Abstract
Evidence for a regulatory role of the miR-34 family in senescence is growing. However, the exact role of miR-34 in aging in vivo remains unclear. Here, we report that a mir-34 loss-of-function mutation in Caenorhabditis elegans markedly delays the age-related physiological decline, extends lifespan, and increases resistance to heat and oxidative stress. We also found that RNAi against autophagy-related genes, atg4, bec-1, or atg9, significantly reversed the lifespan-extending effect of the mir-34 mutants. Furthermore, miR-34a inhibits Atg9A expression at the post-transcriptional level in vitro, and the miR-34a binding sequences in the 3'-UTR of Atg9A contributes to the modulation of Atg9A expression by miR-34a. Our results demonstrate that the C. elegans mir-34 mutation extends lifespan by enhancing autophagic flux in C. elegans, and that miR-34 represses autophagy by directly inhibiting the expression of the autophagy-related proteins Atg9 in mammalian cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s11357-011-9324-3) contains supplementary material, which is available to authorized users.
Keywords: C. elegans, Mir-34, Autophagy, Aging, Lifespan
Introduction
MicroRNAs (miRNAs) are endogenously encoded single-stranded RNAs of about 22 nucleotides in length that are highly conserved and regulate protein expression by interaction with the 3′ untranslated region (3′-UTR) of mRNA (Bartel 2009). Recent research highlights the role of miRNAs in multiple pathways and processes such as development (Boehm and Slack 2005), senescence (Hammond and Sharpless 2008), immunity (Xiao and Rajewsky 2009), cancer (Bussing et al. 2008), and other physiological contexts (Sethupathy and Collins 2008). A number of studies using microRNA microarray analysis both in vitro and in vivo have shown that miR-34 expression increases with age (Maes et al. 2009; Wang et al. 2009; Ibanez-Ventoso et al. 2006; He et al. 2007; Li et al. 2009). Overexpression of miR-34 in many cell lines leads to cell cycle arrest, increased expression of the senescence marker senescence-associated β-galactosidase (SA-β-Gal), and accelerated senescence (Zhao et al. 2010; Ito et al. 2010; He et al. 2007; Christoffersen et al. 2009; Kumamoto et al. 2008; Tazawa et al. 2007). However, the precise biological role of miR-34 in aging in vivo remains elusive.
Autophagy, a lysosome-mediated digestive process involved in protein and organelle degradation, plays a critical role in the regulation of aging and age-related degenerative diseases (Salminen and Kaarniranta 2009a), although the exact molecular mechanisms of this regulation are not known. Recent studies have revealed that distinct stress resistance and longevity signalling pathways, such as SIRT1, p53, NF-kB, and FoxO3 are also potent regulators of autophagic degradation (recently reviewed in (Salminen and Kaarniranta 2009a; Vellai 2009). SIRT1, a mammalian homolog of silent information regulator 2 (Sir2), not only regulates cellular metabolism, cellular survival, stress resistance, and senescence, but is an activator of autophagy (Salminen and Kaarniranta 2009b). In mammals, SIRT1 can interact with and directly deacetylate several components of the mammalian autophagic machinery, such as Atg5, Atg7, and Atg8 proteins, in an NAD-dependent manner (He and Klionsky 2009). There are several similarities in the phenotypes of SIRT1−/− mice and Atg5−/− mice, such as the accumulation of damaged organelles, disruption of energy homeostasis, and early perinatal mortality (Lee et al. 2008). The tumor suppressor protein p53 has been found to strongly influence aging. Recent studies have shown that p53, which modulates DNA damage responses, senescence, oncogene activation, and apoptosis, also plays a major role in the control of autophagy (Green and Kroemer 2009). Deletion, depletion, or inhibition of p53 induces autophagy in human, mouse, and nematode cells (Tasdemir et al. 2008). miR-34, a direct transcriptional target of p53 (Chang et al. 2007), represses SIRT1 expression by binding the 3'-UTR in the SIRT1 gene (Yamakuchi et al. 2008). Furthermore, BCL-2, another direct target gene of miR-34 (Wang et al. 2009), inhibits autophagy by interacting with Beclin 1 (Shimizu et al. 2004), which is the mammalian ortholog of the yeast Atg6/Vps30. These findings suggest that autophagy may be involved in the regulation of aging through miR-34.
The present study was conducted to identify the role of miR-34 in aging in vivo and to examine the mechanisms of autophagy in regulating aging through miR-34. We found that the mir-34 mutation delays aging in Caenorhabditis elegans and that autophagy plays an important role in the process. Furthermore, we found that miR-34 repressed autophagy by directly inhibiting the autophagy-related proteins Atg9 in mammalian cells.
Materials and methods
Animals
Ten healthy male Wistar rats (3 or 24 months old) were used. Each group contained five rats, which were maintained on a 12-h light/12-h dark cycle and fed a standard laboratory diet with free access to water. The procedures used and the care of animals were in accordance with the regulations of the ethics committee of the General Hospital of the People's Liberation Army.
Worms carrying the mir-34 loss-of-function mutant alleles gk437 and n4276 were obtained from the Caenorhabditis Genetics Center and were backcrossed five times to the wild-type N2 strain (gift from Mengqiu Dong). All worms were cultured using standard methods (Brenner 1974). The nematode-rearing temperature was kept at 20°C, unless noted otherwise.
Quantitative reverse-transcriptase polymerase chain reaction
TaqMan miRNA assays were performed to quantify the level of mature miR-34, as described previously (Zhu et al. 2009). Briefly, cDNA was synthesized using Taqman RT reagents (Applied Biosystems, USA), followed by TaqMan-based quantitative PCR using specific mirVanaTM quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) primer sets (Table S6). Primary transcript levels of Atg9A was determined with the SYBR Green Realtime PCR Master Mix (Toyobo, Japan). The qRT-PCR analyses were conducted using an Applied Biosystems ABI Prism 7300 Sequence Detection System, and qRT-PCR reactions were performed using the following parameters: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The relative RNA amounts were calculated using the ΔΔCt method and normalized to U6 snRNA control for rats miR-34a, eft-2 control for C. elegans miR-34, or GAPDH for Hela and HEK293 cells mRNAs. Sequence-specific primers were designed using Primer Premier 5.0 software (PREMIER Biosoft International, USA) for the indicated genes (Table S6).
C. elegans physiological movement studies
Movement assays were performed as described (Huang et al. 2004) with modifications. The physiological studies were begun on day zero by placing L4 hermaphrodites on a Petri dish. Pharyngeal pumping and body movement were assessed by observing animals for 30–60 s using a dissecting microscope. Fast pharyngeal pumping is considered >149 pumps/min, and the time from day 0 to the last day of fast pharyngeal pumping was named the fast pharyngeal pumping span. Animals were classified as “fast moving” if they exhibited well-coordinated sinusoidal body movements and “slow moving” if they displayed less-coordinated or sluggish movements. The time from day zero to the last day of fast movement was referred to as the “fast moving span.”
Intermittent fasting
Intermittent fasting (IF) was performed as described previously (Honjoh et al. 2009), with modifications. Approximately 100 synchronized L4 hermaphrodite worms were raised on NGM plates with live OP50. At day 2 of adulthood, worms were divided into two groups: AL and IF. The AL group of worms was fed ultraviolet-killed OP50 ad libitum throughout their lifespan and transferred to new plates every other day. Worms of the IF group were exposed to intermittent fasting, placed on plates with ultraviolet-killed OP50 every other day, alternating with days without food.
Heat and oxidative stress resistance
Resistance to heat and oxidative stress resistance was assayed as described previously (Honjoh et al. 2009). Briefly, synchronous worms at day 10 of adulthood were raised on NGM plates. Heat stress was applied by incubating worms on NGM plates at 35°C. The surviving worms were scored every 5 h. Hydrogen peroxide (Sigma-Aldrich, USA; cat. no. 316989) was used to induce oxidative stress. Animals were exposed to 2.5 mM hydrogen peroxide on NGM plates at 20°C, and survival was scored every hour.
Plasmids
cDNA fragments of autophagy genes, namely, atg4.1, bec-1, or atg9, were amplified from wild-type N2 worms by PCR. Each fragment was cloned into the pL4440 feeding vector. The 1,091-bp fragment of the Atg9A 3'-UTR containing the miR-34a targeting sequence (ACTGCC) was cloned into the psiCHECKTM-2 dual luciferase reporter plasmid (Promega, USA; cat. no. C8021) to produce psiCHECK-WT-Atg9A using PCR. 5′-ACTGCC-3′ (285–291 bp) in 3′-UTR of Atg9A, the core binding sequence of miR-34a, was substitutely mutated to GACTAT, and subcloned into psiCHECKTM-2, named psiCHECK-MT-Atg9A. To produce the site-directed mutagenesis of the miR34a targeting site, the QuikChange® Site-Directed Mutagenesis Kit (Agilent Technologies, USA; cat. no. 200519) were used. Sequence-specific primers were designed using Primer Premier 5.0 software for the indicated genes (Table S6). The sequences of all constructs were verified.
Lifespan analysis
Lifespan assays were performed at 20°C as described previously (Tavernarakis et al. 2008), with modifications. Synchronous animal populations were generated by hypochlorite treatment of gravid adults to obtain tightly synchronized embryos, which were allowed to develop into adulthood on standard, OP50-seeded, NGM plates. For RNAi lifespan experiments, worms were placed on NGM plates containing 1 mM IPTG and seeded with HT115(DE3) Escherichia coli bacteria transformed with either the pL4440 vector or the target gene RNAi construct. Progeny were grown at 20°C through the L4 larval stage and then transferred to fresh RNAi plates following the L4 moult, groups of 10 worms per plate for a total of 100–150 individuals were assayed per experiment. The day when animals were at the L4 larval stage was considered as t = 0. Animals were transferred to fresh plates every 2 to 4 days thereafter and were examined every day for touch-provoked movement and pharyngeal pumping. The population of animals was followed until death. Worms that died due to internally hatched eggs, an extruded gonad, or desiccation due to crawling on the edge of the plates, were censored and incorporated into the data set. Each survival assay was repeated at least three times, and figures represent typical assays.
Cell culture and transfection method
Human cervical carcinoma Hela cells and human embryonic kidney (HEK) 293 cells were purchased from American Type Culture Collection. All cells were cultured in Dulbecco's modified Eagle medium containing 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. All cell culture products were purchased from Invitrogen Gibco Cell Culture. Cell lines were cultured in a 5% CO2-humidified incubator at 37°C. Medium was refreshed every 3 days during cultivation and every 24 h during experiments. Cells (∼50% confluence) were transfected with miR-34a mimics (Dharmaconcat. no. C-300551-07), miR-34a inhibitors (Dharmacon cat. no. IH-300551-08), miRNA control (Dharmacon cat. no. CN-001000-01), or siRNA control (Dharmacon cat. no. IN-001005-01) using jetPRIME™ (Polyplus-transfection, USA) for 48 h and then treated for 2 h with the lysosomal inhibitors pepstatinA (Sigma-Aldrich cat. no. P5318) and E64d (Sigma-Aldrich cat. no. E8640) (both 10 mg/ml; Pep.A/E64d) or DMSO as control, followed by protein and RNA analysis. The sequence of miR-34a mimics is for hsa-miR-34a reported in miR database (www.mirbase.org). Such mimics can supplement existing endogenous miR-34a levels. Chemical modification technology ensures that only the antisense strand (mature microRNA) enters the microRNA pathway, and thus minimizes interferon response and improves target binding specificity and efficiency. To determine the transfection efficiency of miR-34a mimics or inhibitors, Cy3 labeled negative control miRNA and Cy3 labeled negative control siRNA were transfected into Hela cells and HEK293 cells, respectively. The cells were observed under a fluorescence microscope with a laser scanning confocal imaging system, and the total concentration of viable and nonviable cells was determined and the transfection efficiency are calculated (Rosner et al. 2010; Lingor et al. 2005).
Western blot analysis
Immunoblot analyses were performed as described previously (Gamerdinger et al. 2009). Briefly, 40 μg total protein was subjected to SDS-PAGE gel electrophoresis. Proteins were detected by chemiluminescence using the ECL reagent (Pierce Rockford, USA). Films were scanned with a Molecular Dynamics densitometer, and densitometry was performed using Alphaimager 2200. β-actin was used as a normalization control. Antibodies were purchased from Santa Cruz Biotechnology (anti-SIRT1 rabbit pAb, cat. no. sc-74465; anti-β-actin rabbit pAb, cat. no. sc-130657) and Sigma-Aldrich Inc. (anti-LC3B rabbit pAb, cat. no. L7543; anti-human Atg9A rabbit pAb, cat. no. SAB2100173).
Dual luciferase reporter assay
For the reporter assays, cells were cultured to approximately 80% confluence in a 24-well plate and then cotransfected with either the dual luciferase reporter plasmid (psiCHECK-WT-Atg9A or psiCHECK-WT-Atg9A) and miR-34a mimics (Hela cells) or miR-34a inhibitors (HEK293 cells) for 48 h. Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega cat. no. E1910), and Renilla luciferase activity was normalized to firefly luciferase activity.
Statistical analysis
Survival curves were created using the Kaplan–Meier product-limit method. The log–rank (Mantel–Cox) test was used to evaluate differences between survivals and to determine P values. We used the Prism software package (GraphPad Software Inc., USA) to conduct the statistical analysis and to determine lifespan values.
Measurement data are expressed as mean ± SEM, unless otherwise stated. Statistical analyses were performed using the SPSS 15.0 software package (SPSS, Inc., USA). Statistical differences were evaluated with analysis of variance. Pearson's correlation coefficients were calculated to investigate the association among the indicated parameters. A P value < 0.05 denoted a statistically significant difference.
Results
MiR-34 expression increases with age in vivo
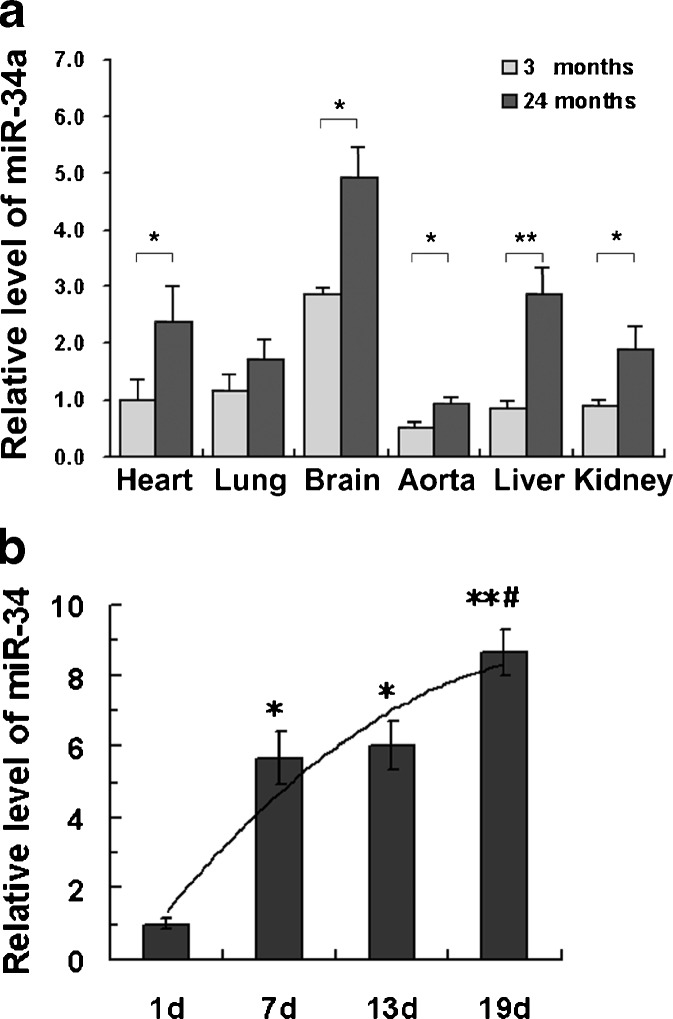
Recent studies have suggested that members of the miR-34 family have a role in senescence (He et al. 2007). To explore miR-34 expression during the adult lifespan, we measured miR-34a in tissues from old (24-month-old) and young (3-month-old) adult Wistar rats by quantitative TaqMan assay. The results showed that miR-34a expression in heart, lung, brain, liver, thoracic aorta, and kidney was 2.36-, 1.49-, 1.71-, 1.85-, 3.45-, and 2.14-fold higher, respectively, in old than in young rats (Fig. 1a). Next, we tested miR-34 expression in C. elegans. A marked increase in the expression of miR-34 was seen in wild-type N2 worms during aging (Fig. 1b), and there was a significant positive correlation between miR-34 expression and lifespan (r = 0.856, P < 0.0001).
Fig. 1.
miR-34 expression increases with age in vivo. a Relative expression of miR-34a in heart, lung, brain, liver, thoracic aorta, and kidney from 3 and 24-month-old rats. Each estimate represents the mean of 3–5 quantitative RT-PCR reactions on independent RNA samples derived from five rats. Values are expressed as mean ± SEM. *P < 0.05, **P < 0.01. b The relative expression of miR-34 in wild-type N2 worms. Results presented are the average values obtained from three replicates. *P < 0.05, **P < 0.01 vs. 1 day adult worms. #P < 0.05 vs. 7-day adult worms
The mir-34 loss-of-function mutation extends C. elegans lifespan
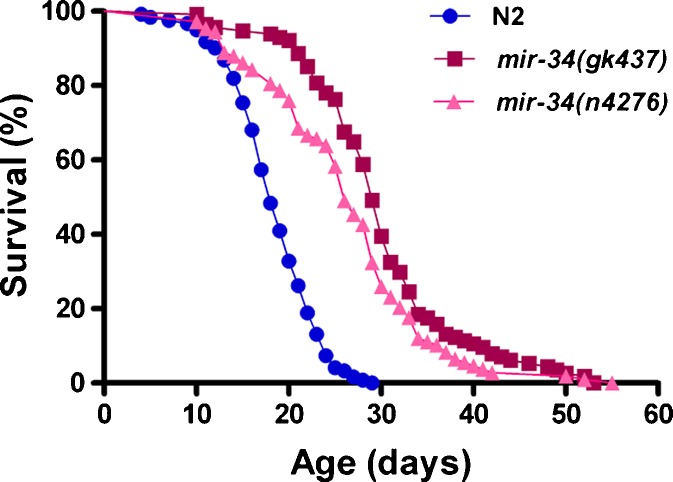
Several studies have indicated that declines in the rate of pharyngeal pumping or the animal movement rate are positively correlated with declines in survival probability and can be used to predict lifespan in C. elegans (Huang et al. 2004). To address the role of miR-34 in the longevity response, we analyzed the two mir-34 loss-of-function mutant alleles (gk437 and n4276). Compared to wild-type N2 worms, the fast pharyngeal pumping span of mir-34(gk437) and mir-34(n4276) mutants was significantly extended by 47% and 38%, and the fast moving span was increased by 78% and 41%, respectively (Table 1). We also measured the lifespan of mir-34 mutants and compared it to that of wild-type N2 animals under standard laboratory conditions. We found a significant increase in the mean lifespan of mir-34 mutants, 29.8 ± 0.8 days in mir-34(gk437) and 26.2 ± 0.8 days in mir-34(n4276) mutants, compared to 18.4 ± 0.4 days in wild-type animals. We also observed an increase in the maximum lifespan (defined as the top 90% percentile) of mir-34 mutants, 45 days in mir-34(gk437) and 40.5 days in mir-34(n4276) mutants, compared to 25 days in the wild-type animals (Fig. 2; Table S1). These results show that eliminating miR-34 extends the lifespan of C. elegans.
Table 1.
Age-related declines in physiological processes of wild-type N2 and mir-34 mutant worms
| Strain | Fast pharyngeal pumping span (days, mean ± SEM)a | Fast body movement span (days, mean ± SEM)a | Nb |
|---|---|---|---|
| N2 | 9.7 ± 0.3 | 9.2 ± 0.2 | 71 |
| mir-34(gk437) | 14.3 ± 0.4** | 16.4 ± 0.3** | 79 |
| mir-34(n4276) | 13.4 ± 0.3* | 13.0 ± 0.3** | 71 |
*P < 0.01, **P < 0.001
aEach mutant was compared to wild-type N2 with the nonparametric log–rank test that compares strain distributions
bThe number of hermaphrodites analyzed
Fig. 2.
The mir-34 loss-of-function extends C. elegans lifespan. Survival curves of wild-type N2 and mir-34 mutant worms. Similar results were obtained in three identical experiments (the details are shown in Table S1)
Autophagy is required for the mir-34 loss-of-function induced longevity
Caloric restriction by dietary restriction (DR) or intermittent fasting (IF) is the most effective and reproducible intervention to extend lifespan in divergent species, ranging from single-celled organisms to mammals (Mair and Dillin 2008; Finch and Ruvkun 2001). To investigate the mechanism of lifespan regulation by miR-34 in C. elegans, we compared the lifespan of mir-34 mutant worms raised under an ad libitum (AL) diet and an under intermittent fasting (IF) diet. Compared with the AL regimen, IF significantly extended the lifespan of wild-type N2 worms by 37% and failed to extend the lifespan of mir-34 mutants (gk437 and n4276) (Fig. 3a–c; Table S2). Moreover, we found that mir-34 mutant alleles displayed a marked increase in resistance to heat and oxidative stress (Fig. 3d, e; Table S3). Recent studies have revealed that autophagy plays an important role in the mediation of longevity by dietary restriction (Salminen and Kaarniranta 2009a) and resistance to stress (Wu et al. 2009) in C. elegans. To further evaluate whether autophagy is also required for lifespan extension in worms with mir-34 mutant alleles, we tested the effects of RNAi knockdown of C. elegans orthologs of the yeast autophagy genes, i.e., atg4.1 (yeast ATG4), bec-1 (yeast ATG6), and atg9 (yeast ATG9), on miR-34 lifespan extension. Synchronized animals that have reached adulthood were fed bacteria expressing the target autophagy gene dsRNA to avoid detrimental effects of decreased autophagy during development (Melendez and Levine 2009). We found that RNAi knockdown of atg4.1, bec-1, or atg9 did not influence the lifespan of wild-type N2 worms, consistent with previous findings (Toth et al. 2008; Hansen et al. 2008; Jia and Levine 2007), but significantly abolished the lifespan extension phenotype of mir-34 mutants (Fig. 3f–h; Table S4). Thus, our results suggest that miR-34 regulates the C. elegans lifespan, possibly through the regulation of the activity of autophagy-related genes.
Fig. 3.
Autophagy mediates mir-34 mutation-induced longevity in C. elegans. (a–c) Effect of ad libitum (AL) and intermittent fasting (IF) on lifespan in N2 (a), mir-34(gk437) (b), and mir-34(n4276) (c) worms. Similar results were obtained in three identical experiments (the details are shown in Table S2). d, e Survival curves of wild-type (N2), mir-34(gk437), and mir-34(n4276) mutant animal populations subjected to heat (d) and oxidative stress (e). Similar results were obtained in three identical experiments (the details are shown in Table S3). f–h Effect of N2 (f), mir-34(gk437) (g), and mir-34(n4276) (h) on lifespan in worms subjected to atg4.1, bec-1, or atg9 RNAi during adulthood. Similar results were obtained in three identical experiments (the details are shown in Table S4)
miR-34 inhibits autophagy in vitro
To elucidate the exact roles of miR-34 in autophagy, we explored whether miR-34 is involved in autophagic processes in vitro. miR-34a expression was low in Hela cells but high in HEK293 cells (Yamakuchi et al. 2008). Hence, miR-34a overexpression was established by transfection with miR-34a mimics in Hela cells, and miR-34a downregulation was established by transfecting miR-34a inhibitors in HEK293 cells. To determine the transfection efficiency of miR-34a mimics or inhibitors, Cy3-labeled negative control miRNA were transfected into Hela cells, and Cy3-labeled negative control siRNA were transfected into HEK293 cells. The transfection efficiency of miR-34a mimics or inhibitors was 98.4% and 94.7%, respectively (Fig. S1). To assess the effectiveness of miR-34a mimics or inhibitors, we detected SIRT1 expression by immunoblot analysis, which had been validated as a direct miR-34a target (Yamakuchi et al. 2008). Transfection of miR-34a mimics (100 nM) into Hela cells or miR-34a inhibitors (100 nM) into HEK293 cells significantly influenced SIRT1 protein expression (Fig. 4a), suggesting that miR-34a function can be modulated by transfection of miR-34a mimics or miR-34a inhibitors. Autophagic flux can be measured by monitoring LC3 conversion from LC3-I to LC3-II, a well-established marker for autophagy, in the presence of lysosomal inhibitors (Kuma et al. 2007). After overexpression of miR-34a was achieved in Hela cells, the conversion to LC3-II upon lysosomal inhibition by pepstatin A and E64d (Pep.A/ E64d) was significantly decreased (Fig. 4b, c). In contrast, after the inhibition of miR-34a expression in HEK293 cells, we detected a significant increase in the accumulation of LC3- II, upon lysosomal inhibition by Pep.A/E64d (Fig. 4d, e).
Fig. 4.
miR-34 inhibits autophagy in vitro. a Hela and HEK293 cells transfected with miR-34a mimics and inhibitors, respectively. SIRT1 protein expression level was detected by immunoblot 24 h later. b Hela cells were transfected with miR-34a mimics or miRNA control for 48 h and then treated for 2 h with the lysosomal inhibitors pepstatinA and E64d (both 10 mg/ml; Pep.A/E64d) or DMSO as a control, followed by immunoblot analysis of the LC3 proteins. c Diagram shows the autophagic flux of Hela cells with different miR-34a levels as described in b. Autophagic flux was determined by the strength of LC3-II accumulation in a 2-h treatment period with Pep.A/E64d. Therefore, normalized LC3-II levels in the absence of inhibitors were subtracted from corresponding levels obtained in the presence of Pep.A/E64d. Values are expressed as mean ± SEM. *P < 0.01 vs. mock, n = 3. d HEK293 cells were transfected with miR-34a inhibitors or siRNA control for 48 h, and the cells were treated as in b, followed by immunoblot analysis of the LC3 proteins. e Diagram shows the autophagic flux of HEK293 cells with different miR-34a levels as described in d. Values are expressed as mean ± SEM. *P < 0.01 vs. mock, n = 3
Atg9A is a direct target of miR-34a
Since mammalian Atg9A is a protein required in the formation of the autophagosome and have been shown to be required at different steps in the autophagic machinery, we then characterized the relationship between miR-34a function and Atg9A expression. We found that miR-34a overexpression in Hela cells by miR-34a mimics transfection decreased Atg9A protein levels (Fig. 5a). In contrast, miR-34a inhibition in HEK293 cells with miR-34a inhibitors increased Atg9A protein expression. Interestingly, miR-34a showed no effect on Atg9A mRNA expression (Fig. 5b), suggesting that miR34a regulates Atg9A expression at the post-transcriptional level.
Fig. 5.
Atg9A is a direct target of miR-34a. a Hela and HEK293 cells transfected with miR-34a mimics and inhibitors, respectively. Atg9A proteins expression level was detected by immunoblot 48 h later. b Quantitative RT-PCR assays for Atg9A. Expression values were normalized to the GAPDH housekeeping gene, and expression in untreated cells was set to 1. Values shown represent means ± SEM of three independent experiments. c Reporter plasmids in which the luciferase coding sequence was fused to the 3′-UTR of Atg9A were transfected into Hela cells in conjunction with miR-34a mimics or HEK293 cells in conjunction with miR-34a inhibitors. Renilla luciferase activity was normalized to firefly luciferase activity. Results shown are the mean ± SEM of triplicate determinations from one of three identical experiments. *P < 0.01
To further elucidate whether miR-34a regulates Atg9A expression via binding to its 3'-UTR sequences, we predicted consensus sequences for miR-34a in the Atg9A 3'-UTR (Table S5). In order to validate these predicted consensus sequences and to determine whether these miR-34a binding sequences directly contributed to the regulation of Atg9A expression, we utilized a dual luciferase reporter gene assay system and finally confirmed that miR-34a regulated Atg9A expression by binding to its 3'-UTR sequences. As shown in Fig. 5c, cotransfection of Hela cells with miR-34a mimics resulted in a 39–48% reduction in the activity of the reporter gene vector containing the wild-type miR-34a targeting sequences (psiCHECK-WT-Atg9A), and cotransfection of HEK293 cells with miR-34a inhibitors resulted in a 51–73% increase in the activity of the reporter gene vector. Notably, miR-34a had no effect on the reporter activity of the mutated vectors (psiCHECK-MT-Atg9A). These results demonstrate that the miR-34a binding sequences in the Atg9A 3'-UTR are required for the miR-34a-mediated inhibition of Atg9A expression.
Discussion
We have demonstrated that miR-34 expression is significantly increased in rat tissues during aging. We have shown that a mir-34 deletion mutation extends the lifespan of C. elegans, and that autophagy genes are required for the mir-34 mutants lifespan extension phenotype. Furthermore, we found that miR-34 inhibits autophagic flux in vitro and can affect the protein expression levels of Atg9. Thus, our results suggest that miR-34 is not only a biomarker of aging, but also an effector of aging, which might be mediated through the inhibition of autophagy genes, at least partly, through Atg9.
A number of studies have investigated the miR-34 family, particularly miR-34a, whose expression increases with age in mouse and model cells in microRNA microarray analyses (Lafferty-Whyte et al. 2009). Recent studies have shown that miR-34 overexpression accelerates the aging process in aging human diploid cells (IMR90) (He et al. 2007) and endothelial progenitor cells (EPCs) (Zhao et al. 2010). Similarly, transient introduction of miR-34a to two human colon cancer cell lines, HCT 116 and RKO, causes the complete suppression of cell proliferation and induces senescence-like phenotypes (Tazawa et al. 2007). These findings add to a growing body of evidence suggesting that miR-34 plays a causal role in aging in vivo. First, as shown in this work, miR-34 expression was tightly linked to aging. miR-34 expression was upregulated during aging in several rat tissues (Fig. 1), and there was a significant positive correlation between miR-34 expression and lifespan in C. elegans (Fig. 2). Furthermore, an mir-34 loss-of-function deletion mutation not only delayed the decline of the age-related physiological changes, such as fast pharyngeal pumping span or fast moving span, but also extended the lifespan by 42–62% in C. elegans.
He et al. (2007) found that miR-34 up-regulation in IMR-90 cells led to substantial growth inhibition by suppressing CDK4, CCNE2, and MET expression, increasing SA-β-Gal expression, and accelerating cellular senescence. In this study, we found that the mir-34 mutation prevents the lifespan extension that results from an intermittent fasting (IF) regimen and markedly increased resistance to heat and oxidative stress. Autophagy is required for dietary restriction-mediated lifespan extention and resistance to stress in C. elegans. Melendez et al. (2003) was the first to report that RNAi knockdown of the autophagy gene bec-1 shortened the lifespan of daf-2 mutants. Hars et al. (2007) also found that two other autophagy genes, atg-7 and atg-12, were also essential for lifespan extension in daf-2 mutants. Furthermore, recent studies have shown that autophagy is required for the longevity phenotype of the C. elegans dietary restriction mutants (eat-2(ad1113)) (Jia and Levine 2007; Hansen et al. 2008; Toth et al. 2008) and p53 ortholog cep-1 mutant animals (Tavernarakis et al. 2008). These studies clearly emphasize autophagy as an important factor for the regulation of longevity in C. elegans. We found that RNAi knockdown of autophagy-related genes had no effect on the lifespan of wild-type N2 worms but significantly decreased the lifespan extension of mir-34 mutant animals (Fig. 3). Thus, autophagy may be involved in the regulation of the lifespan through miR-34.
Interestingly, the latest research has shown that some of the miR-34 target genes, such as SIRT1 and BCL-2, are also important regulators of autophagic degradation (Zhang et al. 2009; Zalckvar et al. 2009; Swerdlow et al. 2008; Lee et al. 2008). All of these studies support the hypothesis that miR-34 may have a role in regulating autophagy. Hela and HEK293 cells were transfected with miR-34a mimics and inhibitors in vitro, respectively, and the results showed that miR-34a reduced autophagic flux. Autophagy involves the bulk degradation of cytoplasmic components by the lysosomal/vacuolar system. More than 31 autophagy-related (Atg) proteins are involved in the process (He and Klionsky 2009). Atg9 is the only transmembrane protein required for autophagy among the current 31 characterized Atg proteins and is present both at the pre-autophagosomal structure (PAS) and multiple peripheral structures, such as mitochondria and the Golgi complex, both of which may provide membranes to the forming autophagosomes (Reggiori et al. 2005). Atg9 also promotes lipid transport from the membrane origins to the PAS and helps assemble the intact phagophore membrane (Reggiori et al. 2004). We have shown that miR-34a regulates the Atg9A expression in cells culture experiments. We showed that expression of the miR-34a mimics decreased the protein expression levels of Atg9A and reduced the activity of a reporter gene plasmid containing the miR-34a consensus sequences (Fig. 5). In contrast, the influence of miR-34a on reporter activity was abolished in the absence of the miR-34a consensus sequences. Furthermore, we have shown that miR-34a modulates the protein expression of Atg9A but has no effect on the mRNA expression of Atg9A. Our findings indicate that miR-34a may regulate the expression of Atg9A, thereby modulating the autophagic activity.
In summary, our results have demonstrated that the depletion of miR-34 activity extends the lifespan by enhancement of autophagic flux in C. elegans and that miR-34 repression of autophagy may occur through the direct inhibition of the expression of autophagy-related proteins Atg9 in mammalian cells. It is intriguing that miR-34 might be considered a partial biomarker of aging. Future studies will have to show whether miR-34 may contribute to the regulation of aging and aging-associated diseases.
Electronic supplementary materials
(PDF 108 kb)
(PDF 22 kb)
(PDF 23 kb)
(PDF 23 kb)
(PDF 25 kb)
(PDF 26 kb)
(PDF 29 kb)
Acknowledgments
We thank Dr. Mengqiu Dong (National Institute of Biological Science, Beijing,China) for providing helpful discussions and materials, Dr. Nektarios Tavernarakis (Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology-Hellas, Crete, Greece) for providing the pL4440-bec-1 plasmids, and Dr. Christian Mϋnz (Institute of Experimental Immunology, Department of Pathology, University Hospital of Zurich, Zurich, Switzerland) for the donation of the pEGFP-HLC3 plasmids.
This work was supported by a grant (2007CB507400) from the National Basic Research Program of China to XMC.
Footnotes
Jurong Yang and Dapeng Chen contributed equally to this work.
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310(5756):1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14(9):400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH (2009) p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. doi:10.1038/cdd.2009.109 [DOI] [PubMed]
- Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28(7):889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Sharpless NE. HMGA2, microRNAs, and stem cell aging. Cell. 2008;135(6):1013–1016. doi: 10.1016/j.cell.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C (2008) A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4 (2):e24. doi:10.1371/journal.pgen.0040024 [DOI] [PMC free article] [PubMed]
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. Autophagy regulates ageing in C. elegans. Autophagy. 2007;3(2):93–95. doi: 10.4161/auto.3636. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457(7230):726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101(21):8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5(3):235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398(4):735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3(6):597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- Kuma A, Matsui M, Mizushima N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007;3(4):323–328. doi: 10.4161/auto.4012. [DOI] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68(9):3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafferty-Whyte K, Cairney CJ, Jamieson NB, Oien KA, Keith WN. Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta. 2009;1792(4):341–352. doi: 10.1016/j.bbadis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Bates DJ, An J, Terry DA, Wang E (2009) Up-regulation of key microRNAs, and inverse down-regulation of their predicted oxidative phosphorylation target genes, during aging in mouse brain. Neurobiol Aging. doi:10.1016/j.neurobiolaging.2009.04.020 [DOI] [PubMed]
- Lingor P, Koeberle P, Kugler S, Bahr M. Down-regulation of apoptosis mediators by RNAi inhibits axotomy-induced retinal ganglion cell death in vivo. Brain. 2005;128(Pt 3):550–558. doi: 10.1093/brain/awh382. [DOI] [PubMed] [Google Scholar]
- Maes OC, Sarojini H, Wang E (2009) Stepwise up-regulation of MicroRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol. doi:10.1002/jcp.21834 [DOI] [PubMed]
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–754. doi: 10.1146/annurev.biochem.77.061206.171059. [DOI] [PubMed] [Google Scholar]
- Melendez A, Levine B (2009) Autophagy in C. elegans. WormBook:1–26. doi:10.1895/wormbook.1.147.1 [DOI] [PubMed]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;Science 301(5638):1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/S1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1(2):101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M, Siegel N, Fuchs C, Slabina N, Dolznig H, Hengstschlager M. Efficient siRNA-mediated prolonged gene silencing in human amniotic fluid stem cells. Nat Protoc. 2010;5(6):1081–1095. doi: 10.1038/nprot.2010.74. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15(5):217–224. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. SIRT1: regulation of longevity via autophagy. Cell Signal. 2009;21(9):1356–1360. doi: 10.1016/j.cellsig.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24(10):489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6(12):1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Swerdlow S, McColl K, Rong Y, Lam M, Gupta A, Distelhorst CW. Apoptosis inhibition by Bcl-2 gives way to autophagy in glucocorticoid-treated lymphocytes. Autophagy. 2008;4(5):612–620. doi: 10.4161/auto.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10(6):676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavernarakis N, Pasparaki A, Tasdemir E, Maiuri MC, Kroemer G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008;4(7):870–873. doi: 10.4161/auto.6730. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104(39):15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16(1):94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C (2009) miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits bcl2 translation. Brain Res Bull. doi:10.1016/j.brainresbull.2009.08.006 [DOI] [PubMed]
- Wu JJ, Quijano C, Chen E, Liu H, Cao L, Fergusson MM, Rovira II, Gutkind S, Daniels MP, Komatsu M, Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging Albany NY. 2009;1(4):425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105(36):13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Eisenstein M, Kimchi A. Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy. 2009;5(5):720–722. doi: 10.4161/auto.5.5.8625. [DOI] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Wu JC, Lin F, Han R, Han F, Fukunaga K, Qin ZH (2009) Down-regulation of Bcl-2 enhances autophagy activation and cell death induced by mitochondrial dysfunction in rat striatum. J Neurosci Res. doi:10.1002/jnr.22152 [DOI] [PubMed]
- Zhao T, Li J, Chen AF. MicroRNA-34a induces endothelial progenitor cell senescence and impedes its angiogenesis via suppressing silent information regulator 1. Am J Physiol Endocrinol Metab. 2010;299(1):E110–E116. doi: 10.1152/ajpendo.00192.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5(6):816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 108 kb)
(PDF 22 kb)
(PDF 23 kb)
(PDF 23 kb)
(PDF 25 kb)
(PDF 26 kb)
(PDF 29 kb)