A great truth is a truth whose opposite is also a great truth.” In particular, this should apply to processes which display paradoxically opposing natures. This thought came to mind when reading the article by Rello-Varona et al.1 concerning the autophagic removal of micronuclei (MN), published recently in Cell Cycle. Here, macroautophagy (hereon termed autophagy) was convincingly shown to be involved in the degradation of MN induced in osteosarcoma U2OS cells after cell cycle disruption. The authors suggested that this process may contribute to genome stabilization, however were surprised that the proportion of MN undergoing autophagic processing (~2–5%) was so small. A marginal contribution of autophagy to MN elimination or a rapid turnover of the autophagic MN were suggested as possible causes. Here we discuss several alternative possibilities and more generally consider the various mechanisms involved in the genome stabilization and chromatin autophagy of tumor cells undergoing reversible polyploidy.
Cell-cycle disruptors, such as those used by Rello-Varona et al.,1 alongside irradiation and other genotoxic insults can induce reversible polyploidy in TP53 function defective cells which in turn is able to contribute to cell survival. Reversible polyploidy has been observed in various tumor cell lines of mesenchymal and epithelial origin after genotoxic treatment (irradiation, doxorubicin, etoposide, nocodazole, paclitaxel, and cisplatin), (for review and more recent work see refs. 2-6). It is also observed at low frequency in nontreated lymphoma and HeLa cell lines.7,8 The generality of this process for tumor evolution (also termed ‘neosis’2,9 or the ‘cancer life cycle’10,11) was suggested previously, although still remains to be shown for human tumors in vivo.
Tetraploidy is associated with genome instability, aneuploidy, and increased mutability serving as a driver of carcinogenesis.12 Concurrently, aneuploidy has adverse effects on proliferation.13 Therefore it is expected that the reversible polyploidy caused by anti-cancer drugs should elicit mechanisms which mitigate aneuploidy, reduce mutation load and the associated genotoxic/metabolic stress, thus favoring cell fitness for tumor progression and treatment resistance.11 As autophagy has been found to reduce DNA damage and stabilize the genome,14 we therefore propose that chromatin autophagy is involved intimately in this process.
It is well established that TP53 function deficiency favors endopolyploidy, micronucleation, and resistance in response to genotoxic damage and spindle disrupters. Rello-Varona et al.1 used U2OS cells which display wild-type TP53 and RB1, but which have lost expression of CDKN2A/p16INK4a. This genotype is equivalent to mutant TP53, as the downstream cell cycle suppressors CDKN1A/p21 and CDKN2A are linked with a positive feedback loop.15 Therefore, as would be expected, all four cell-cycle disruptors employed by the authors displayed enhanced MN, tetraploidy, and higher level polyploidy, followed by a return to diploidy.
MN are routinely observed as a hallmark of genotoxicity and chromosome instability, and result from aberrant mitosis often leading to mitotic catastrophe or mitotic slippage and endopolyploidy. The U2OS cells releasing autophagic MN are seen after mitotic slippage as judged by their nuclear morphology. In these cells autophagic MN are observed alongside clearly non-autophagic MN. Why the autophagic MN were observed so rarely and alongside non-autophagic MN in the same polyploid cell requires further discussion. First, we should consider the link between MN autophagy and DNA repair.
Release of Rad51-enriched MN was previously reported in irradiated HeLa cells by Haaf et al.16 and for the first time was suggested as being involved in sorting damaged DNA through repair. DNA repair by homologous recombination (HR) is typically enhanced in TP53-function deficient polyploid tumor cells and protects them from apoptosis.17-19 However, although DNA repair by HR is antagonistic to apoptosis, it is still compatible with simultaneous chromatin sorting by autophagic buds from the same cells.8 In the chromatin buds of endopolyploid cells undergoing intense HR, the DNA recombinases Rad51 and Rad52 are either organized into repair foci or are seen in a disorganized pattern8,19 (exemplified on Fig. 1A and B). In the latter case the DNA in the buds is degraded (low DAPI staining and γH2AFX/γH2AX-positivity) suggestive of failed repair by HR and selective autophagy of that particular chromatin material.8,20 Support for this association between failed repair and autophagy comes from Robert et al.21 who recently showed the direct link between the processing of double-strand breaks by repair enzymes and autophagy.
Figure 1. (A and B) Characterization of the association between MN, homologous recombination repair and DNA integrity in TP53 mutant lymphoma cells after γ-irradiation insult. (A) A polyploid nucleus undergoing intensive DNA repair by HR (testified by multiple repair foci positive for Rad51 and γH2AFX/γH2AX). A MN (arrow) is being released which involves a large repair focus. The corresponding DAPI gray-scale image (insert below) shows no loss of DNA content in this MN. (B) A polyploid lymphoma cell undergoing both DNA repair by HR (as testified by the presence of multiple Rad52-positive foci) and simultaneous release of two MN (arrows) containing large aggregates of Rad52 and degrading DNA, as seen by the reduced DAPI content (shown in the insert to the left); (C and D) release of whole sub-nuclei from polyploid lymphoma cells showing (C) selective chromatin degradation of sub-nuclei seen using the acridine orange in situ denaturation test [where red fluorescence indicates degraded DNA (arrowed) and green fluorescence indicates intact DNA] and on (D) sequestration of large amounts of DNA (DAPI stained blue) in a perinuclear vacuole accumulating CTSB/Cathepsin B, indicative of autophagy (red, arrowed). Bars: 20 μm. Figures are republished with new annotation: (A) from reference 19 and (C) from reference 20, with permission of Elsevier; (B and D) were originally published in reference 8, copyright holder Portland Press.
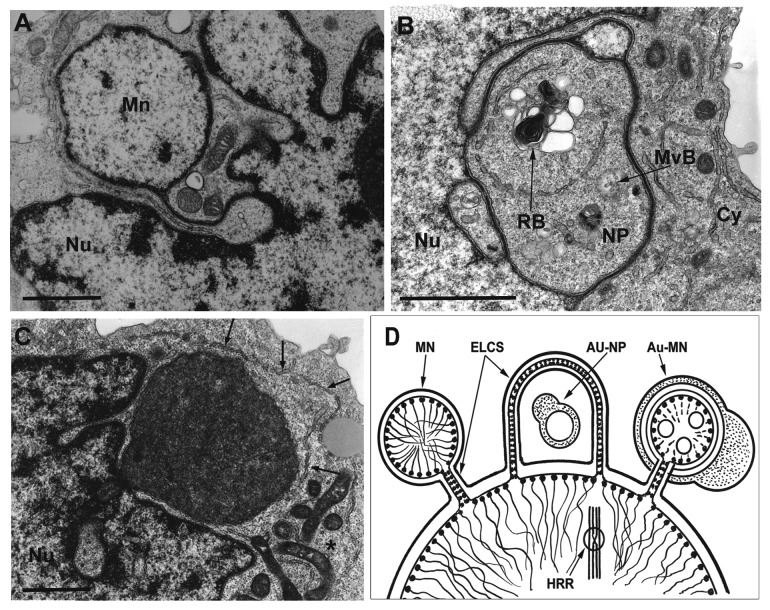
As the autophagic and non-autophagic MN in most cases are seen in close vicinity to the main nuclei of polyploid cells, or are presented as nuclear buds, their relationship with the nuclear envelope (NE) should also be considered. In particular, envelope limited chromatin sheets (ELCS) appear to connect some MN to the main nucleus (exemplified on Fig. 2A). ELCS are flat folds of the inner NE attached with ~30 nm heterochromatin fibrils. These folds project into the perinuclear cistern and into the cytoplasm forming nuclear pockets (NP), which then fuse with the nucleus again (for review see refs. 23 and 24; Fig. 2D). Therefore, some MN found near the main nucleus may be connected to it by thin ELCS, while some nuclear buds may in fact represent NPs of ELCS with cytoplasmic content. Like MN, ELCS have long been acknowledged as a cytological marker of aneuploidy and associated with poor prognosis in lymphoma and other tumors.23,24 Also like MN, ELCS appear as a result of aberrant mitosis and/or slippage caused by genotoxic treatment or spindle perturbation.22
Figure 2. The relationship between MN, NE, ELCS, DNA repair and chromatin autophagy in endopolyploid cells undergoing genotoxic stress. (A) A MN with normal chromatin structure is linked to the nucleus (NU) by ELCS. (B) In the nuclear pocket (NP) the dark organelles with convoluted membranes represent the residual body (RB), which usually results from autophagic lysosomal activity. The sequestration by a double membrane and the presence of a multivesicular body (MvB) nearby are also indicative of cytoplasmic autophagy within the NP; Nu, nucleus; Cy, cytoplasm. (C) Extrusion of a large membrane enclosed MN containing degraded chromatin via the nuclear pore. Note the sequestration of the cytoplasmic territory around it by a double membrane (arrows) and assembly of activated mitochondria nearby (asterisk). Bars: 1 μm. Figures are republished with new annotation: (A and B) from reference 22, with permission of Springer and (C) from reference 20, with permission of Elsevier. (D) A schematic showing a cross-section of a nucleus undergoing micronucleation and chromatin autophagy. Interphase chromosomes are joined to the NE by heterochromatin rows attached to the LBR of the inner nuclear membrane, which form the chromatin band of the ELCS. The left micronucleus (MN) is connected to the nucleus by ELCS and is not autophagic; it can be reunited with the nuclear DNA or alternatively form an ELCS nuclear pocket bridging to the nucleus at another site. The process may favor the search for homology for recombination repair of double-strand breaks in the foci (HRR). The cytoplasmic content of the nuclear pocket often undergoes autophagy (Au-NP) (designated by sequestration of the double membrane fusing with a lysosome). If DNA repair has failed (and DNA remains fragmented), a signal for budding of autophagic MN (Au-MN) may be obtained from unbound repair factors (illustrated as free rings OOO) for execution of selective autophagy (designated by sequestration of the double membrane fusing with a lysosome).
ELCS enclosing chromatin bound to LBR/Lamin B receptors24 can apparently transfer chromatin.23 Although at this point purely conjecture, we therefore propose that some MN which are connected with the main nucleus by ELCS, can be further translocated as an ELCS loop to the NE at a new site to facilitate HR (exemplified in Fig. 2A and B and depicted in the scheme in Fig. 2D). The translocation of the chromatin by ELCS may occur through nuclear rotation,25,26 and is apparently enhanced in lymphoma and HeLa cells after DNA damage (unpublished observations). When DNA fails to find homology and bind recombinase, it should signal for selective degradation and budding of the MN for autophagy (as exemplified on Fig. 2C and designated in Fig. 2D). As a result both non-autophagic and autophagic MN may collaborate in facilitating genome stabilization in endopolyploid tumor cells. This model would explain why most MN are not seen in the process of autophagic degradation—because they are instead engaged in DNA repair and chromatin resorting or are otherwise disconnected from the nucleus.
As noted by Rello-Varona et al.1 the MN autophagy caused by cell-cycle disrupters occurs on a background of ongoing cytoplasmic autophagy. In addition, activated mitochondria and cytoplasmic autophagy are frequently observed in the cytoplasmic pockets of ELCS which are internally digested in endopolyploid lymphoma cells after genotoxic treatment22 as shown in Figure 2B and designated in Figure 2D. Therefore, cytoplasmic autophagy may partner with chromatin autophagy in the maintenance of genome stability by at least two mechanisms: (1) by supporting and extending the viability of the cell as it undergoes protracted DNA repair; and (2) more directly, by providing the energy needed for nuclear rotation by microtubules at the site of the ELCS pockets.
Somewhat similar autophagic digestion of chromatin by nuclear envelope buds and formation of perinuclear autophagic vacuoles has previously been described in several disease envelopathies and corresponding in vitro models.27 It follows then that these phenomena are induced by genome instability on one hand, and nuclear envelope instability, on the other, highlighting the role of the chromatin-nuclear envelope relationship in maintaining genome integrity and order. Another phenomenon of nuclear autophagy related to the NE has been described in yeast; named piecemeal microautophagy of the nucleus (PMN).28 Here tiny pieces of the nucleus are invaginated in the apposed perinuclear vacuole with subsequent scission and digestion of the enclosed material; the terminal stage of the process requires the core macroautophagy genes.29 During this process, excess pre-ribosomal material from the nucleolus is mostly eliminated, alongside chromatin. Amplified rDNA may also be removed and if so, PMN would serve to contribute to the stabilization of the genome that we propose occurs in mammalian cells.
As detailed earlier, chromatin autophagy is related not only to MN. Selective degradation, autophagic digestion and expulsion of whole sub-nuclei also occurs in endopolyploid tumor cells8,20 (as shown on Fig. 1C and Fig. 1D). This activity was seen in several lymphoma and also HeLa cells after different treatments (irradiation, paclitaxel, nocodazole, etoposide) and coincides with de-polyploidization by a-cytotomic multi-polar mitoses. In contrast to autophagy of the MN during the polyploidization phase where DNA synthesis can be concurrent,20,22 whole sub-nuclei prepared for autophagy selectively halt DNA synthesis and become degraded and extruded.8,20 Subsequently, a proportion of the retained sub-nuclei not only maintain DNA synthesis but also accumulate the self-renewal factors POU5F1/Oct4 and Nanog, sequester their own cytoplasm and then dissipate as mitotic descendents.8,20,30 These features associated with the elimination of whole sub-nuclei, namely cessation of DNA synthesis, TUNEL-positivity, involvement of lysosomal enzymes, and active expulsion are somewhat in common with those described for the elimination of the vegetative macronucleus in unicellular Tetrahymena,31 perhaps indicating an evolutionary origin for this process in tumors. Presumably such autophagic processes also provide the nutrients and energy for the surviving secondary subcells thus favoring the ultimate escape from genotoxic damage. A similar nutritive role was suggested for the autophagic elimination of whole nuclei in syncytial filamentous fungi.32
In conclusion, the role of autophagy in the maintenance of genome stability is well established by genetic and other mechanistic studies,14,21,28 however, the understanding of the specific contribution made by chromatin autophagy is only just beginning. Here, we underline the significance of polyploidy, and reversible endopolyploidy in particular, for chromatin autophagy, highlight the regulatory role of DNA repair and regulation by the nuclear envelope and discuss the differing kinds of chromatin autophagy, by MN and release of whole sub-nuclei in this process. Exactly how all of these various facets are coordinated to regulate genome stability and ultimately tumor cell fate will clearly require further research.
Acknowledgments
The authors would like to thank Profs. V. Groma and V. Osse for discussion of EM images. Research was funded by the “Latvian National Research Programme 2010–2013 BIOMEDICINE” and European Social Fund within the project “Support for Doctoral Studies at University of Latvia.” T.R.J. was supported by a studentship from the BBSRC. Exchange visits between Riga and Southampton were supported by the Royal Society of London. The publishing costs associated with this article are provided by the ERDF project no. 2DP/2.1.1.2.0/10/ APIA/VIAA/004.
Glossary
Abbreviations:
- MN
micronucleus
- NE
nuclear envelope
- ELCS
envelope-limited chromatin sheets
- NP
nuclear pockets
- EM
electron microscopy
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/21610
References
- 1.Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Mariño G, et al. Autophagic removal of micronuclei. Cell Cycle. 2012;11:170–6. doi: 10.4161/cc.11.1.18564. [DOI] [PubMed] [Google Scholar]
- 2.Rajaraman R, Guernsey DL, Rajaraman MM, Rajaraman SR. Stem cells, senescence, neosis and self-renewal in cancer. Cancer Cell Int. 2006;6:25. doi: 10.1186/1475-2867-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–77. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zybina T, Zybina E. Cell cycle modification in trophoblast cell populations in the course of placenta formation. In: Kusic-Tisma J, ed. DNA replication and related cellular processes. Rijeka, InTech, 2011:227-58. [Google Scholar]
- 5.Puig PE, Guilly MN, Bouchot A, Droin N, Cathelin D, Bouyer F, et al. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int. 2008;32:1031–43. doi: 10.1016/j.cellbi.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Vitale I, Senovilla L, Jemaà M, Michaud M, Galluzzi L, Kepp O, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–84. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erenpreisa J, Ivanov A, Wheatley SP, Kosmacek EA, Ianzini F, Anisimov AP, et al. Endopolyploidy in irradiated p53-deficient tumour cell lines: persistence of cell division activity in giant cells expressing Aurora-B kinase. Cell Biol Int. 2008;32:1044–56. doi: 10.1016/j.cellbi.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erenpreisa J, Salmina K, Huna A, Kosmacek EA, Cragg MS, Ianzini F, et al. Polyploid tumour cells elicit paradiploid progeny through depolyploidizing divisions and regulated autophagic degradation. Cell Biol Int. 2011;35:687–95. doi: 10.1042/CBI20100762. [DOI] [PubMed] [Google Scholar]
- 9.Sundaram M, Guernsey DL, Rajaraman MM, Rajaraman R. Neosis: a novel type of cell division in cancer. Cancer Biol Ther. 2004;3:207–18. doi: 10.4161/cbt.3.2.663. [DOI] [PubMed] [Google Scholar]
- 10.Erenpreisa J, Cragg MS. Cancer: a matter of life cycle? Cell Biol Int. 2007;31:1507–10. doi: 10.1016/j.cellbi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Erenpreisa J, Cragg MS. MOS, aneuploidy and the ploidy cycle of cancer cells. Oncogene. 2010;29:5447–51. doi: 10.1038/onc.2010.310. [DOI] [PubMed] [Google Scholar]
- 12.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Mohanna MA, Al-Khalaf HH, Al-Yousef N, Aboussekhra A. The p16INK4a tumor suppressor controls p21WAF1 induction in response to ultraviolet light. Nucleic Acids Res. 2007;35:223–33. doi: 10.1093/nar/gkl1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haaf T, Raderschall E, Reddy G, Ward DC, Radding CM, Golub EI. Sequestration of mammalian Rad51-recombination protein into micronuclei. J Cell Biol. 1999;144:11–20. doi: 10.1083/jcb.144.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raderschall E, Bazarov A, Cao J, Lurz R, Smith A, Mann W, et al. Formation of higher-order nuclear Rad51 structures is functionally linked to p21 expression and protection from DNA damage-induced apoptosis. J Cell Sci. 2002;115:153–64. doi: 10.1242/jcs.115.1.153. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov A, Cragg MS, Erenpreisa J, Emzinsh D, Lukman H, Illidge TM. Endopolyploid cells produced after severe genotoxic damage have the potential to repair DNA double strand breaks. J Cell Sci. 2003;116:4095–106. doi: 10.1242/jcs.00740. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov A, Ivanova M, Erenpreisa J, Gloushen SV, Freivalds T, Cragg MS. Image analysis of DNA repair and apoptosis in tumor cells with differing sensitivity to DNA damage. IFMBE Proc. 2008;20:524–7. doi: 10.1007/978-3-540-69367-3_140. [DOI] [Google Scholar]
- 20.Erenpreisa JA, Cragg MS, Fringes B, Sharakhov I, Illidge TM. Release of mitotic descendants by giant cells from irradiated Burkitt’s lymphoma cell line. Cell Biol Int. 2000;24:635–48. doi: 10.1006/cbir.2000.0558. [DOI] [PubMed] [Google Scholar]
- 21.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–9. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erenpreisa J, Ivanov A, Cragg M, Selivanova G, Illidge T. Nuclear envelope-limited chromatin sheets are part of mitotic death. Histochem Cell Biol. 2002;117:243–55. doi: 10.1007/s00418-002-0382-6. [DOI] [PubMed] [Google Scholar]
- 23.Ghadially FN. Ultrastructural pathology of the cell and matrix. Boston, Butterworth-Heinemann, 1997. [Google Scholar]
- 24.Olins DE, Olins AL. Nuclear envelope-limited chromatin sheets (ELCS) and heterochromatin higher order structure. Chromosoma. 2009;118:537–48. doi: 10.1007/s00412-009-0219-3. [DOI] [PubMed] [Google Scholar]
- 25.Paddock SW, Albrecht-Buehler G. Rigidity of the nucleus during nuclear rotation in 3T3 cells. Exp Cell Res. 1988;175:409–13. doi: 10.1016/0014-4827(88)90205-4. [DOI] [PubMed] [Google Scholar]
- 26.Gerashchenko MV, Chernoivanenko IS, Moldaver MV, Minin AA. Dynein is a motor for nuclear rotation while vimentin IFs is a “brake”. Cell Biol Int. 2009;33:1057–64. doi: 10.1016/j.cellbi.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 27.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5:795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 28.Roberts P, Moshitch-Moshkovitz S, Kvam E, O’Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–41. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen E-L, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010;316:2099–112. doi: 10.1016/j.yexcr.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 31.Lu E, Wolfe J. Lysosomal enzymes in the macronucleus of Tetrahymena during its apoptosis-like degradation. Cell Death Differ. 2001;8:289–97. doi: 10.1038/sj.cdd.4400807. [DOI] [PubMed] [Google Scholar]
- 32.Shoji JY, Kikuma T, Arioka M, Kitamoto K. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS ONE. 2010;5:15650. doi: 10.1371/ journal.pone.0015650. [DOI] [PMC free article] [PubMed] [Google Scholar]