Triglcyerides must be degraded by an enzymatic process called lipolysis before they can cross cellular membranes. This wide-ranging and authoritative review by Young and Zechner surveys the current state of affairs regarding intravascular and cellular lipolysis and focuses on the recently discovered key proteins involved, including CPIHBPI, adipose triglyceride lipase, and GCI-58. Extensive discussion is also given over to the role of lipolysis in human disease.
Keywords: adipose triglyceride lipase, CGI-58, GPIHBP1, hormone-sensitive lipase, lipolysis, lipoprotein lipase, monoglyceride lipase
Abstract
All organisms use fatty acids (FAs) for energy substrates and as precursors for membrane and signaling lipids. The most efficient way to transport and store FAs is in the form of triglycerides (TGs); however, TGs are not capable of traversing biological membranes and therefore need to be cleaved by TG hydrolases (“lipases”) before moving in or out of cells. This biochemical process is generally called “lipolysis.” Intravascular lipolysis degrades lipoprotein-associated TGs to FAs for their subsequent uptake by parenchymal cells, whereas intracellular lipolysis generates FAs and glycerol for their release (in the case of white adipose tissue) or use by cells (in the case of other tissues). Although the importance of lipolysis has been recognized for decades, many of the key proteins involved in lipolysis have been uncovered only recently. Important new developments include the discovery of glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1), the molecule that moves lipoprotein lipase from the interstitial spaces to the capillary lumen, and the discovery of adipose triglyceride lipase (ATGL) and comparative gene identification-58 (CGI-58) as crucial molecules in the hydrolysis of TGs within cells. This review summarizes current views of lipolysis and highlights the relevance of this process to human disease.
Fatty acids (FAs) are essential molecules with multiple biological functions; they are integral components of membrane lipids, efficient energy substrates, and potent second messengers. The limited water solubility of FAs and their relatively low critical micellar concentration cause them to form micelles in an aqueous environment. Micelle formation and the avid association of FAs with cellular membranes can adversely affect membrane and cell function. To prevent deleterious effects of high extracellular or intracellular FA concentrations, long chain FAs are “detoxified” by esterification to glycerol, a trivalent alcohol, forming relatively inert triglycerides (TGs). A large fraction of FAs in TG-rich lipoproteins (TRLs; chylomicrons and very-low-density lipoproteins [VLDL]) and in cytoplasmic lipid droplets (LDs) are found in the form of TGs. In response to metabolic demand, TGs can be hydrolyzed to FAs and glycerol by a process called lipolysis, which involves the enzymatic activity of TG hydrolases (lipases).
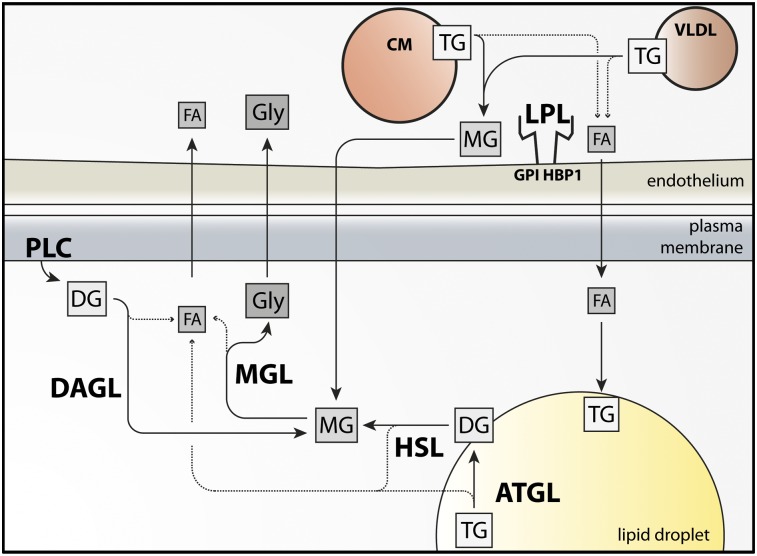
The importance of lipolysis for virtually all organisms became clear with the realization that TGs cannot move across cell membranes without first being degraded by lipases. More than a century ago, it was observed that pancreatic juice contains a “fat-splitting” activity and that this activity is essential for the uptake of dietary fat by the intestine (Bernard 1856; Balser 1882; Whitehead 1909). Later, in the 1950s, it was shown that plasma TGs must be hydrolyzed intravascularly (Korn 1955a,b) to facilitate uptake of FAs and monoglycerides (MGs) by parenchymal cells (Fig. 1). Also, intracellular hydrolysis of the TGs stored in white adipose tissue (WAT) facilitates the release of FAs and glycerol from cells, a process that provides metabolic intermediates to tissues during fasting (Fig. 1; Vaughan et al. 1964).
Figure 1.
Schematic of intravascular lipolysis and intracellular lipolysis. Intravascular lipolysis: LPL hydrolyzes TGs in VLDLs and chylomicrons (CM) on the luminal face of capillaries, principally in adipose tissue and striated muscle. LPL is bound to the surface of capillaries by a glycosylphosphatidylinositol-anchored glycoprotein, GPIHBP1. LPL products, FAs and MGs, traverse the endothelium and are taken up by the underlying parenchymal cells (adipocytes and myocytes). MGs are further catabolized by monoacylglycerol lipase (MGL), generating FAs and glycerol. LPL-generated FAs are re-esterified to TG or oxidized. Glycerol is resecreted and transported to the liver. Intracellular lipolysis: Hormonal stimulation of lipolysis activates ATGL and HSL, which hydrolyze TGs and diglycerides (DGs), respectively, in cellular LDs. MGs are hydrolyzed by MGL. Lipolytic FAs enter oxidative or biosynthetic pathways in most tissues. WAT releases FAs into the bloodstream, where they bind to albumin and eventually enter peripheral tissues. Hydrolysis of glycerolphospholipids by phospholipase C (PLC) and diacylglycerol lipase (DAGL) also generates MGs.
Intravascular and cellular lipolysis has been studied for many decades, but the last few years have witnessed important new discoveries regarding lipolytic enzymes, enzyme mechanisms, and enzyme transport. This review summarizes these new results and discusses their importance for human disease (Table 1).
Table 1.
Diseases and phenotypes related to genes involved in lipolysis

Lipolysis within blood vessels
Vertebrates have a complex and finely tuned system for delivering lipid nutrients to parenchymal cells. The central element of this system is the intravascular hydrolysis of TGs carried within the plasma lipoproteins (Havel 1977, 1980). Intestinal enterocytes package dietary lipids into chylomicrons, which enter the lymphatics and then quickly reach the bloodstream. Chylomicrons transport TGs to adipose tissue for storage and to vital tissues (e.g., the heart and skeletal muscle) for use as fuel. The liver secretes VLDL, which serves to redistribute TG to adipose tissue, heart, and muscle. Chylomicrons and VLDL are spherical particles containing a core of neutral lipids, mainly TGs; the surface of these particles contains polar lipids (e.g., phospholipids and free cholesterol), apolipoprotein B (either apo-B48 or apo-B100), and several exchangeable apolipoproteins (e.g., apo-CI, apo-CII, apo-CIII, apo-E, apo-AI, apo-AII, apo-AIV, and apo-AV).
The large size of chylomicron (∼100–1200 nm in diameter) and VLDL (30–80 nm) particles means that they have little capacity to move across capillaries; hence, the movement of lipids to parenchymal cells depends on hydrolysis of the TGs within these particles, a process that is carried out by lipoprotein lipase (LPL) along the luminal surface of capillary endothelial cells (Fig. 1; Brunzell and Deeb 2001). Intravascular lipolysis removes ∼90% of the TGs from the particles (Bergman et al. 1971), generating remnant lipoproteins that are largely taken up and removed by hepatocytes. In the liver, the fenestrated capillaries allow remnant lipoproteins to interact directly with hepatocytes.
The recognition that plasma TGs undergo intravascular hydrolysis began with the discovery that fat particles in the blood are cleared more rapidly after an injection of heparin and that the clearing of opalescence continues after the plasma is placed in a test tube (Hahn 1943). Later, Korn (1955a,b) showed that the clearing factor was LPL, which converts the TGs in plasma to less turbid products (FAs and glycerol). In addition, given that heparin is not capable by itself of reducing the turbidity of lipemic plasma, Korn (1955a,b) concluded that heparin must release LPL from tissues into the plasma. Within a few years, Havel and Gordon (1960) characterized a family in which three siblings had creamy plasma and showed that their post-heparin plasma was deficient in TG hydrolase activity. The three siblings were homozygous for LPL deficiency, the first example of an inherited disorder of plasma lipid metabolism (Havel 2010).
The importance of LPL in the processing of TRLs led to considerable excitement among physiologists, biochemists, and physicians. Even before the advent of molecular biology, several laboratories showed that intravascular lipolysis by LPL depends on a cofactor (later identified as apo-CII) that is carried on the surface of TRLs (Kinnunen et al. 1977; Bengtsson and Olivecrona 1979). The importance of LPL in lipolysis was underscored by Bensadoun and coworkers (Kompiang et al. 1976), who purified the enzyme, generated antibodies, and showed that those antibodies, when injected into animals, prevented the lipolytic processing of TRLs.
LPL
The cDNA for LPL was cloned by several groups (Enerbäck et al. 1987; Kirchgessner et al. 1987; Wion et al. 1987). Full-length human LPL is 474 amino acids in length, 448 amino acids after removal of the signal peptide. The N-terminal portion of the mature protein (the first 312 amino acids) encodes the catalytic domain, while the C-terminal domain is required for binding TG substrates (Wong et al. 1994). Substrate binding requires the presence of three tryptophans in LPL's C terminus (Lookene et al. 1997a). When those residues are mutated, hydrolysis of triolein is markedly reduced, but tributyrin hydrolysis is minimally affected, indicating that soluble TGs with very short acyl chains can be processed without the lipid-binding sequences. The same tryptophans are also essential for LPL's ability to bind to TRLs and TG emulsion particles (Lookene et al. 1997a). Consistent with these findings, an LPL-specific monoclonal antibody, 5D2, which binds to the tryptophan-rich domain (Chang et al. 1998), blocks triolein but not tributyrin hydrolysis (Chang et al. 1998).
The fact that LPL could be released into the plasma by heparin (Korn 1955a,b) implied that LPL might contain positively charged heparin-binding sequences, and indeed, this was the case (Kirchgessner et al. 1987). A strong heparin-binding domain is located within the C terminus of the protein (residues 403–407) (Ma et al. 1994a; Sendak et al. 1998; Lookene et al. 2000), just a few residues downstream from the lipid-binding sequences; another is located between residues 292 and 304 (Hata et al. 1993).
LPL is secreted and enzymatically active as a homodimer (Iverius and Ostlund-Lindqvist 1976; Garfinkel et al. 1983). The ability of cells to produce and secrete LPL homodimers depends on a membrane protein of the endoplasmic reticulum (ER), lipase maturation factor 1 (LMF1) (Peterfy et al. 2007). In the absence of LMF1, LPL cannot form homodimers and is degraded intracellularly (Doolittle and Peterfy 2010). LPL homodimers are formed by two monomers oriented in a head-to-tail fashion. When an expression vector containing two LPL cDNAs, arranged head-to-tail in a single ORF, is expressed in cultured cells, a catalytically active enzyme is produced (Wong et al. 1997); the “single-molecule LPL” has the same size as native LPL homodimers, as judged by density gradient ultracentrifugation. The LPL monomers probably act cooperatively to hydrolyze TGs, with the lipid-binding domain of one monomer providing substrates to the catalytic domain of the partner monomer. In support of this idea, transfection of cultured cells with two catalytically inactive LPL constructs—one with a mutation in the C-terminal lipid-binding motif and a second with a mutation in the N-terminal catalytic domain—results in the production of catalytically active LPL (Kobayashi et al. 2002).
LPL contains N-linked glycans at Asn 43 and Asn 359. Asn 43 is crucial for LPL secretion, while Asn 359 is less so (Semenkovich et al. 1990; Ben-Zeev et al. 1994; Busca et al. 1995). N-linked glycans may be crucial for the stability of LPL homodimers, but a variety of other sequences are required for stable homodimers (Mailly et al. 1997; Keiper et al. 2001). The sequences that participate directly in interactions between the two monomers are not known.
The rationale for LPL being a homodimer is not understood, but one possible explanation is that this structure facilitates regulation of LPL activity levels. The Olivecrona laboratory (Sukonina et al. 2006) showed that angiopoietin-related protein 4 (ANGPTL4), a physiological inhibitor of LPL, reduces LPL activity by converting catalytically active homodimers to inactive monomers. In adipose tissue, ANGPTL4 plays a key role in changing LPL activity levels in response to metabolic cues. The homodimeric structure could also be important in other ways. It is possible, for example, that the two monomers serve distinct roles (i.e., that LPL is a functional heterodimer). In the case of another enzyme homodimer, cyclooxygenase, the binding of substrate by one monomer alters the structure and function of the partner monomer such that one monomer plays a catalytic role, while the partner monomer serves an allosteric role (Yuan et al. 2006, 2009). Whether LPL is a “functional heterodimer,” in terms of either binding substrates or partner proteins, is unknown and needs investigation.
Without apo-CII, LPL is able to bind to lipoproteins, but catalytic activity is minimal (Olivecrona and Beisiegel 1997). Sequences within the C-terminal portion of apo-CII are essential for its interaction with LPL (Shen et al. 2010). Sequences within the N-terminal portion of LPL have been implicated in apo-CII binding (Davis et al. 1992), but C-terminal sequences also could play a role (Hill et al. 1998). The exact nature of apo-CII–LPL interactions and how apo-CII binding promotes enzymatic activity are incompletely understood.
Binding of LPL by endothelial cells
The fact that LPL is required for the processing of TRLs has been known for decades. Until recently, however, several elements of LPL physiology remained mysteries. One was LPL's binding site on the luminal face of capillaries. For years, dogma held that LPL was bound to capillary endothelial cells by electrostatic interactions between its heparin-binding motifs and negatively charged heparan sulfate proteoglycans (HSPGs) (Cryer 1989; Merkel et al. 2002). This view made some sense, given that LPL is readily released into the bloodstream by heparin, a low-molecular-weight sulfated glycosaminoglycan. Also, LPL binds to HSPGs in vitro (Klinger et al. 1985; Lookene et al. 1997b), and LPL binding to cultured cells can be reduced by reducing the sulfation of HSPGs or removing them enzymatically (Shimada et al. 1981; Hoogewerf et al. 1991). On the other hand, the “HSPG model” was never satisfying, mainly because it lacked biochemical specificity. HSPGs are produced by many cell types, and it was never clear why LPL would end up binding to HSPGs in the capillary lumen rather than to the HSPGs surrounding the cells where LPL is made (myocytes and adipocytes). Another nagging mystery was how LPL reached the capillary lumen. LPL is produced by myocytes and adipocytes and secreted into the interstitial spaces, yet its site of action is within the capillary lumen. One study suggested that the VLDL receptor might play a role in moving LPL to the capillary lumen (Obunike et al. 2001), but again, this explanation was not satisfying because VLDL receptor deficiency in mice has minimal effects on plasma lipid metabolism (Frykman et al. 1995).
Insights into both of these mysteries—LPL's binding site in capillaries and how LPL manages to reach the capillary lumen—came with the discovery of glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1), a GPI-anchored member of the lymphocyte antigen 6 (Ly6) family of proteins (Ioka et al. 2003; Beigneux et al. 2007). GPIHBP1's relevance to lipolysis was obvious with the development of Gpihbp1 knockout mice (Gpihbp1−/−), which manifest severe hypertriglyceridemia, even when fed a low-fat chow diet (Beigneux et al. 2007). Additional studies showed that GPIHBP1, when expressed on cultured cells, binds LPL avidly and that the bound LPL can be released with heparin (Beigneux et al. 2007). Also, GPIHPB1 is expressed at high levels in capillaries of heart and brown adipose tissue (BAT), two tissues that produce large amounts of LPL (Beigneux et al. 2007; Davies et al. 2010). GPIHBP1 is absent from the capillaries of the brain, an organ that depends largely on glucose for fuel (Beigneux et al. 2007). It is also absent from the endothelial cells of venules, arterioles, and larger blood vessels (Davies et al. 2010).
GPIHBP1 contains two principal domains: an N-terminal acidic domain (highly enriched in aspartic and glutamic acids) and a cysteine-rich Ly6 domain (∼80 amino acids in length) (Ioka et al. 2003; Beigneux et al. 2007). The Ly6 domain contains 10 cysteines, all arranged in a characteristic spacing pattern and all disulfide-linked, resulting in a three-fingered structural motif (Fletcher et al. 1994).
Mutation of the acidic domain impairs LPL binding (Gin et al. 2008). The Ly6 domain is equally important; mutation of any of the 10 cysteines markedly reduces GPIHPB1's ability to bind LPL (Beigneux et al. 2009c). Subsequent alanine-scanning mutagenesis studies uncovered 12 residues—most of which are in the “second finger” of the three-fingered motif—that are important for LPL binding (Beigneux et al. 2011). Whether those residues participate directly in LPL binding or are simply required for folding of the three-fingered structure is not clear. The Ly6 domain in mouse GPIHBP1 contains one N-linked glycosylation site, which is important for the trafficking of GPIHBP1 from the ER to the cell surface (Beigneux et al. 2008).
The discovery that GPIHBP1 domains are important for LPL binding prompted Beigneux et al. (2009c) to propose that LPL might contain two distinct GPIHBP1-binding domains. It makes sense that GPIHBP1's acidic domain might interact with one of LPL's heparin-binding domains, and in support of this idea, mutation of LPL's main C-terminal heparin-binding domain (residues 403–407) inhibits binding of LPL to GPIHBP1 (Gin et al. 2008). The portion of the LPL molecule that interacts with GPIHBP1's Ly6 domain has not been identified with certainty but likely includes sequences located further downstream (amino acids ∼416–435). Mutating that region abolishes binding of human LPL to GPIHBP1 and does so without significant effects on heparin binding (Voss et al. 2011). Recent studies showed that the C terminus of LPL (residues 298–448), free of the N-terminal catalytic domain, binds avidly and specifically to GPIHBP1 (Gin et al. 2012).
Site-directed mutagenesis studies and immunochemical studies have yielded important insights regarding sequences relevant to LPL–GPIHBP1 interactions (Beigneux et al. 2009c; Beigneux et al. 2011; Voss et al. 2011; Gin et al. 2012); however, the residues that participate directly in these interactions are not known. Determining structures for GPIHBP1, LPL, and GPIHBP1–LPL complexes remains an important objective for the field.
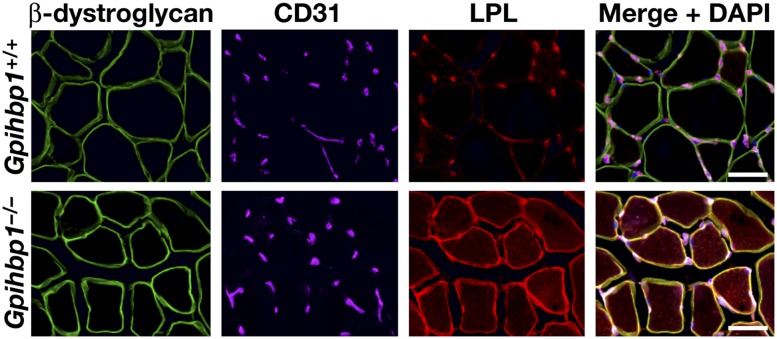
The fact that GPIHBP1 binds LPL and is located in capillaries suggested that it might be a “platform for lipolysis” on the luminal side of capillary endothelial cells (Beigneux et al. 2007). Subsequent studies revealed that GPIHBP1 has a second and more intriguing function—picking up LPL from the interstitial spaces and shuttling it across capillaries to the capillary lumen (Davies et al. 2010). In Gpihbp1−/− mice, LPL never reaches the luminal side of capillaries and remains mislocalized in the interstitial spaces surrounding parenchymal cells (Fig. 2) (Davies et al. 2010). GPIHBP1's transport function has been documented in both cultured cells and live mice. GPIHBP1-expressing endothelial cells, when cultured on a porous grid, transport both LPL and GPIHBP1-specific monoclonal antibodies from the basolateral to the apical side of cells (Davies et al. 2010). Also, when GPIHBP1-specific monoclonal antibodies are injected into the interstitium of skeletal muscle or BAT, they quickly associate with endothelial cells and move to the capillary lumen (Davies et al. 2010, 2012). GPIHBP1 and LPL movement across endothelial cells can be inhibited with dynasore and genistein, consistent with a vesicular transport process. By immunogold electron microscopy, GPIHBP1 and LPL can be found in invaginations in the plasma membrane of endothelial cells and in transcytotic vesicles (Davies et al. 2012) .
Figure 2.
Confocal immunofluorescence microscopy showing that LPL is tightly bound within the interstitial spaces in skeletal muscle of Gpihbp1−/− mice. Images show β-dystroglycan (a marker of skeletal myocytes) (green), CD31 (a marker of endothelial cells) (purple), and LPL (red) in muscle from a wild-type (Gpihbp1+/+) mouse and a Gpihbp1−/− mouse. The vast majority of the LPL in the skeletal muscle of the wild-type mouse is bound to capillaries. In the Gpihbp1−/− mouse, the LPL is mislocalized to the interstitial spaces surrounding myocytes. (Reprinted from Davies et al. 2010, © 2010, with permission from Elsevier.)
The “mislocalized LPL” in the muscle and adipose tissue of GPIHBP1-deficient mice remains tightly bound in the interstitial spaces, presumably bound to HSPGs near the surface of myocytes and adipocytes (Davies et al. 2010). Even when GPIHBP1 is expressed normally, it seems likely that the LPL secreted by myocytes and adipocytes would bind first to HSPGs surrounding parenchymal cells. It is tempting to speculate that GPIHBP1's acidic domain may act as a lasso, weakening LPL–HSPG interactions and then promoting the binding of LPL to endothelial cells—with high affinity and lock-and-key specificity—by dual interactions with GPIHBP1's acidic and Ly6 domains. In this way, newly secreted LPL would be driven from the interstitial spaces to the surface of endothelial cells. Also, LPL is a homodimer and therefore could have two binding sites for GPIHBP1. The stoichiometry of LPL–GPIHBP1 binding has not been determined, but the binding of LPL by two GPIHBP1 molecules would presumably greatly increase the affinity of LPL binding to endothelial cells.
In addition to LPL, GPIHBP1 binds apo-AV (Beigneux et al. 2007; Gin et al. 2007, 2008, 2011), another heparin-binding protein with a key role in plasma TG metabolism (Pennacchio et al. 2001, 2002). This interaction is mediated by interactions between apo-AV's heparin-binding motif and GPIHBP1’s acidic domain (the binding does not appear to involve the Ly6 domain) (Gin et al. 2011). GPIHBP1 does not bind apo-B or apo-E, both of which contain strong heparin-binding domains (Gin et al. 2011).
The discovery of GPIHBP1 solved some lingering mysteries in lipolysis, but others remain. The molecular basis for LPL–GPIHBP1 interactions, how LPL is activated by apo-CII, and the mechanism for the inhibition of LPL activity by ANGPTL4 need to be elucidated. Also, no one understands mechanisms for the margination of TRLs along the surface of capillaries so that lipolysis can proceed. The prevailing view has been that TRLs bind to HSPGs on the endothelial cells (Cryer 1989; Merkel et al. 2002), but this idea needs testing. A second possibility is that GPIHBP1 (or GPIHBP1-bound LPL) is responsible for TRL margination. No one knows whether TRLs would stop along capillaries in the absence of GPIHBP1. Another mystery is how the products of lipolysis move across capillaries to parenchymal cells.
For adipose tissue and striated muscle, high levels of LPL and GPIHBP1 expression are closely matched (Beigneux et al. 2007), but there are two tissues where the levels of GPIHBP1 and LPL expression are quite different. First, GPIHBP1 transcripts are present in high levels in the lung, but there are only trace levels of LPL transcripts in the lung (Olafsen et al. 2010). The explanation for this discrepancy is not understood with certainty, but several lines of evidence have suggested that the GPIHBP1 in lung capillaries serves to scavenge LPL produced by other tissues (Olafsen et al. 2010). Thus, GPIHBP1 could serve to “sop up” any LPL that escapes from the interstitial spaces, preventing it from reaching the systemic circulation. Second, certain regions of the brain (for example, the hippocampus) produce substantial amounts of LPL transcripts (Goldberg et al. 1989; Ben-Zeev et al. 1990; Vilaro et al. 1990; Bessesen et al. 1993), but GPIHBP1 transcripts in the brain are extremely low (Beigneux et al. 2007). The production of LPL by neurons has significant effects on adiposity and body weight (Wang et al. 2011a), but the mechanisms are not clear. Given the absence of GPIHBP1 in brain capillaries, it seems possible that LPL in the brain functions in the interstitial spaces surrounding neurons and glia rather than having a role in intravascular lipolysis. More studies are required.
GPIHBP1 is expressed at a low level in the liver but is still easily detectable on endothelial cells by immunohistochemistry, where it presumably serves to capture LPL. LPL transcripts in the liver are low but easily detectable, and LPL transcript levels increase significantly on a high-cholesterol diet (Zhang et al. 2001). Under certain circumstances—for example, fibroblast growth factor 21 (FGF21) overexpression or the induction of torpor—a close relative of LPL, pancreatic lipase, is induced in the liver and presumably plays a role in providing free FAs to the cells of the liver (Inagaki et al. 2008). Pancreatic lipase expression has also been observed in adipose tissue in the setting of FGF1 deficiency (Jonker et al. 2012). These observations are intriguing, but a link to GPIHBP1 seems unlikely because, unlike LPL, pancreatic lipase lacked the ability to bind to GPIHBP1 in either cell-free or cell-based binding assays (Jonker et al. 2012).
One final point about GPIHBP1 and LPL expression patterns deserves discussion: LPL is expressed in all vertebrates, but GPIHBP1 is found only in mammals (including the egg-laying platypus) (Beigneux et al. 2007, 2009a). GPIHBP1 presumably originated from another Ly6 gene by a gene duplication event, and there has been speculation that GPIHBP1's acidic domain came from another gene (Holmes and Cox 2012). The absence of GPIHBP1 in other vertebrates (e.g., birds and fish) poses intriguing questions (Young et al. 2011). Do the endothelial cells of those species have a distinct protein for shuttling LPL across capillaries? Is it possible that the LPL in those species functions within the interstitial spaces? Do these species transport TRLs outside of capillaries into the interstitial space where LPL exists rather than transporting LPL to the capillary lumen where TRLs exist? Answers to these questions are not known. In any case, it seems possible that the appearance of GPIHBP1 in mammals might somehow be related to a hallmark of mammals—nursing the young (Young et al. 2011). LPL is expressed at high levels in lactating mammary glands, and it seems possible that GPIHBP1 and intravascular lipolysis evolved to facilitate the movement of lipid nutrients from plasma TRLs to the mammary epithelium.
Hepatic lipase (HL) and endothelial lipase (EL)
LPL is not the only lipase with a role in intravascular lipolysis. Two other enzymes of the same family, HL and EL, participate in the remodeling of plasma lipoproteins. Like LPL, HL and EL are secreted as homodimers (Hill et al. 1997; Griffon et al. 2009), require LMF1 for secretion, and bind avidly to heparin (Peterfy et al. 2007; Ben-Zeev et al. 2010). In vivo, both are presumed to bind HSPGs and can be released from their binding sites with a bolus of heparin (Krauss et al. 1973; Fuki et al. 2003; Badellino et al. 2006). Neither HL nor EL binds to GPIHBP1 (Gin et al. 2011).
Human HL is ∼40% identical to LPL at the amino acid level (Komaromy and Schotz 1987; Datta et al. 1988; Martin et al. 1988). Unlike LPL, it is not sensitive to inhibition by high concentrations of sodium chloride and is not activated by apo-CII. HL is expressed highly in hepatocytes and is located mainly at the surface of those cells and surrounding endothelial cells (Breedveld et al. 1997; Sanan et al. 1997). HL exhibits low TG hydrolase activity against chylomicron substrates and instead hydrolyzes TGs in chylomicron remnants, intermediate density lipoproteins (IDL), and TG-rich high-density lipoproteins (HDLs) (Hegele et al. 1993; Connelly 1999; Santamarina-Fojo et al. 2004). Inhibition of HL with specific antibodies interferes with the conversion of IDL to LDL and leads to an accumulation of remnant particles (Goldberg et al. 1982). HL variants associated with reduced enzymatic activity lead to higher HDL cholesterol levels (Cohen et al. 1994, 1999; Guerra et al. 1997; Vega et al. 1998). The basis for HL's lipoprotein substrate specificities has been investigated with LPL–HL chimeras and likely involves sequences in both the N-terminal and C-terminal portions of the protein (Davis et al. 1992; Dugi et al. 1995).
EL is made primarily by endothelial cells and is 44% identical to LPL and 41% identical to HL at the amino acid level (Jaye et al. 1999; Cohen 2003). EL is primarily a phospholipase A1, hydrolyzing phospholipids in HDL particles, which reduces HDL size and accelerates their removal from the plasma (Duong et al. 2003; Ishida et al. 2003; Jin et al. 2003a). Overexpression of EL in transgenic mice lowers HDL cholesterol levels, while knocking out the gene for EL has the opposite effects (Ishida et al. 2003; Jin et al. 2003a). The physiologic significance of EL-mediated changes in HDL levels is unclear, particularly since the altered HDL levels do not appear to affect the efficiency of reverse cholesterol transport in vivo (RJ Brown et al. 2010).
The specificities of HL and EL for different lipids and lipoproteins (Jaye et al. 1999; Duong et al. 2003; Griffon et al. 2006) and the consequences of HL and EL deficiencies for plasma lipoprotein levels have been studied in depth (Ishida et al. 2003; Jin et al. 2003a). Nevertheless, the physiologic purpose of both enzymes is less well defined than for LPL. One can confidently state that LPL plays a central role in processing dietary lipids so that they can move to parenchymal cells—a clear and undeniably important function that has been conserved through evolution. HL and EL affect the concentration of lipids and lipoproteins in the plasma, but there is no comparable clarity of physiologic purpose. It would appear that HL and EL fine-tune a complex system of lipoprotein processing and thereby affect the delivery of lipid nutrients to specific cell types. The consequences of the “fine tuning” of nutrient delivery are not necessarily trivial, particularly since genome-wide association studies have uncovered a link between the gene for HL and macular degeneration (Neale et al. 2010; Cipriani et al. 2012). EL is regulated by cytokines and could influence both the inflammatory pathway and changes in lipoprotein levels and properties during inflammation (Jin et al. 2003b; Yasuda et al. 2010). Interestingly, HL and EL protein sequences have changed much more rapidly in vertebrate evolution than LPL sequences (Holmes et al. 2011a,b,c).
LPL, HL, and EL have functions independent of catalysis. All interact with members of the LDL receptor family and HSPGs (Mulder et al. 1993; Merkel et al. 1998; Fuki et al. 2003; Dichek et al. 2004; Yasuda et al. 2007). In the case of LPL and HL, this “bridging” function is thought to promote the uptake and clearance of remnant lipoproteins.
Intracellular lipolysis
Lipolysis of TGs in cytoplasmic LDs (Fig. 1) fulfills two important functions. In WAT, lipolysis generates FAs and glycerol that are released into the blood for transport to other tissues; for example, cardiac muscle, skeletal muscle, liver, or BAT. This release of FAs and glycerol during fasting is unique to WAT; other tissues do not release the products of TG hydrolysis and instead use them in a cell-autonomous manner for lipid synthesis or oxidation. The main enzymes for TG catabolism are the same in adipose and nonadipose tissues.
Adipose TG lipase (ATGL)
The initial step of TG hydrolysis is catalyzed by ATGL (Zechner et al. 2009). Surprisingly, the enzyme was only uncovered recently, in 2004 (Jenkins et al. 2004; Villena et al. 2004; Zimmermann et al. 2004). Previously, hormone-sensitive lipase (HSL) was considered the only neutral TG hydrolase in adipose and nonadipose tissues. Consequently, it came as a surprise when the targeted deletion of the Hsl gene in mice did not lead to TG accumulation and obesity (Osuga et al. 2000). Instead, HSL-deficient mice were lean and exhibited TG hydrolysis rates in WAT that were reduced by only 50% compared with wild-type WAT, arguing for the existence of at least one other neutral TG hydrolase. ATGL fulfilled the requirements for the “missing lipase.” ATGL cleaves TGs with high efficiency and specificity (Zimmermann et al. 2004; Eichmann et al. 2012). The preferred substrates for ATGL are saturated or unsaturated medium and long and very long chain esters in TGs.
Human ATGL is 506 amino acids in length. The N-terminal region of ATGL contains a patatin domain (186 amino acids) named after a related motif in patatin, the most abundant soluble protein in the potato tuber (Rydel et al. 2003). The human and the mouse genomes code for nine and eight patatin domain-containing proteins, respectively. In 2006, the gene family was named patatin-like phospholipase domain containing A 1–9 (PNPLA1–9), with ATGL being PNPLA2 (Wilson et al. 2006). ATGL and other patatin domain-containing proteins are found in all eukaryotes, including fungi, plants, and animals. Patatin-like proteins are also common in bacteria and may affect disease pathogenesis (Banerji and Flieger 2004). PNPLA1–9 act as TG hydrolases, phospholipases, retinyl esterases, and acyltransferases (Kienesberger et al. 2009b; Rajakumari and Daum 2010; Kumari et al. 2012). In mammals, ATGL (PNPLA2) is most closely related to PNPLA3 (Baulande et al. 2001). PNPLA3 exhibits TG hydrolase and acyltransferase activity (He et al. 2010; Huang et al. 2011; Kumari et al. 2012; Li et al. 2012) and has recently attracted major interest because a single amino acid sequence polymorphism (I148M) is strongly associated with hepatic steatosis. Individuals homozygous for the Met-148 allele exhibit increased susceptibility to alcoholic and nonalcoholic fatty liver disease (Romeo et al. 2008).
The structure of ATGL has not yet been determined, but sequence homologies with patatin domain proteins that have already been crystallized (Pat17 and cPLA2) suggest that the active site of ATGL is assembled around a catalytic dyad involving Ser 47 and Asp 166 (Rydel et al. 2003). Replacement of either of those residues with alanine leads to loss of enzymatic activity (Schweiger et al. 2008; Duncan et al. 2010). The nucleophilic Ser 47 is located within a GXSXG consensus sequence commonly found in serine hydrolases. Structural modeling revealed that the active site is located within the patatin domain (amino acids 10–185). The patatin domain itself is embedded in a 250-amino-acid α–β–α sandwich structure in the N-terminal half of the protein. The C-terminal half of the protein consists of α-helical and loop regions with a presumed hydrophobic lipid-binding domain between amino acids 315 and 360. The functional importance of this region for LD binding was proven by the finding that ATGL lacking this region is catalytically active against artificial substrate emulsions but is unable to bind to LDs within cells (Schweiger et al. 2008; Duncan et al. 2010).
ATGL activity is regulated by multiple mechanisms on the transcriptional and post-transcriptional level (for review, see Lass et al. 2011). The most important post-translational mechanisms for ATGL regulation include its delivery to LDs by vesicular transport (Beller et al. 2008; Guo et al. 2008; Soni et al. 2009; Ellong et al. 2011) and its activation by comparative gene identification-58 (CGI-58, also named α/β hydrolase domain-containing 5, ABHD5) (Lass et al. 2006). CGI-58 was originally found by comparison between the human and the Caernohabditis elegans proteome. Mutations in the CGI-58 gene are causative for a rare human disease known as neutral lipid storage disease (NLSD) with ichthyosis (NLSDI) (discussed further below). Human CGI-58 is a 349-amino-acid protein that belongs to the large family of α/β hydrolase domain-containing proteins (Oberer et al. 2011). The α/β hydrolase domain of CGI-58 contains a pseudocatalytic triad consisting of Asn 155, His 329, and Asp 303. Asn 155 is situated within a GXNXG consensus sequence and apparently replaces the nucleophilic serine normally found in catalytically active serine hydrolases. Given the absence of that serine, it is not surprising that CGI-58 exhibits no measurable hydrolase activity against a variety of lipid esters.
The activation of TG hydrolases by protein cofactors is not unusual. As noted earlier, LPL requires apo-CII for efficient hydrolysis of TGs within TRLs, and pancreatic lipase requires colipase for the hydrolysis of dietary lipids. It is widely assumed that these cofactors change the properties of the lipid–water interphase; however, a detailed molecular mechanism for coactivation is lacking for any lipase, and it is quite possible that mechanisms are substantially different for different lipases. For ATGL activation, it is conceivable that CGI-58 affects substrate presentation, changes the conformation of ATGL, or is involved in product removal. Mutagenesis studies have demonstrated that both LD binding and ATGL binding are required for efficient ATGL activation by CGI-58 (Gruber et al. 2010). A tryptophan-rich stretch within the first 20 amino acids of CGI-58 mediates its binding to LDs. A naturally occurring splice variant of CGI-58 lacking this region is unable to activate ATGL (Yang et al. 2010a).
Elegant studies by Granneman et al. (2007, 2009) demonstrated that the activation step of ATGL is hormone-dependent. In nonstimulated cells, CGI-58 is unavailable for ATGL activation because it binds to perilipin-1. β-Adrenergic stimulation leads to perilipin-1 phosphorylation by protein kinase-A (PKA) and dissociation of CGI-58, allowing CGI-58 to bind to ATGL and activate its catalytic activity. Mutations in perilipin-1 that prevent CGI-58 binding lead to unrestrained lipolysis and partial lipodystrophy in humans (Gandotra et al. 2011). Importantly, CGI-58 also broadens the substrate specificity of ATGL (Eichmann et al. 2012). Generally, ATGL is specific for TG substrates and exhibits only weak diglyceride (DG) lipase, phospholipase, and transacylase activity (in which two DG molecules generate one TG and one MG molecule). In the hydrolysis of TGs, ATGL preferably cleaves fatty acyl esters at the sn-2 position of the glycerol backbone, resulting in the production of sn-1,3 DGs. This is unusual for TG lipases, which typically hydrolyze the ester bonds in the sn-1 or sn-3 positions. The specificity of ATGL for sn-2 position esters may indicate an evolutionary relationship to phospholipase A2 enzymes of the PNPLA family. Upon hormonal stimulation and enzyme activation by CGI-58, the substrate specificity extends to the sn-1 position of TGs (Eichmann et al. 2012). Thus, fully activated ATGL generates both sn-1,3 DGs and sn-2,3 DGs but no measurable amounts of sn-1,2 DGs.
In addition to its ability to activate ATGL, CGI-58 acts as an acyl-CoA lysophosphatidic acid acyltransferase (LPAAT) (Ghosh et al. 2008; Montero-Moran et al. 2009). Compared with classical LPAAT enzymes, the specific activity of CGI-58 is low, and the structural basis for the enzymatic activity remains obscure.
Recently, Liu and colleagues (Yang et al. 2010b) identified a peptide inhibitor of ATGL, which is encoded by the G0/G1 switch gene 2 and called G0S2. Both human and murine G0S2 contain 103 amino acids. G0S2 was first discovered >20 years ago in cultured mononuclear cells during the cell cycle transition from G0 to G1 (Russell and Forsdyke 1991). The protein has been assigned diverse biological activities, including roles in cell cycle, cell proliferation, apoptosis, inflammation, and carcinogenesis (Heckmann et al. 2013). The best-studied function of G0S2, however, relates to its role in lipid metabolism (Yang et al. 2010b; Schweiger et al. 2012). G0S2 is expressed highly in WAT, BAT, and liver. Only very low levels of the protein can be detected in skeletal and cardiac muscle. In in vitro studies, G0S2 binds to the patatin domain of ATGL and suppresses ATGL hydrolytic activity independently of CGI-58. Consistent with its ability to inhibit ATGL, overexpression of G0S2 in cultured cells leads to an accumulation of TGs, whereas silencing of G0S2 causes TG depletion. Whether G0S2 regulates ATGL-mediated lipolysis in vivo is not known.
When ATGL was discovered, it was observed that the enzyme was phosphorylated (Zimmermann et al. 2004). Later, at least two phosphorylation sites were identified in murine ATGL: Ser 406 and Ser 430 (Bartz et al. 2007). Phosphorylation of Ser 406 increases the catalytic activity of ATGL in hormone-stimulated lipolysis; however, the protein kinase responsible for enzyme phosphorylation is controversial. Watt and colleagues (Mason et al. 2012; Pagnon et al. 2012) reported that PKA phosphorylates Ser 406, but Ahmadian et al. (2012) found that AMP-kinase was responsible for this modification.
HSL
Originally described in 1964 (Vaughan et al. 1964), HSL was long considered to be the only enzyme for the hydrolysis of cellular TG depots. As noted earlier, this view changed when Osuga et al. (2000) found that HSL-deficient mice are not obese and showed no signs of TG accumulation in either adipose or nonadipose tissues. Instead, HSL-deficient mouse tissues retained TG hydrolase activity and accumulated large amounts of DG in multiple tissues (Haemmerle et al. 2002), suggesting that HSL was more important as a DG hydrolase than as a TG hydrolase. This conclusion was also consistent with the original finding that HSL hydrolyzed DG 10-fold more efficiently than TG (Vaughan et al. 1964). In addition to DG and TG, HSL also cleaves FA esters in MGs, cholesterol esters, and retinyl esters; the highest esterase activities are observed when short chain carboxylic esters are used as substrates. Thus, HSL possesses a broad substrate specificity (Holm et al. 2000). HSL has a preference for primary ester bonds in the sn-3 position of the glycerol backbone but can also hydrolyze sn-1 esters (Rodriguez et al. 2010). Accordingly, it hydrolyzes both DG stereoisomers generated by ATGL (sn-1,3 and sn-2,3 DGs) with high efficiency. In the absence of HSL, ATGL products (sn-1,3 and sn-2,3 DGs) accumulate in adipose and nonadipose tissues (Haemmerle et al. 2002; Eichmann et al. 2012). Despite massive DG accumulation, hyperactivation of conventional or novel protein kinase-C (PKC) isoenzymes has not been reported, and HSL knockouts do not develop insulin resistance (Mulder et al. 2003; Voshol et al. 2003; Park et al. 2005). These findings may be due to the fact that the most potent activators of PKCs among DG isoforms are sn-1,2 DGs (Boni and Rando 1985; Sanchez-Pinera et al. 1999), which are not formed in appreciable amounts during ATGL-mediated lipolysis (Eichmann et al. 2012). Thus, determination of total DG concentrations without knowledge of stereoisomer distribution and cellular localization may be inadequate to predict PKC activation and effects on insulin signaling. This conclusion may explain the inconsistent correlation between tissue DG concentrations and development of insulin resistance in various mouse models (Farese et al. 2012; Sun and Lazar 2013).
The absence of ATGL in mice reduces FA release from adipose tissue by ∼70% (Haemmerle et al. 2006), and experiments with a highly specific HSL inhibitor showed that FAs released from ATGL-deficient adipose tissue result from TG (and DG) hydrolysis by HSL (Schweiger et al. 2006). Although these findings demonstrated that HSL is capable of hydrolyzing cellular TGs in WAT (in addition to DGs), it is not clear how important HSL-mediated TG hydrolysis is when ATGL is expressed normally. HSL-deficient mice exhibit an interesting adipose tissue phenotype with hypertrophic white and brown adipocytes but reduced total WAT mass (Osuga et al. 2000; Wang et al. 2001; Haemmerle et al. 2002). Adipose mass decreases with age and is virtually undetectable in mice >12 mo of age. WAT loss was attributed to reduced lipid synthesis as a result of defective peroxisome proliferator-activated receptor γ (PPARγ) signaling (Zimmermann et al. 2003). A crucial role of HSL in PPARγ signaling was later confirmed (Shen et al. 2011) by suggesting that HSL-mediated lipolysis may provide FA and retinoic acid as ligands for PPARγ and retinoic X receptor (RXR), respectively. Another reason for WAT loss in HSL-deficient mice may be cell death from increased inflammation, macrophage infiltration, and the formation of “crown-like structures” characterized by macrophages surrounding hypertrophic adipocytes (Cinti et al. 2005).
The importance of HSL in cholesterol ester and retinyl ester hydrolysis was also addressed in HSL-deficient mice. Interestingly, retinyl ester hydrolase activity was blunted in HSL-deficient WAT, leading to increased retinyl ester concentrations, decreased retinol and retinal levels, and increased “browning” of adipocytes, as judged by increased uncoupling protein-1 (UCP-1) expression (Strom et al. 2008, 2009). Similarly, HSL-deficient mice lack cholesterol ester hydrolase activity in various tissues and accumulate cholesterol esters (Sekiya et al. 2008; Obrowsky et al. 2012). Cholesterol ester and retinyl ester hydrolase activities may be important in steroidogenesis and retinoic acid formation, respectively (Kraemer et al. 2002; Strom et al. 2009). Lack of these activities in steroid hormone-producing tissues such as testes could underlie the defective spermiogenesis and sterility phenotype in male HSL-deficient mice (Vallet-Erdtmann et al. 2004).
The human gene for HSL codes for protein isoforms that differ in length between 775 to 1076 amino acids, a consequence of alternative splicing involving the first few exons of the gene (Laurin et al. 2000). Interestingly, HSL is unique in the genome of mammals with essentially no related homologous proteins. HSL is most prominently expressed in WAT and BAT, but lower levels can be detected in many nonadipose tissues. Although the structure of HSL has not been determined, mutagenesis studies have uncovered three functional domains (Holm et al. 2000). The N-terminal domain, comprising amino acids 1–300 of adipose HSL, is responsible for LD binding. The C-terminal domain harbors the catalytic triad (S-424, D-693, and H-723) within an α/β hydrolase fold. The nucleophilic serine is located within a GXSXG consensus sequence, which is typical for serine hydrolases. The third domain is a regulatory module located between amino acids 521 and 669, with at least five serine residues that can be phosphorylated (Holm 2003). In particular, the reversible phosphorylation of Ser 659 and Ser 660 regulates enzyme activity. The most important endocrine effectors of HSL are catecholamines, atrial natriuretic peptide, growth hormone, and insulin (Wang et al. 2008). Kinases involved in HSL phosphorylation include PKA, cGMP-activated kinase, AMP-activated kinase, extracellular signal-regulated kinase, glycogen synthase kinase-4, and Ca2+/calmodulin-dependent kinase. HSL dephosphorylation is catalyzed by protein phosphatase 2B (Holm 2003).
MG lipase (MGL)
The products of the ATGL and HSL reactions are sn-1 and sn-2 MGs. Additionally, the intravascular hydrolysis of lipoprotein-associated TGs by LPL generates sn-3 MGs that enter adipocytes and myocytes (Morley and Kuksis 1972; Rogalska et al. 1993). Thus, at the step of MG hydrolysis, the pathways of intravascular and intracellular lipolysis converge. A third source of MGs derives from lipolysis of plasma membrane-associated glycerophospholipids by membrane-bound phospholipases C and DG lipases. The main enzyme responsible for the catabolism of MGs from all three sources is MGL, originally identified in 1964 (Vaughan et al. 1964). The protein contains 303 amino acids and belongs to the large group of α/β hydrolase fold hydrolases and esterases. The enzyme crystallizes as a homodimer with an apolar helix lid covering the active site (Bertrand et al. 2010; Labar et al. 2010). The active site contains a catalytic triad (Ser 122, Asp 239, and His 269) in which the nucleophilic serine is located within a GXSXG consensus sequence.
MGL is ubiquitously expressed, with the highest levels found in adipose tissue and testis. The MGL gene encodes various mRNA splice variants coding for proteins that are between 283 and 313 amino acids long. The enzyme localizes to the plasma membrane, ER, LDs, and cytoplasm. Compared with TG hydrolases, MGL has a much higher specific activity and does not exhibit significant substrate specificity, hydrolyzing all MG stereoisomers with similar efficiency (Tornqvist and Belfrage 1976). The physiological importance of MGL became evident when it was found to be the major enzyme in the catabolism of 2-arachidonylglycerol (2-AG), an important endocannabinoid regulating multiple aspects of metabolism and energy homeostasis (Dinh et al. 2002). 2-AG binds to G-protein-coupled cannabinoid receptor 1 (CBR-1) and CBR-2, affecting a large number of physiological processes, including motor function, pain, appetite, cognition, emotional behavior, and immunity (Di Marzo 2009). The classical pathway of 2-AG synthesis involves the conversion of phosphatidylinositols to DGs by phospholipase C and the subsequent hydrolysis of DGs to 2-AG by diacylglycerol lipase within the plasma membrane. Whether hydrolysis of LD-associated TGs by ATGL and HSL also contributes to 2-AG production and endocannabinoid signaling is unknown.
Lipolysis—three steps, three enzymes, or more?
The current concept of classical hormone-stimulated lipolysis by β-adrenergic agonists in adipocytes can be summarized as follows (Fig. 3). First, hormone (e.g., catecholamine) binding to the Gs-protein-coupled β-adrenergic receptors stimulates adenylate cyclase. Increased cAMP levels activate PKA, which phosphorylates perilipin-1 and HSL. Perilipin-1 phosphorylation facilitates ATGL activation by CGI-58 and the translocation of phosphorylated and activated HSL from the cytoplasm to LDs. Together, these enzymes hydrolyze TGs and DGs to MGs, which are subsequently converted to glycerol and FA by MGL. Hormonal stimulation of lipolysis activates FA and glycerol release from adipose tissue by >100-fold. ATGL, HSL, and MGL are responsible for >90% of the total lipolytic activity in WAT (Schweiger et al. 2006).
Figure 3.
Lipolytic processing of LDs in adipose tissue under basal and hormone-stimulated conditions. In unstimulated adipocytes, TG hydrolysis is low because HSL and perilipin-1 are not phosphorylated. This results in the cytoplasmic localization of HSL and CGI-58 binding to perilipin-1 (PLIN-1). Consequently, CGI-58 is unavailable for ATGL activation. Hormonal stimulation results in a fully active lipolytic cascade with efficient TG hydrolysis. Activated PKA phosphorylates HSL and PLIN-1. This causes dissociation of the PLIN-1/CGI-58 complex and stimulation of ATGL by CGI-58. At the same time, HSL translocates to the LD, interacting with PLIN-1. The combination of ATGL and HSL generates DGs and, ultimately, FAs and glycerol (Gly).
Much less is known about enzymes and mechanisms that control lipolysis in nonadipose tissues. Although ATGL, HSL, and MGL are also observed in these tissues, their activities are much lower than in adipose tissue. Additionally, other TG hydrolases may also contribute to TG hydrolysis in liver, muscle, or macrophages (Quiroga and Lehner 2012). Many nonadipose tissues express perilipin-2 and perilipin-5 but not perilipin-1. Perilipin-2 and perilipin-5 regulate ATGL activity in the liver and cardiac muscle by restricting access of ATGL to LDs (Listenberger et al. 2007; Bell et al. 2008; Granneman et al. 2011; Wang et al. 2011b; Kuramoto et al. 2012). However, detailed mechanisms underlying ATGL activity in these tissues are not entirely clear and require more characterization. A number of other LD-binding proteins (e.g., pigment epithelium-derived factor) have been shown to affect lipolysis in adipose and nonadipose tissues (Chung et al. 2008; Borg et al. 2011), but again, mechanisms underlying their actions are unclear.
Lipolysis and disease
Considering the central role of lipases in lipid and energy metabolism, it is not surprising that they have proven to be relevant to human disease. Monogenetic deficiencies of LPL or ATGL lead to severe alterations in lipid metabolism (Table 1) and can be lethal if not treated properly. In addition, changes in intravascular and intracellular lipolysis affect numerous polygenic metabolic diseases such as atherosclerosis, obesity, and type 2 diabetes mellitus. Here, we summarize recent studies in which altered lipolysis has been linked to human disease.
Intravascular lipolysis and disease
LPL deficiency
The loss of LPL activity leads to familial chylomicronemia syndrome, a syndrome characterized by creamy plasma and markedly elevated plasma TG levels (>1000 mg/dL) (Table 1; Havel and Gordon 1960). This disease is associated with a high risk of pancreatitis, a characteristic skin rash (eruptive xanthomas), and, occasionally, neurological manifestations (Brunzell and Deeb 2001). The absence of intravascular TRL processing is accompanied by reduced delivery of lipid nutrients to tissues, as demonstrated by deficiencies of essential FAs in adipose tissue (Ullrich et al. 2001; Weinstein et al. 2012) and breast milk (Steiner et al. 1985; Ginzinger et al. 1999). LPL-deficient mice die within a day of birth with hypoglycemia and plasma TG levels >20,000 mg/dL (Weinstock et al. 1995) but can be rescued by transient expression of LPL with an adenovirus (Strauss et al. 2001). The plasma TG levels in “rescued” Lpl knockout mice are quite high (>5.000 mg/dL), even on a low-fat chow diet. In mice, heterozygosity for LPL deficiency results in mildly elevated plasma TG levels (Weinstock et al. 1995). In humans, a single defective LPL allele often leads to mild or moderate hypertriglyceridemia (Miesenbock et al. 1993; Nordestgaard et al. 1997) but occasionally to severe hypertriglyceridemia, particularly during pregnancy (Ma et al. 1994b; Henderson et al. 1998).
For years, it was presumed that chylomicronemia did not predispose to atherosclerosis because the large size of TRLs prevented them from entering the arterial intima (Nordestgaard and Zilversmit 1988). However, some patients with chylomicronemia due to LPL deficiency have developed significant atherosclerosis (Benlian et al. 1996). This clinical observation in humans has been bolstered by the observation that rescued LPL-deficient mice (Zhang et al. 2008) and GPIHBP1-deficient mice (Weinstein et al. 2010) develop spontaneous atherosclerosis even on a low-fat chow diet.
LPL deficiency is associated with an absence of LPL activity in postheparin plasma (Havel and Gordon 1960). Over the past two decades, dozens of clinically significant LPL mutations have been described. Some abolish the production of LPL transcripts or lead to unstable transcripts; certain missense mutations prevent secretion of LPL from cells, others interfere with the stability of LPL homodimers, and others yield LPL homodimers that are stable yet catalytically inactive (Hayden et al. 1991). In recent years, new mechanisms of disease have been uncovered. For example, a pair of LPL missense mutations, C418Y and E421K, first identified in chylomicronemia patients (Henderson et al. 1996, 1998), do not affect LPL secretion or catalytic activity but abolish LPL's ability to bind to GPIHBP1 and be transported across endothelial cells to the capillary lumen (Fig. 4; Voss et al. 2011). Also, missense mutations at residues 409 and 410, long recognized to abolish LPL activity (Previato et al. 1992; Ginzinger et al. 1996), were recently shown to enhance LPL susceptibility to furin-mediated cleavage at residue 297 (Gin et al. 2012). Although the latter mutations were located in close proximity to LPL's GPIHBP1-binding domain, they did not appear to interfere with binding to GPIHBP1 (Gin et al. 2012).
Figure 4.
Schematic of a cross-section of a capillary depicting the location of GPIHBP1 and LPL and their role in the lipolytic processing of TRLs in health and disease. The top panel depicts a normal capillary illustrating the role of GPIHBP1 in capturing LPL in the interstitial space and moving it to the capillary lumen, where it hydrolyzes TGs in TRLs, converting them to smaller remnant lipoproteins. The bottom left panel depicts impaired delivery of LPL to the capillary lumen in the setting of a mutant GPIHBP1 (e.g., GPIHBP1-Q115P) (Beigneux et al. 2009b) lacking the ability to bind LPL. The bottom right panel depicts impaired delivery of LPL to the capillary lumen in the setting of an LPL mutant (e.g., LPL-C418Y) (Voss et al. 2011) that lacks the ability to bind GPIHBP1. When LPL does not reach the capillary lumen, there is a striking accumulation of TRLs in the plasma.
Apo-CII deficiency
Homozygous loss of apo-CII also leads to familial chylomicronemia (Breckenridge et al. 1978), but these mutations are less common and tend to be less severe than LPL mutations (Table 1). Heterozygous apo-CII deficiency has no effect on lipid levels. A variety of nonsense, frameshift, and missense mutations have been uncovered (Connelly et al. 1987; Cox et al. 1988; Crecchio et al. 1990; Reina et al. 1992; Inadera et al. 1993; Tuzgol et al. 1994). Some of the missense mutations impair binding of apo-CII to TRLs and are associated with extremely low levels of apo-CII in the plasma. One of the missense mutations, Leu72Pro, underscored the importance of apo-CII's C terminus in LPL activation (Lam et al. 2006). The diagnosis of apo-CII deficiency can be suspected when low LPL activity levels in the postheparin plasma are normalized with apo-CII-containing plasma (Breckenridge et al. 1978). Transfusing apo-CII-deficient patients with donor plasma lowers plasma TG levels (Breckenridge et al. 1978).
LMF1 deficiency
Lmf1 was initially identified as the mutant gene (Peterfy et al. 2007) in a naturally occurring mutant strain of mice (“combined lipase deficiency” or cld mice) lacking both LPL and HL activity (Olivecrona et al. 1986; Langner et al. 1989). LMF1 is also important for the secretion of EL (Ben-Zeev et al. 2010). LMF1 is critical for the formation and secretion of lipase homodimers (Peterfy et al. 2007). A human subject homozygous for a nonsense mutation in LMF1 (Y439X) had markedly elevated plasma TG levels; postheparin LPL and HL levels were reduced by 91% and ∼50%, respectively (Peterfy et al. 2007). A second homozygous nonsense mutation, W464X, was identified in another patient with severe hypertriglyceridemia; postheparin LPL activity in that patient was reduced by 76% (Cefalu et al. 2009). Like apo-CII deficiency, LMF1 deficiency in humans is a recessive syndrome (Peterfy et al. 2007), and clinically significant mutations appear to be quite rare (Surendran et al. 2012).
GPIHBP1 deficiency
During the past few years, homozygous GPIHBP1 mutations have been uncovered in patients with severe chylomicronemia (Table 1; Beigneux et al. 2009b; Franssen et al. 2010; Olivecrona et al. 2010; Charriere et al. 2011; Coca-Prieto et al. 2011; Rios et al. 2011). Most of the mutations described thus far are missense mutations within the Ly6 domain that interfere with the ability of GPIHBP1 to bind LPL. Impaired binding means that LPL cannot associate with endothelial cells and cannot reach the capillary lumen (Fig. 4). Many of the mutations involve conserved cysteines within the Ly6 domain (Franssen et al. 2010; Olivecrona et al. 2010; Charriere et al. 2011; Coca-Prieto et al. 2011). That cysteine mutations would cause disease is not surprising, since these residues are required for producing the three-fingered structure of the Ly6 domain and are essential for LPL binding and transport across endothelial cells (Beigneux et al. 2009c, 2011).
Other GPIHBP1 missense mutations (for example, Q115P) have occurred in residues located near conserved cysteines (Beigneux et al. 2009b; Surendran et al. 2012). The Q115P mutation markedly impairs LPL binding (Beigneux et al. 2009b, 2011).
Recently, Charriere et al. (2011) identified a 35-year-old man with chylomicronemia and a homozygous G175R mutation. That mutation, located in GPIHBP1's C-terminal domain, reduces the amount of GPIHBP1 at the cell surface and likely interferes with the addition of the GPI anchor. Hobbs and Cohen (Rios et al. 2011) identified a homozygous 17.5-kb deletion that included the entire GPIHBP1 gene in a family with chylomicronemia. Charriere et al. (2011) also identified a large deletion in a patient with chylomicronemia.
Humans with heterozygous GPIHBP1 mutations appear to be normolipidemic (Franssen et al. 2010; Charriere et al. 2011; Rios et al. 2011). In mice, heterozygosity for the Gpihbp1 knockout mutation lowers GPIHBP1 protein levels in tissues by 50% but has no effect on plasma lipid levels (Beigneux et al. 2007). Presumably, half-normal amounts of GPIHBP1 are sufficient to transport LPL to the capillary lumen.
In Gpihbp1 knockout mice, an injection of heparin detaches LPL from its binding sites in the interstitial spaces, allowing the enzyme to enter the plasma (Beigneux et al. 2007; Weinstein et al. 2008). Once this occurs, the plasma TG levels fall (Weinstein et al. 2008). The situation is different in humans. At the doses of heparin that are given to humans, the amount of LPL that enters the plasma compartment is very low (Franssen et al. 2010; Olivecrona et al. 2010; Rios et al. 2011), and the effect on plasma TG levels is minimal or absent (Franssen et al. 2010; Rios et al. 2011).
Other LPL regulators
Mutations in LPL regulatory factors also alter plasma TG levels. Heterozygosity for a nonsense mutation in the gene for apo-CIII, a negative regulator of lipolysis, lowers plasma TG and cholesterol levels and is associated with reduced coronary artery calcification (Pollin et al. 2008). Defects in apo-AV lead to increased plasma TG levels and, occasionally, severe chylomicronemia (Grosskopf et al. 2005; Dorfmeister et al. 2008). Missense mutations in the gene for ANGPTL4, a lipase inhibitor (Yoshida et al. 2002; Koster et al. 2005), result in lower plasma TG levels (Romeo et al. 2007, 2009; Yin et al. 2009). Nonsense mutations in ANGPTL3, an inhibitor of both LPL and EL, lead to very low plasma lipid levels (Musunuru et al. 2010).
HL and EL deficiency
Mutations in HL and EL result in modest changes in plasma lipid levels, and given the variability in plasma lipid levels within human populations, these deficiencies are unlikely to be diagnosed without genetic screening. Individuals with HL deficiency have increased levels of TGs in the HDL and IDL fractions and more remnant lipoproteins and appear to be more susceptible to coronary artery disease (Hegele et al. 1991, 1993; Brand et al. 1996; Connelly and Hegele 1998; Santamarina-Fojo et al. 2004). Subjects with an inactivating mutation in EL have higher HDL cholesterol levels (Edmondson et al. 2009). The latter finding raised considerable interest and suggested that EL inhibitors might be useful for increasing plasma HDL levels and reducing coronary disease risk (Jin et al. 2007; O'Connell et al. 2012). However, a large study has recently shown that an EL missense mutation that lowers EL activity and raises HDL cholesterol levels does not change susceptibility to coronary artery disease (Voight et al. 2012).
Intracellular lipolysis and disease
NLSD
NLSD is a rare nonlysosomal, autosomal-recessive lipid storage disorder (Table 1; Chanarin et al. 1975). Affected individuals accumulate TG-containing LDs in multiple tissues, including skin, skeletal muscle, heart, liver, the CNS, and blood leukocytes. The finding of lipid-filled vacuoles in leukocytes was first discovered by Jordans (1953) and is pathognomonic for the disease. More variable clinical features include liver steatosis and hepatomegaly, skeletal and cardiac myopathy, growth retardation, cataracts, hearing loss, and/or loss of mental acuity (Dorfman et al. 1974; Chanarin et al. 1975). The most obvious phenotype in a subset of NLSD patients is a severe skin defect known as nonbullous congenital ichthyosiform erythroderma (Dorfman et al. 1974; Chanarin et al. 1975), which is associated with a defect in the epidermal water barrier. Previously called Chanarin-Dorfman syndrome, this disease is now called NLSDI. Igal et al. (1997) categorized >40 patients with NLSD into two groups according to the presence or absence of ichthyosis and observed that NLSDI patients were more likely to develop liver steatosis, whereas the group without ichthyosis was more likely to develop skeletal and cardiac myopathy. The NLSD variant without ichthyosis is now called NLSD with myopathy (NLSDM). Mutations in the gene for ATGL (PNPLA2) cause NLSDM, whereas mutations in the gene for CGI-58 cause NLSDI.
ATGL deficiency and NLSDM
In 2007, Fischer's group (Fischer et al. 2007) reported that ATGL mutations cause NLSDM, and since then, 30 human cases of NLSDM have been recorded with at least 15 different mutations. Mutations include single-, di- and tetranucleotide deletions, nucleotide transversions, and splice site alterations (Schweiger et al. 2009). Aside from a single missense mutation in ATGL, all other mutations lead to premature stop codons due to frameshift or nonsense mutations. Interestingly, the clinical presentation varies among patients, and it is not clear whether the differences relate to the intrinsic properties of the mutant protein or are due to gene–gene or gene–environment interactions. Generally, phenotypes in humans resemble those in ATGL-deficient mice but are less dramatic. Affected patients typically experience muscle weakness during their teenage years. By the third decade, patients often develop cardiac steatosis and dilated cardiomyopathy. This condition can be severe and often makes cardiac transplantation inevitable (Hirano et al. 2008).
Cardiac death in patients (and in the knockout mice) may not be due entirely to the accumulation of TG or the lack of energy substrates. The reason is that hearts of Atgl knockout mice exhibit a severe defect in mitochondrial oxidation of FAs and glucose, a consequence of defective PPARα/PPARγ coactivator 1 (PGC-1) signaling (Haemmerle et al. 2011). This metabolic defect leads to lipid deposition in the heart and cardiomyopathy. Treatment of ATGL-deficient mice with PPARα agonists improves the metabolic defect, reverses cardiomyopathy, and prolongs life. ATGL activity is also important for PPARα signaling in the liver and BAT (Sapiro et al. 2009; Ong et al. 2011; Ahmadian et al. 2012; Mottillo et al. 2012). The mechanism of how ATGL regulates PPARα-mediated gene expression is not known, but since PPARs require lipid ligands for activation, it is conceivable that the ATGL reaction provides these ligands (or their precursors). Whether a similar mechanism is involved in the development of cardiomyopathy in patients with NLSDM and whether treating them with PPAR agonists would have beneficial effects remain to be investigated. LPL also regulates PPARα signaling (Ziouzenkova et al. 2003). Transgenic mice overexpressing LPL in the heart exhibit increased PPARα target gene expression and peroxisome proliferation.
CGI-58 deficiency and NLSDI
Homozygosity mapping, linkage disequilibrium analysis, and candidate gene sequencing showed that mutations in the gene for CGI-58 cause NLSDI (Lefevre et al. 2001). This finding predated the discovery of ATGL, and accordingly, the mechanism for lipid accumulation was initially mysterious. The discovery that CGI-58 activates the TG hydrolase activity of ATGL (Lass et al. 2006) eventually provided an explanation for the lipid accumulation phenotype. However, the ichthyosis phenotype was not readily explained because ATGL-deficient humans and mice exhibit no skin abnormalities. This suggested that CGI-58 has an ATGL-independent function in the skin (and possibly in other tissues as well). Of note, the skin defect in CGI-58-deficient mice was far more severe than in humans with NLSDI (Radner et al. 2010). Cgi-58 knockout mice die soon after birth from water loss, a consequence of a severe defect in the permeability barrier of the epidermis. CGI-58 deficiency causes a marked reduction of epidermal acylceramides in mice and humans (Radner et al. 2010; Uchida et al. 2010). Since acylceramides are essential lipids in the formation of the corneocyte lipid envelope, their absence in CGI-58-deficient skin is most likely responsible for the epidermal barrier defect. How CGI-58 affects acylceramide synthesis is currently not clear.
HSL deficiency
A recent study uncovered two Amish subjects with homozygous loss of HSL due to a deletion mutation. According to a meeting abstract (Albert et al. 2011), these patients express no HSL protein, have a 50% reduction in adipocyte size, and accumulate DG in fat cells, reminiscent of findings in Hsl knockout mice (Wang et al. 2001; Haemmerle et al. 2002). Decreased fat mass in HSL-deficient mice is due to decreased PPARγ activity, resulting in decreased lipogenesis (Zimmermann et al. 2003). In the future, it will be interesting to determine whether human HSL deficiency also affects PPARγ-regulated lipid synthesis. Male Hsl knockout mice are also infertile (Osuga et al. 2000). Currently, no information exists on fertility in male HSL-deficient humans. However, a recent study reported an increased frequency of a single-nucleotide polymorphism in the human HSL gene in infertile males (Vatannejad et al. 2011), suggesting that genetic variation in HSL may be a risk factor for male infertility.
ATGL, CGI-58, HSL, and metabolic disease
A potential role of lipolysis in the development of obesity and insulin resistance in humans has been discussed extensively (for review, see Arner 2005; Jocken and Blaak 2008; Lafontan and Langin 2009; Kolditz and Langin 2010). TG accumulation in tissues is commonly, but not always, associated with the development of insulin resistance (Sun and Lazar 2013). Interestingly, despite neutral lipid accumulation in multiple tissues, no major alterations in glucose tolerance or insulin sensitivity have been reported in ATGL- and CGI-58-deficient humans (Kobayashi et al. 2008). The dissociation of TG storage and insulin resistance has also been observed in mice (Haemmerle et al. 2006; Kienesberger et al. 2009a; Hoy et al. 2011; Turpin et al. 2011), supporting the concept that TG accumulation per se is not sufficient to cause insulin resistance. In mice, ATGL also affects pancreatic insulin secretion, leading to reduced plasma insulin levels (Peyot et al. 2009). One study reported decreased plasma insulin levels in NLSDM patients (Akiyama et al. 2007), but additional studies of affected humans are needed to conclude that ATGL is necessary for normal insulin production.
Hepatic ATGL and CGI-58 expression levels have a pronounced effect on liver TG levels. While liver-specific knockouts of Atgl or Cgi-58 cause severe hepatosteatosis, overexpression of ATGL results in low hepatic TG stores (Reid et al. 2008; JM Brown et al. 2010; Wu et al. 2011). These changes in ATGL expression and hepatic TG levels, however, do not seem to affect hepatic VLDL synthesis or plasma TG concentrations, suggesting that the ATGL reaction may not be important in providing FAs for VLDL assembly (Wu et al. 2011). Similarly, there are no apparent defects in plasma lipoprotein metabolism in patients with NLSDM or NLSDI (Akiyama et al. 2007; Fischer et al. 2007).
The severe defects in brown adipocyte function and thermogenesis in ATGL-deficient mice have highlighted the importance of lipolysis in these processes (Haemmerle et al. 2006; Ahmadian et al. 2012). The presence and functional importance of brown adipocytes have also been demonstrated in humans (van Marken Lichtenbelt et al. 2009; Virtanen et al. 2009). To date, it is not known whether patients with NLSDM exhibit a BAT defect that affects thermogenesis.
MGL and disease
The discovery that 2-AG acts as an endocannabinoid signaling molecule ignited great interest in its synthesis pathways and its degradation by MGL. Thus far, mutations that inactivate MGL have not been reported in humans. However, experiments in mutant mice suggest that MGL could prove to be a target for treating various disorders. MGL deficiency in mice causes massive accumulation of MGs, specifically 2-AG, in multiple tissues (Chanda et al. 2010; Schlosburg et al. 2010; Taschler et al. 2011). In the brain, 2-AG levels increase >20-fold; however, chronic exposure to increased 2-AG concentrations leads to desensitization of endocannabinoid signaling (Chanda et al. 2010; Schlosburg et al. 2010), with no significant effects on food intake, energy expenditure, and other metabolic parameters. In contrast, acute treatment of mice with a MGL inhibitor provoked the expected cannabimimetic effects of analgesia, hypothermia, and hypomotility (Long et al. 2009). Conversely, when MGL activity was increased in the forebrain by transgenic overexpression, the animals were lean and had reduced adipose mass and improved glucose tolerance (Jung et al. 2012). Unexpectedly, the animals were hyperphagic and consumed this additional energy for thermogenesis. 2-AG acts in concert with another lipid mediator, anandamide (arachidonylethanolamide), in the activation of CBRs and endocannabinoid signaling. To induce analgesia in patients with chronic pain, it may be necessary to inhibit both MGL and FA amide hydrolase (FAAH), the main catabolic enzyme for anadamide.
MGL is not only important in inactivating 2-AG for endocannabinoid signaling, it also provides arachidonic acid for prostaglandin synthesis. Recently, Nomura et al. (2012) demonstrated that MGL deficiency leads to reduced arachidonic acid availability, reduced levels of neuroinflammatory prostaglandins in the brain, and neuroprotection in a mouse model of Parkinson's disease. Thus, inhibition of MGL could represent an anti-inflammatory treatment strategy.
MGL deficiency leads to MG accumulation in the liver, reduced plasma glycerol levels, decreased VLDL synthesis, and lower plasma TG concentrations (Taschler et al. 2011). Interestingly, MGL-deficient mice are more sensitive to insulin than wild-type mice, but the mechanism is currently unclear. One possibility involves the role of MGs as ligands for the G-protein-coupled receptor 119 (GPR119) (Hansen et al. 2012). This receptor acts as a fat sensor, with the highest levels of expression in the pancreas and the intestine. It is activated by a variety of oleic acid-containing lipids, including 2-oleylglycerol (2-OG), and regulates glucagon-like peptide-1 (GLP-1) and insulin secretion in enteroendocrine cells and pancreatic β cells. 2-OG is a major product of dietary fat hydrolysis by pancreatic lipase and induces GLP-1 secretion in intestinal cells through GPR119. This activity is also consistent with the finding that MGL overexpression in the small intestine reduces intestinal MG levels (including 2-OG), causing hyperphagia and obesity (Chon et al. 2012). In the pancreas, 2-OG and other MGs are products of both intravascular LPL activity and intracellular ATGL/HSL activity. All three lipases affect insulin secretion from pancreatic islet β cells (Roduit et al. 2001; Pappan et al. 2005; Peyot et al. 2009), and it is conceivable that, depending on the activity of MGL, the local MG concentration regulates pancreatic insulin secretion through GPR119.
Lipolysis in cancer and cancer cachexia
Metabolism in cancer cells has witnessed a revival in recent years (Vander Heiden et al. 2009). This includes renewed interest in lipid metabolism in cancer biology. In addition to numerous lipid signals and mediators that affect tumor cell proliferation, TG anabolic and catabolic pathways are instrumental in providing energy, membrane lipids, and other lipid intermediates for cancer cell growth (Menendez 2009). LPL expression by rapidly proliferating cancer cells could affect the delivery and uptake of lipid nutrients to those cells. Immunohistochemical studies have demonstrated the presence of LPL and CD36 (a FA transporter) in the majority of breast cancer, liposarcoma, and prostate cancer samples (Kuemmerle et al. 2011). Interestingly, LPL is frequently overexpressed in invasive cervical squamous cell carcinomas (Carter et al. 2012). Most impressively, patients with chronic lymphocytic leukemia (CLL) whose leukemia cells express high levels of LPL transcripts have a lower survival rate (Van Bockstaele et al. 2007; Maloum et al. 2009; Kaderi et al. 2011). One study indicated that high LPL transcript levels in CLL are associated with increased LPL protein production; however, enzymatic activity assays suggested that much of the LPL could be inactive (Mansouri et al. 2010). At this point, it is unclear whether LPL levels are simply a useful prognostic indicator for CLL or whether the high levels of LPL expression directly affect the behavior of the tumor cells. This topic needs further investigation.
Little is known about the role of intracellular lipases in carcinogenesis and tumor proliferation, but results in model organisms suggest a potential involvement. For example, in yeast (Saccharomyces cerevisiae), the ATGL ortholog Tgl4 participates in the regulation of cell cycle progression (Kurat et al. 2006). Tgl4 is activated by phosphorylation through the cyclin-dependent kinase Cdk1/Cdc28 (orthologous to mammalian Cdc2) at the G1/S transition and provides FAs during periods of high demand for membrane lipid synthesis. Deletion of Tgl4 (together with Tgl3) abolishes TG hydrolysis and delays cell division.
Cravatt and colleagues (Nomura et al. 2010) found that MGL has potent protumorigenic activity in mice and that MGL expression is induced in many aggressive human cancers. Interestingly, the tumor-promoting role of MGL appears to rely on both its metabolic function in providing FAs for cell membranes and its role as an endocannabinoid signaling antagonist (Nomura et al. 2011). However, not all studies agree on the role of MGL in cancer cell proliferation. One study found a tumor-promoting role for MGL in colorectal cancer (Ye et al. 2011), while another reported a tumor-suppressive role (Sun et al. 2013).
Although it is unknown whether ATGL/CGI-58 and HSL affect the development and proliferation of tumors, a crucial role for these proteins in cancer-associated cachexia was recently uncovered. Cachexia is defined as an unwanted loss of adipose tissue and muscle mass that cannot be corrected by dietary measures. It occurs in the end stages of many chronic diseases and is most commonly observed in cancer. Arner and colleagues (Agustsson et al. 2007; Ryden et al. 2008) noted that patients with cancer-associated cachexia exhibit increased lipolysis and FA release from adipose tissue. In a recent study, Das et al. (2011) found that ATGL deficiency in mice can prevent the loss of adipose tissue and muscle in animals with Lewis lung carcinoma, suggesting that intracellular lipolysis could play a central role in the physiology of cachexia. The mechanisms linking lipolysis with the pathogenesis of cachexia are currently under investigation, and many questions remain unanswered. For example, it is not known whether ATGL/HSL-catalyzed lipolysis is important in the pathogenesis of all forms of cachexia or only in cachexia associated with certain cancers. It will also be important to tease out the relative contributions of adipose or muscle lipolysis to the development of cachexia. Clarification of these issues will reveal whether ATGL and/or HSL are appropriate targets for preventing cachexia in humans.
Lipolysis and infectious disease
Given the conservation of LPL and GPIHBP1 in mammalian evolution, one might have imagined that endothelial cell pathogens would have evolved strategies to hijack these proteins and use them as vehicles for infecting endothelial cells or moving across capillaries. Thus far, however, no one has found evidence for these processes, but sooner or later, we suspect that examples will be uncovered. Certain meningococcal proteins are known to bind to heparin or negatively charged proteoglycans on the surface of cells (de Vries et al. 1998; Serruto et al. 2010). Similarly, the dengue fever virus is thought to infect endothelial cells by binding to negatively charged HSPGs (Dalrymple and Mackow 2011). Interestingly, the uptake of the dengue fever virus can be reduced with heparin. The major locus for mouse adenovirus susceptibility maps to a Ly6 gene locus containing GPIHBP1 (Spindler et al. 2010), and the infection of some adenoviral strains can be inhibited with heparin (Dechecchi et al. 2001). Finally, red blood cells, when infected with different strains of Plasmodium, are thought to bind to HSPGs on endothelial cells (Vogt et al. 2003; Chotivanich et al. 2012). This binding can be inhibited with heparin. At this point, whether any of these pathogens make use of GPIHBP1 as a binding site is unknown but certainly plausible.
In some cases, LPL could play a role in protecting the host from infection (Thomssen and Bonk 2002; Andreo et al. 2007; Shimizu et al. 2010; Maillard et al. 2011). Hepatitis C virus (HCV) is known to associate with plasma lipoproteins, and lipoprotein receptors could play a role in virus uptake by cells. Treatment of lipoproteins with catalytically active LPL reduces infectivity of the virus, likely by “tying up” the virus on the surface of cells and preventing productive infection within cells (Maillard et al. 2011).
Active lipid metabolism and intracellular LD turnover are also connected to the life cycle of various bacteria, viruses, and parasites. For example, Mycobacterium tuberculosis, an obligate intracellular bacterium, infects macrophages in the lung and uses host TGs as its main energy source (Neyrolles et al. 2006). The bacilli import FAs derived from host TGs, store them as TGs in intracellular lipid inclusions, and rehydrolyze them using their own lipases to meet metabolic demands. Chlamydia trachomatis, an intracellular pathogen that causes conjunctivitis, urethritis, and pelvic inflammatory disease, also depends on TG and cholesterol esters from host LDs. In contrast to M. tuberculosis, Chlamydia lacks proteins with homology with known TG hydrolases. Thus, it is assumed that TGs used by that organism are processed by host lipases such as ATGL. Bacterial proteins (e.g., Lda3) may facilitate this process by removing perilipins from the surface of LDs, thereby enabling access of lipases to TG substrates (Cocchiaro et al. 2008).
Viruses have also been shown to use elements of host TG metabolism. The HCV life cycle depends on—and interferes with—host cell LD formation and degradation. A viral nucleocapsid protein named core translocates from the ER to the LD surface, which provides a platform for viral assembly (Boulant et al. 2007; Miyanari et al. 2007; Herker et al. 2010). This process relies on the TG-synthesizing enzyme acyl-CoA diacylglycerol acyltransferase-1 (DGAT1) (Herker et al. 2010). The absence of DGAT1 or LDs in hepatocytes leads to defective HCV assembly. The dependence of virus assembly on DGAT1 and LDs is also consistent with the observation that one of the major clinical manifestations of HCV infection is liver steatosis (Asselah et al. 2006). Recent data suggest that core causes LD accumulation by inhibiting intracellular TG hydrolysis and reducing TG catabolism (Harris et al. 2011). Dengue fever virus is closely related to HCV and displays a similar dependency on host cell LDs for viral replication and assembly. Moreover, dengue fever virus infection induces both host cell lipogenesis and autophagy for TG degradation (Heaton and Randall 2010; Heaton et al. 2010). The released FAs subsequently undergo mitochondrial β oxidation, and the resulting energy is used for viral RNA replication. Moreover, the degradation of LDs by autophagy may serve as an immune evasion strategy (Heaton and Randall 2010). Host LDs or lipid metabolites are also important for replication of rotavirus (Cheung et al. 2010), cytomegalovirus, (Zhu et al. 2002), and influenza virus (Munger et al. 2008), but the role of host lipolytic processes or lipases in these processes is insufficiently understood.
Acknowledgments
We thank Dr. Martina Schweiger, Dr. Thomas Eichmann, Dr. Achim Lass, and Dr. Anne Beigneux for their help in preparing this manuscript. This work was supported by the Leducq Transatlantic Network TNT 12CVD04 (to R.Z. and S.G.Y.), and the National Heart, Lung, and Blood Institute HL090553 and HL087228 (to S.G.Y.). R.Z. was supported by grants P21296 and SFB LIPOTOX F30, the Doktoratskolleg Molecular Enzymology W901, and the Wittgenstein Award Z136, which are funded by the Austrian Science Fund (FWF). Additional funding was obtained from “GOLD: Genomics of lipid-associated disorders,” which was part of the Austrian Genome Project “GEN-AU: Genome Research in Austria” funded by the Austrian Ministry of Science and Research and the FFG.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.209296.112.
References
- Agustsson T, Ryden M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, Isaksson B, Permert J, Arner P 2007. Mechanism of increased lipolysis in cancer cachexia. Cancer Res 67: 5531–5537 [DOI] [PubMed] [Google Scholar]
- Ahmadian M, Abbott MJ, Tang T, Hudak CS, Kim Y, Bruss M, Hellerstein MK, Lee HY, Samuel VT, Shulman GI, et al. 2012. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab 13: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Sakai K, Ogawa M, McMillan JR, Sawamura D, Shimizu H 2007. Novel duplication mutation in the patatin domain of adipose triglyceride lipase (PNPLA2) in neutral lipid storage disease with severe myopathy. Muscle Nerve 36: 856–859 [DOI] [PubMed] [Google Scholar]
- Albert J, Horenstein R, Yerges-Armstrong L, Sreenivasan U, Snitker S, O'Connell J, McLenithan JC, Shuldiner A, Sztalryd C, Damcott C 2011. A human hormone sensitive lipase (HSL) knockout: Window into mechanisms of dyslipidemia and diabetes. 61st Annual Meeting of the American Society of Human Genetics, Program No. 81. http://www.ichg2011.org/cgi-bin/showdetail.pl?absno=11253 [Google Scholar]
- Andreo U, Maillard P, Kalinina O, Walic M, Meurs E, Martinot M, Marcellin P, Budkowska A 2007. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol 9: 2445–2456 [DOI] [PubMed] [Google Scholar]
- Arner P 2005. Human fat cell lipolysis: Biochemistry, regulation and clinical role. Best Pract Res Clin Endocrinol Metab 19: 471–482 [DOI] [PubMed] [Google Scholar]
- Asselah T, Rubbia-Brandt L, Marcellin P, Negro F 2006. Steatosis in chronic hepatitis C: Why does it really matter? Gut 55: 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badellino KO, Wolfe ML, Reilly MP, Rader DJ 2006. Endothelial lipase concentrations are increased in metabolic syndrome and associated with coronary atherosclerosis. PLoS Med 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balser WA 1882. Ueber Fettnekrose, eine zuweilen tödtliche Krankheit des Menschen. Virchows Arch 90: 520–535 [Google Scholar]
- Banerji S, Flieger A 2004. Patatin-like proteins: A new family of lipolytic enzymes present in bacteria? Microbiology 150: 522–525 [DOI] [PubMed] [Google Scholar]
- Bartz R, Zehmer JK, Zhu M, Chen Y, Serrero G, Zhao Y, Liu P 2007. Dynamic activity of lipid droplets: Protein phosphorylation and GTP-mediated protein translocation. J Proteome Res 6: 3256–3265 [DOI] [PubMed] [Google Scholar]
- Baulande S, Lasnier F, Lucas M, Pairault J 2001. Adiponutrin, a transmembrane protein corresponding to a novel dietary- and obesity-linked mRNA specifically expressed in the adipose lineage. J Biol Chem 276: 33336–33344 [DOI] [PubMed] [Google Scholar]
- Beigneux AP, Davies B, Gin P, Weinstein MM, Farber E, Qiao X, Peale P, Bunting S, Walzem RL, Wong JS, et al. 2007. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab 5: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Gin P, Davies BS, Weinstein MM, Bensadoun A, Ryan RO, Fong LG, Young SG 2008. Glycosylation of Asn-76 in mouse GPIHBP1 is critical for its appearance on the cell surface and the binding of chylomicrons and lipoprotein lipase. J Lipid Res 49: 1312–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Davies BS, Bensadoun A, Fong LG, Young SG 2009a. GPIHBP1, a GPI-anchored protein required for the lipolytic processing of triglyceride-rich lipoproteins. J Lipid Res 50: S57–S62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Franssen R, Bensadoun A, Gin P, Melford K, Peter J, Walzem RL, Weinstein MM, Davies BS, Kuivenhoven JA, et al. 2009b. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol 29: 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Gin P, Davies BSJ, Weinstein MM, Bensadoun A, Fong LG, Young SG 2009c. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J Biol Chem 284: 30240–30247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigneux AP, Davies BS, Tat S, Chen J, Gin P, Voss CV, Weinstein MM, Bensadoun A, Pullinger CR, Fong LG, et al. 2011. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J Biol Chem 286: 19735–19743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M, Wang H, Chen H, McLenithan JC, Gong DW, Yang RZ, Yu D, Fried SK, Quon MJ, Londos C, et al. 2008. Consequences of lipid droplet coat protein downregulation in liver cells: Abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes 57: 2037–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol 6: e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson G, Olivecrona T 1979. Apolipoprotein CII enhances hydrolysis of monoglycerides by lipoprotein lipase, but the effect is abolished by fatty acids. FEBS Lett 106: 345–348 [DOI] [PubMed] [Google Scholar]
- Benlian P, De Gennes JL, Foubert L, Zhang H, Gagné SE, Hayden M 1996. Premature atherosclerosis in patients with familial chylomicronemia caused by mutations in the lipoprotein lipase gene. N Engl J Med 335: 848–854 [DOI] [PubMed] [Google Scholar]
- Ben-Zeev O, Doolittle MH, Singh N, Chang CH, Schotz MC 1990. Synthesis and regulation of lipoprotein lipase in the hippocampus. J Lipid Res 31: 1307–1313 [PubMed] [Google Scholar]
- Ben-Zeev O, Stahnke G, Liu G, Davis RC, Doolittle MH 1994. Lipoprotein lipase and hepatic lipase: The role of asparagine-linked glycosylation in the expression of a functional enzyme. J Lipid Res 35: 1511–1523 [PubMed] [Google Scholar]
- Ben-Zeev O, Hosseini M, Lai CM, Ehrhardt N, Wong H, Cefalu AB, Noto D, Averna MR, Doolittle MH, Peterfy M 2010. Lipase maturation factor 1 is required for endothelial lipase activity. J Lipid Res 52: 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bergman EN, Havel RJ, Wolfe BM, Bøhmer T 1971. Quantitative studies of the metabolism of chylomicron triglycerides and cholesterol by liver and extrahepatic tissues of sheep and dogs. J Clin Invest 50: 1831–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C. 1856. Mémoire sur le pancréas et sur la role du suc pancréatique dans les phénomèmes digestifs particulièrement dans la digestion des matières grasses neutres. Nabu Press, BiblioBazaar, Charleston, SC. [Google Scholar]
- Bertrand T, Auge F, Houtmann J, Rak A, Vallee F, Mikol V, Berne PF, Michot N, Cheuret D, Hoornaert C, et al. 2010. Structural basis for human monoglyceride lipase inhibition. J Mol Biol 396: 663–673 [DOI] [PubMed] [Google Scholar]
- Bessesen DH, Richards CL, Etienne J, Goers JW, Eckel RH 1993. Spinal cord of the rat contains more lipoprotein lipase than other brain regions. J Lipid Res 34: 229–238 [PubMed] [Google Scholar]
- Boni LT, Rando RR 1985. The nature of protein kinase C activation by physically defined phospholipid vesicles and diacylglycerols. J Biol Chem 260: 10819–10825 [PubMed] [Google Scholar]
- Borg ML, Andrews ZB, Duh EJ, Zechner R, Meikle PJ, Watt MJ 2011. Pigment epithelium-derived factor regulates lipid metabolism via adipose triglyceride lipase. Diabetes 60: 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S, Targett-Adams P, McLauchlan J 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J Gen Virol 88: 2204–2213 [DOI] [PubMed] [Google Scholar]
- Brand K, Dugi KA, Brunzell JD, Nevin DN, Santamarina-Fojo S 1996. A novel A → G mutation in intron I of the hepatic lipase gene leads to alternative splicing resulting in enzyme deficiency. J Lipid Res 37: 1213–1223 [PubMed] [Google Scholar]
- Breckenridge WC, Little JA, Steiner G, Chow A, Poapst M 1978. Hypertriglyceridemia associated with deficiency of apolipoprotein C-II. N Engl J Med 298: 1265–1273 [DOI] [PubMed] [Google Scholar]
- Breedveld B, Schoonderwoerd K, Verhoeven AJM, Willemsen R, Jansen H 1997. Hepatic lipase is localized at the parenchymal cell microvilli in rat liver. Biochem J 321: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Betters JL, Lord C, Ma Y, Han X, Yang K, Alger HM, Melchior J, Sawyer J, Shah R, et al. 2010. CGI-58 knockdown in mice causes hepatic steatosis but prevents diet-induced obesity and glucose intolerance. J Lipid Res 51: 3306–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RJ, Lagor WR, Sankaranaravanan S, Yasuda T, Quertermous T, Rothblat GH, Rader DJ 2010. Impact of combined deficiency of hepatic lipase and endothelial lipase on the metabolism of both high-density lipoproteins and apolipoprotein B-containing lipoproteins. Circ Res 107: 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunzell JD, Deeb SS 2001. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In The metabolic and molecular bases of inherited disease (ed. Scriver CR, et al.), pp. 2789–2816. McGraw-Hill, New York [Google Scholar]
- Busca R, Pujana MA, Pognonec P, Auwerx J, Deeb SS, Reina M, Vilaro S 1995. Absence of N-glycosylation at asparagine 43 in human lipoprotein lipase induces its accumulation in the rough endoplasmic reticulum and alters this cellular compartment. J Lipid Res 36: 939–951 [PubMed] [Google Scholar]
- Carter SA, Foster NA, Scarpini CG, Chattopadhyay A, Pett MR, Roberts I, Coleman N 2012. Lipoprotein lipase is frequently overexpressed or translocated in cervical squamous cell carcinoma and promotes invasiveness through the non-catalytic C terminus. Br J Cancer 107: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cefalu AB, Noto D, Arpi ML, Yin F, Spina R, Hilden H, Barbagallo CM, Carroccio A, Tarugi P, Squatrito S, et al. 2009. Novel LMF1 nonsense mutation in a patient with severe hypertriglyceridemia. J Clin Endocrinol Metab 94: 4584–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanarin I, Patel A, Slavin G, Wills EJ, Andrews TM, Stewart G 1975. Neutral-lipid storage disease: A new disorder of lipid metabolism. BMJ 1: 553–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda PK, Gao Y, Mark L, Btesh J, Strassle BW, Lu P, Piesla MJ, Zhang MY, Bingham B, Uveges A, et al. 2010. Monoacylglycerol lipase activity is a critical modulator of the tone and integrity of the endocannabinoid system. Mol Pharmacol 78: 996–1003 [DOI] [PubMed] [Google Scholar]
- Chang S-F, Reich B, Brunzell JD, Will H 1998. Detailed characterization of the binding site of the lipoprotein lipase-specific monoclonal antibody 5D2. J Lipid Res 39: 2350–2359 [PubMed] [Google Scholar]
- Charriere S, Peretti N, Bernard S, Di Filippo M, Sassolas A, Merlin M, Delay M, Debard C, Lefai E, Lachaux A, et al. 2011. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J Clin Endocrinol Metab 96: E1675–E1679 [DOI] [PubMed] [Google Scholar]
- Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U 2010. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol 84: 6782–6798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon SH, Douglass JD, Zhou YX, Malik N, Dixon JL, Brinker A, Quadro L, Storch J 2012. Over-expression of monoacylglycerol lipase (MGL) in small intestine alters endocannabinoid levels and whole body energy balance, resulting in obesity. PLoS ONE 7: e43962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotivanich K, Udomsangpetch R, Suwanarusk R, Pukrittayakamee S, Wilairatana P, Beeson JG, Day NP, White NJ 2012. Plasmodium vivax adherence to placental glycosaminoglycans. PLoS ONE 7: e34509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Doll JA, Gattu AK, Shugrue C, Cornwell M, Fitchev P, Crawford SE 2008. Anti-angiogenic pigment epithelium-derived factor regulates hepatocyte triglyceride content through adipose triglyceride lipase (ATGL). J Hepatol 48: 471–478 [DOI] [PubMed] [Google Scholar]
- Cinti S, Mitchell G, Barbatelli G, Murano I, Ceresi E, Faloia E, Wang S, Fortier M, Greenberg AS, Obin MS 2005. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 46: 2347–2355 [DOI] [PubMed] [Google Scholar]
- Cipriani V, Leung HT, Plagnol V, Bunce C, Khan JC, Shahid H, Moore AT, Harding SP, Bishop PN, Hayward C et al. 2012. Genome-wide association study of age-related macular degeneration identifies associated variants in the TNXB–FKBPL–NOTCH4 region of chromosome 6p21.3. Hum Mol Genet 21: 4138–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca-Prieto I, Kroupa O, Gonzalez-Santos P, Magne J, Olivecrona G, Ehrenborg E, Valdivielso P 2011. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: Identification of a novel GPIHBP1 mutation. J Intern Med 270: 224–228 [DOI] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci 105: 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC 2003. Endothelial lipase: Direct evidence for a role in HDL metabolism. J Clin Invest 111: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Wang Z, Grundy SM, Stoesz MR, Guerra R 1994. Variation at the hepatic lipase and apolipoprotein AI/CIII/AIV loci is a major cause of genetically determined variation in plasma HDL cholesterol levels. J Clin Invest 94: 2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Vega GL, Grundy SM 1999. Hepatic lipase: New insights from genetic and metabolic studies. Curr Opin Lipidol 10: 259–267 [DOI] [PubMed] [Google Scholar]
- Connelly PW 1999. The role of hepatic lipase in lipoprotein metabolism. Clin Chim Acta 286: 243–255 [DOI] [PubMed] [Google Scholar]
- Connelly PW, Hegele RA 1998. Hepatic lipase deficiency. Crit Rev Clin Lab Sci 35: 547–572 [DOI] [PubMed] [Google Scholar]
- Connelly PW, Maguire GF, Little JA 1987. Apolipoprotein CIISt. Michael. Familial apolipoprotein CII deficiency associated with premature vascular disease. J Clin Invest 80: 1597–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DW, Wills DE, Quan F, Ray PN 1988. A deletion of one nucleotide results in functional deficiency of apolipoprotein CII (apo CII Toronto). J Med Genet 25: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crecchio C, Capurso A, Pepe G 1990. Identification of the mutation responsible for a case of plasmatic apolipoprotein CII deficiency (Apo CII-Bari). Biochem Biophys Res Commun 168: 1118–1127 [DOI] [PubMed] [Google Scholar]
- Cryer A 1989. The role of the endothelium in myocardial lipoprotein dynamics. Mol Cell Biochem 88: 7–15 [DOI] [PubMed] [Google Scholar]
- Dalrymple N, Mackow ER 2011. Productive dengue virus infection of human endothelial cells is directed by heparan sulfate-containing proteoglycan receptors. J Virol 85: 9478–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, Gorkiewicz G, Tamilarasan KP, Kumari P, Trauner M, et al. 2011. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science 333: 233–238 [DOI] [PubMed] [Google Scholar]
- Datta S, Luo C-C, Li W-H, VanTuinen P, Ledbetter DH, Brown MA, Chen S-H, Liu S-W, Chan L 1988. Human hepatic lipase. Cloned cDNA sequence, restriction fragment length polymorphisms, chromosomal localization, and evolutionary relationships with lipoprotein lipase and pancreatic lipase. J Biol Chem 263: 1107–1110 [PubMed] [Google Scholar]
- Davies BSJ, Beigneux AP, Barnes RH II, Tu Y, Gin P, Weinstein MM, Nobumori C, Nyrén R, Goldberg IJ, Olivecrona G, et al. 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab 12: 42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BS, Goulbourne CN, Barnes RH II, Turlo KA, Gin P, Vaughan S, Vaux DJ, Bensadoun A, Beigneux AP, Fong LG, et al. 2012. Assessing mechanisms of GPIHBP1 and lipoprotein lipase movement across endothelial cells. J Lipid Res 53: 2690–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RC, Wong H, Nikazy J, Wang K, Han Q, Schotz MC 1992. Chimeras of hepatic lipase and lipoprotein lipase. Domain localization of enzyme-specific properties. J Biol Chem 267: 21499–21504 [PubMed] [Google Scholar]
- Dechecchi MC, Melotti P, Bonizzato A, Santacatterina M, Chilosi M, Cabrini G 2001. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J Virol 75: 8772–8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries FP, Cole R, Dankert J, Frosch M, van Putten JP 1998. Neisseria meningitidis producing the Opc adhesin binds epithelial cell proteoglycan receptors. Mol Microbiol 27: 1203–1212 [DOI] [PubMed] [Google Scholar]
- Dichek HL, Qian K, Agrawal N 2004. Divergent effects of the catalytic and bridging functions of hepatic lipase on atherosclerosis. Arterioscler Thromb Vasc Biol 24: 1696–1702 [DOI] [PubMed] [Google Scholar]
- Di Marzo V 2009. The endocannabinoid system: Its general strategy of action, tools for its pharmacological manipulation and potential therapeutic exploitation. Pharmacol Res 60: 77–84 [DOI] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D 2002. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci 99: 10819–10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle MH, Peterfy M 2010. Mechanisms of lipase maturation. Clin Lipidol 5: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman ML, Hershko C, Eisenberg S, Sagher F 1974. Ichthyosiform dermatosis with systemic lipidosis. Arch Dermatol 110: 261–266 [PubMed] [Google Scholar]
- Dorfmeister B, Zeng WW, Dichlberger A, Nilsson SK, Schaap FG, Hubacek JA, Merkel M, Cooper JA, Lookene A, Putt W, et al. 2008. Effects of six APOA5 variants, identified in patients with severe hypertriglyceridemia, on in vitro lipoprotein lipase activity and receptor binding. Arterioscler Thromb Vasc Biol 28: 1866–1871 [DOI] [PubMed] [Google Scholar]
- Dugi KA, Dichek HL, Santamarina-Fojo S 1995. Human hepatic and lipoprotein lipase: The loop covering the catalytic site mediates lipase substrate specificity. J Biol Chem 270: 25396–25401 [DOI] [PubMed] [Google Scholar]
- Duncan RE, Wang Y, Ahmadian M, Lu J, Sarkadi-Nagy E, Sul HS 2010. Characterization of desnutrin functional domains: Critical residues for triacylglycerol hydrolysis in cultured cells. J Lipid Res 51: 309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong M, Psaltis M, Rader DJ, Marchadier D, Barter PJ, Rye KA 2003. Evidence that hepatic lipase and endothelial lipase have different substrate specificities for high-density lipoprotein phospholipids. Biochemistry 42: 13778–13785 [DOI] [PubMed] [Google Scholar]
- Edmondson AC, Brown RJ, Kathiresan S, Cupples LA, Demissie S, Manning AK, Jensen MK, Rimm EB, Wang J, Rodrigues A, et al. 2009. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest 119: 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann TO, Kumari M, Haas JT, Farese RV Jr, Zimmermann R, Lass A, Zechner R 2012. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J Biol Chem 287: 41446–41457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellong EN, Soni KG, Bui QT, Sougrat R, Golinelli-Cohen MP, Jackson CL 2011. Interaction between the triglyceride lipase ATGL and the Arf1 activator GBF1. PLoS ONE 6: e21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerbäck S, Semb H, Bengtsson-Olivecrona G, Carlsson P, Hermansson M-L, Olivecrona T, Bjursell G 1987. Molecular cloning and sequence analysis of cDNA encoding lipoprotein lipase of guinea pig. Gene 58: 1–12 [DOI] [PubMed] [Google Scholar]
- Farese RV Jr, Zechner R, Newgard CB, Walther TC 2012. The problem of establishing relationships between hepatic steatosis and hepatic insulin resistance. Cell Metab 15: 570–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Lefevre C, Morava E, Mussini JM, Laforet P, Negre-Salvayre A, Lathrop M, Salvayre R 2007. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet 39: 28–30 [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Harrison RA, Lachmann PJ, Neuhaus D 1994. Structure of a soluble, glycosylated form of the human complement regulatory protein CD59. Structure 2: 185–199 [DOI] [PubMed] [Google Scholar]
- Franssen R, Young SG, Peelman F, Hertecant J, Sierts JA, Schimmel AWM, Bensadoun A, Kastelein JJP, Fong LG, Dallinga-Thie GM, et al. 2010. Chylomicronemia with sow postheparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ Cardiovasc Genet 3: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frykman PK, Brown MS, Yamamoto T, Goldstein JL, Herz J 1995. Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc Natl Acad Sci 92: 8453–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuki IV, Blanchard N, Jin W, Marchadier DH, Millar JS, Glick JM, Rader DJ 2003. Endogenously produced endothelial lipase enhances binding and cellular processing of plasma lipoproteins via heparan sulfate proteoglycan-mediated pathway. J Biol Chem 278: 34331–34338 [DOI] [PubMed] [Google Scholar]
- Gandotra S, Lim K, Girousse A, Saudek V, O'Rahilly S, Savage DB 2011. Human frame shift mutations affecting the carboxyl terminus of perilipin increase lipolysis by failing to sequester the adipose triglyceride lipase (ATGL) coactivator, AB-hydrolase containing 5 (ABHD5). J Biol Chem 286: 34998–35006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel AS, Kempner ES, Ben-Zeev O, Nikazy J, James SJ, Schotz MC 1983. Lipoprotein lipase: Size of the functional unit determined by radiation inactivation. J Lipid Res 24: 775–780 [PubMed] [Google Scholar]
- Ghosh AK, Ramakrishnan G, Rajasekharan R 2008. YLR099C (ICT1) encodes a soluble Acyl-CoA-dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae. J Biol Chem 283: 9768–9775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gin P, Beigneux AP, Davies B, Young MF, Ryan RO, Bensadoun A, Fong LG, Young SG 2007. Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim Biophys Acta 1771: 1464–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gin P, Yin L, Davies BS, Weinstein MM, Ryan RO, Bensadoun A, Fong LG, Young SG, Beigneux AP 2008. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J Biol Chem 284: 29554–29562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gin P, Beigneux AP, Voss C, Davies BS, Beckstead JA, Ryan RO, Bensadoun A, Fong LG, Young SG 2011. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler Thromb Vasc Biol 31: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gin P, Goulbourne CN, Adeyo O, Beigneux AP, Davies BS, Tat S, Voss CV, Bensadoun A, Fong LG, Young SG 2012. Chylomicronemia mutations yield new insights into interactions between lipoprotein lipase and GPIHBP1. Hum Mol Genet 21: 2961–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger DG, Lewis ME, Ma Y, Jones BR, Liu G, Jones SD 1996. A mutation in the lipoprotein lipase gene Is the molecular basis of chylomicronemia in a colony of domestic cats. J Clin Invest 97: 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzinger DG, Clee SM, Dallongeville J, Lewis ME, Henderson HE, Bauje E, Rogers QR, Jensen DR, Eckel RH, Dyer R, et al. 1999. Lipid and lipoprotein analysis of cats with lipoprotein lipase deficiency. Eur J Clin Invest 29: 17–26 [DOI] [PubMed] [Google Scholar]
- Goldberg IJ, Le NA, Paterniti JR Jr, Ginsberg HN, Lindgren FT, Brown WV 1982. Lipoprotein metabolism during acute inhibition of hepatic triglyceride lipase in the cynomolgus monkey. J Clin Invest 70: 1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg IJ, Soprano DR, Wyatt ML, Vanni TM, Kirchgessner TG, Schotz MC 1989. Localization of lipoprotein lipase mRNA in selected rat tissues. J Lipid Res 30: 1569–1577 [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Granneman RL, Greenberg AS, Obin MS, Zhu Z 2007. Analysis of lipolytic protein trafficking and interactions in adipocytes. J Biol Chem 282: 5726–5735 [DOI] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Krishnamoorthy R, Rathod M 2009. Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J Biol Chem 284: 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman JG, Moore HP, Mottillo EP, Zhu Z, Zhou L 2011. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J Biol Chem 286: 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffon N, Budreck EC, Long CJ, Broedl UC, Marchadier DH, Glick JM, Rader DJ 2006. Substrate specificity of lipoprotein lipase and endothelial lipase: Studies of lid chimeras. J Lipid Res 47: 1803–1811 [DOI] [PubMed] [Google Scholar]
- Griffon N, Jin W, Petty TJ, Millar J, Badellino KO, Saven JG, Marchadier DH, Kempner ES, Billheimer J, Glick JM, et al. 2009. Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J Biol Chem 284: 23322–23330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf I, Baroukh N, Lee SJ, Kamari Y, Harats D, Rubin EM, Pennacchio LA, Cooper AD 2005. Apolipoprotein A-V deficiency results in marked hypertriglyceridemia attributable to decreased lipolysis of triglyceride-rich lipoproteins and removal of their remnants. Arterioscler Thromb Vasc Biol 25: 2573–2579 [DOI] [PubMed] [Google Scholar]
- Gruber A, Cornaciu I, Lass A, Schweiger M, Poeschl M, Eder C, Kumari M, Schoiswohl G, Wolinski H, Kohlwein SD, et al. 2010. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J Biol Chem 285: 12289–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra R, Wang J, Grundy SM, Cohen JC 1997. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci 94: 4532–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haemmerle G, Zimmermann R, Hayn M, Theussl C, Waeg G, Wagner E, Sattler W, Magin TM, Wagner EF, Zechner R 2002. Hormone-sensitive lipase deficiency in mice causes diglyceride accumulation in adipose tissue, muscle, and testis. J Biol Chem 277: 4806–4815 [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Lass A, Zimmermann R, Gorkiewicz G, Meyer C, Rozman J, Heldmaier G, Maier R, Theussl C, Eder S, et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312: 734–737 [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, Hesselink M, Jaeger D, Kienesberger PC, Zierler K, et al. 2011. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat Med 17: 1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn PF 1943. Abolishment of alimentary lipemia following injection of heparin. Science 98: 19–20 [DOI] [PubMed] [Google Scholar]
- Hansen HS, Rosenkilde MM, Holst JJ, Schwartz TW 2012. GPR119 as a fat sensor. Trends Pharmacol Sci 33: 374–381 [DOI] [PubMed] [Google Scholar]
- Harris C, Herker E, Farese RV Jr, Ott M 2011. Hepatitis C virus core protein decreases lipid droplet turnover: A mechanism for core-induced steatosis. J Biol Chem 286: 42615–42625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Ridinger DN, Sutherland S, Emi M, Shuhua Z, Myers RL, Ren K, Cheng T, Inoue I, Wilson DE, et al. 1993. Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J Biol Chem 268: 8447–8457 [PubMed] [Google Scholar]
- Havel RJ 1977. The origin and fate of chylomicrons and VLDL. Adv Exp Med Biol 82: 406–412 [DOI] [PubMed] [Google Scholar]
- Havel RJ 1980. Lipoprotein biosynthesis and metabolism. Ann N Y Acad Sci 348: 16–29 [DOI] [PubMed] [Google Scholar]
- Havel RJ 2010. Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler Thromb Vasc Biol 30: 9–19 [DOI] [PubMed] [Google Scholar]
- Havel RJ, Gordon RS Jr 1960. Idiopathic hyperlipemia: Metabolic studies in an affected family. J Clin Invest 39: 1777–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MR, Ma Y, Brunzell J, Henderson HE 1991. Genetic variants affecting human lipoprotein and hepatic lipases. Curr Opin Lipidol 2: 104–109 [Google Scholar]
- He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, Hobbs HH 2010. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 285: 6706–6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Randall G 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8: 422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci 107: 17345–17350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Zhang X, Xie X, Liu J 2013. The G0/G1 switch gene 2 (G0S2): Regulating metabolism and beyond. Biochim Biophys Acta 1831: 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Little JA, Connelly PW 1991. Compound heterozygosity for mutant hepatic lipase in familial hepatic lipase deficiency. Biochem Biophys Res Commun 179: 78–84 [DOI] [PubMed] [Google Scholar]
- Hegele RA, Little JA, Vezina C, Maguire GF, Tu L, Wolever TS, Jenkins DJ, Connelly PW 1993. Hepatic lipase deficiency. Clinical, biochemical, and molecular genetic characteristics. Arterioscler Thromb 13: 720–728 [DOI] [PubMed] [Google Scholar]
- Henderson HE, Hassan F, Marais D, Hayden MR 1996. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem Biophys Res Commun 227: 189–194 [DOI] [PubMed] [Google Scholar]
- Henderson H, Leisegang F, Hassan F, Hayden M, Marais D 1998. A novel Glu421Lys substitution in the lipoprotein lipase gene in pregnancy-induced hypertriglyceridemic pancreatitis. Clin Chim Acta 269: 1–12 [DOI] [PubMed] [Google Scholar]
- Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med 16: 1295–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JS, Davis RC, Yang D, Schotz MC, Wong H 1997. Hepatic lipase: High-level expression and subunit structure determination. Methods Enzymol 284: 232–246 [DOI] [PubMed] [Google Scholar]
- Hill JS, Yang D, Nikazy J, Curtiss LK, Sparrow JT, Wong H 1998. Subdomain chimeras of hepatic lipase and lipoprotein lipase. Localization of heparin and cofactor binding. J Biol Chem 273: 30979–30984 [DOI] [PubMed] [Google Scholar]
- Hirano K, Ikeda Y, Zaima N, Sakata Y, Matsumiya G 2008. Triglyceride deposit cardiomyovasculopathy. N Engl J Med 359: 2396–2398 [DOI] [PubMed] [Google Scholar]
- Holm C 2003. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Biochem Soc Trans 31: 1120–1124 [DOI] [PubMed] [Google Scholar]
- Holm C, Osterlund T, Laurell H, Contreras JA 2000. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Annu Rev Nutr 20: 365–393 [DOI] [PubMed] [Google Scholar]
- Holmes RS, Cox LA 2012. Comparative studies of glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1: Evidence for a eutherian mammalian origin for the GPIHBP1 gene from an LY6-like gene. 3 Biotech 2: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS, Vandeberg JL, Cox LA 2011a. Comparative studies of vertebrate lipoprotein lipase: A key enzyme of very low density lipoprotein metabolism. Comp Biochem Physiol Part D Genomics Proteomics 6: 224–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS, Vandeberg JL, Cox LA 2011b. Vertebrate endothelial lipase: Comparative studies of an ancient gene and protein in vertebrate evolution. Genetica 139: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RS, Vandeberg JL, Cox LA 2011c. Vertebrate hepatic lipase genes and proteins: A review supported by bioinformatic studies. Open access bioinformatics 2011: 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogewerf AJ, Cisar LA, Evans DC, Bensadoun A 1991. Effect of chlorate on the sulfation of lipoprotein lipase and heparan sulfate proteoglycans. Sulfation of heparan sulfate proteoglycans affects lipoprotein lipase degradation. J Biol Chem 266: 16564–16571 [PubMed] [Google Scholar]
- Hoy AJ, Bruce CR, Turpin SM, Morris AJ, Febbraio MA, Watt MJ 2011. Adipose triglyceride lipase-null mice are resistant to high-fat diet-induced insulin resistance despite reduced energy expenditure and ectopic lipid accumulation. Endocrinology 152: 48–58 [DOI] [PubMed] [Google Scholar]
- Huang Y, Cohen JC, Hobbs HH 2011. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem 286: 37085–37093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igal RA, Rhoads JM, Coleman RA 1997. Neutral lipid storage disease with fatty liver and cholestasis. J Pediatr Gastroenterol Nutr 25: 541–547 [DOI] [PubMed] [Google Scholar]
- Inadera H, Hibino A, Kobayashi J, Kanzaki T, Shirai K, Yukawa S, Saito Y, Yoshida S 1993. A missense mutation (Trp 26 → Arg) in exon 3 of the apolipoprotein CII gene in a patient with apolipoprotein CII deficiency (apo CII-Wakayama). Biochem Biophys Res Commun 193: 1174–1183 [DOI] [PubMed] [Google Scholar]
- Inagaki T, Lin VY, Goetz R, Mohammadi M, Mangelsdorf DJ, Kliewer SA 2008. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab 8: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioka RX, Kang M-J, Kamiyama S, Kim D-H, Magoori K, Kamataki A, Ito Y, Takei YA, Sasaki M, Suzuki T, et al. 2003. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J Biol Chem 278: 7344–7349 [DOI] [PubMed] [Google Scholar]
- Ishida T, Choi S, Kundu RK, Hirata K, Rubin EM, Cooper AD, Quertermous T 2003. Endothelial lipase is a major determinant of HDL level. J Clin Invest 111: 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverius PH, Ostlund-Lindqvist AM 1976. Lipoprotein lipase from bovine milk. Isolation procedure, chemical characterization, and molecular weight analysis. J Biol Chem 251: 7791–7795 [PubMed] [Google Scholar]
- Jaye M, Lynch KJ, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader DJ 1999. A novel endothelial-derived lipase that modulates HDL metabolism. Nat Genet 21: 424–428 [DOI] [PubMed] [Google Scholar]
- Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 279: 48968–48975 [DOI] [PubMed] [Google Scholar]
- Jin W, Millar JS, Broedl U, Glick JM, Rader DJ 2003a. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J Clin Invest 111: 357–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Sun GS, Marchadier D, Octtaviani E, Glick JM, Rader DJ 2003b. Endothelial cells secrete triglyceride lipase and phospholipase activities in response to cytokines as a result of endothelial lipase. Circ Res 92: 644–650 [DOI] [PubMed] [Google Scholar]
- Jin W, Wang X, Millar JS, Quertermous T, Rothblat GH, Glick JM, Rader DJ 2007. Hepatic proprotein convertases modulate HDL metabolism. Cell Metab 6: 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocken JW, Blaak EE 2008. Catecholamine-induced lipolysis in adipose tissue and skeletal muscle in obesity. Physiol Behav 94: 219–230 [DOI] [PubMed] [Google Scholar]
- Jonker JW, Suh JM, Atkins AR, Ahmadian M, Li P, Whyte J, He M, Juguilon H, Yin YQ, Phillips CT, et al. 2012. A PPARγ–FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis. Nature 485: 391–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordans GH 1953. The familial occurrence of fat containing vacuoles in the leukocytes diagnosed in two brothers suffering from dystrophia musculorum progressiva (ERB.). Acta Med Scand 145: 419–423 [DOI] [PubMed] [Google Scholar]
- Jung KM, Clapper JR, Fu J, D'Agostino G, Guijarro A, Thongkham D, Avanesian A, Astarita G, DiPatrizio NV, Frontini A, et al. 2012. 2-Arachidonoylglycerol signaling in forebrain regulates systemic energy metabolism. Cell Metab 15: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaderi MA, Kanduri M, Buhl AM, Sevov M, Cahill N, Gunnarsson R, Jansson M, Smedby KE, Hjalgrim H, Jurlander J, et al. 2011. LPL is the strongest prognostic factor in a comparative analysis of RNA-based markers in early chronic lymphocytic leukemia. Haematologica 96: 1153–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper T, Schneider JG, Dugi KA 2001. Novel site in lipoprotein lipase (LPL415;-438) essential for substrate interaction and dimer stability. J Lipid Res 42: 1180–1186 [PubMed] [Google Scholar]
- Kienesberger PC, Lee D, Pulinilkunnil T, Brenner DS, Cai L, Magnes C, Koefeler HC, Streith IE, Rechberger GN, Haemmerle G, et al. 2009a. Adipose triglyceride lipase deficiency causes tissue-specific changes in insulin signaling. J Biol Chem 284: 30218–30229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger PC, Oberer M, Lass A, Zechner R 2009b. Mammalian patatin domain containing proteins: A family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res 50: S63–S68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen PKJ, Jackson RL, Smith LC, Gotto AM Jr, Sparrow JT 1977. Activation of lipoprotein lipase by native and synthetic fragments of human plasma apolipoprotein C-II. Proc Natl Acad Sci 74: 4848–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgessner TG, Svenson KL, Lusis AJ, Schotz MC 1987. The sequence of cDNA encoding lipoprotein lipase. A member of a lipase gene family. J Biol Chem 262: 8463–8466 [PubMed] [Google Scholar]
- Klinger MM, Margolis RU, Margolis RK 1985. Isolation and characterization of the heparan sulfate proteoglycans of brain. Use of affinity chromatography on lipoprotein lipase-agarose. J Biol Chem 260: 4082–4090 [PubMed] [Google Scholar]
- Kobayashi Y, Nakajima T, Inoue I 2002. Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl-terminal domain. Eur J Biochem 269: 4701–4710 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Inoguchi T, Maeda Y, Nakashima N, Kuwano A, Eto E, Ueno N, Sasaki S, Sawada F, Fujii M, et al. 2008. The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J Clin Endocrinol Metab 93: 2877–2884 [DOI] [PubMed] [Google Scholar]
- Kolditz CI, Langin D 2010. Adipose tissue lipolysis. Curr Opin Clin Nutr Metab Care 13: 377–381 [DOI] [PubMed] [Google Scholar]
- Komaromy MC, Schotz MC 1987. Cloning of rat hepatic lipase cDNA: Evidence for a lipase gene family. Proc Natl Acad Sci 84: 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kompiang IP, Bensadoun A, Yang MW 1976. Effect of an anti-lipoprotein lipase serum on plasma triglyceride removal. J Lipid Res 17: 498–505 [PubMed] [Google Scholar]
- Korn ED 1955a. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J Biol Chem 215: 1–14 [PubMed] [Google Scholar]
- Korn ED 1955b. Clearing factor, a heparin-activated lipoprotein lipase. II. Substrate specificity and activation of coconut oil. J Biol Chem 215: 15–26 [PubMed] [Google Scholar]
- Koster A, Chao YB, Mosior M, Ford A, Gonzalez-DeWhitt PA, Hale JE, Li D, Qiu Y, Fraser CC, Yang DD, et al. 2005. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: Regulation of triglyceride metabolism. Endocrinology 146: 4943–4950 [DOI] [PubMed] [Google Scholar]
- Kraemer FB, Shen WJ, Natu V, Patel S, Osuga J, Ishibashi S, Azhar S 2002. Adrenal neutral cholesteryl ester hydrolase: Identification, subcellular distribution, and sex differences. Endocrinology 143: 801–806 [DOI] [PubMed] [Google Scholar]
- Krauss RM, Windmueller HG, Levy RI, Fredrickson DS 1973. Selective measurement of two different triglyceride lipase activities in rat postheparin plasma. J Lipid Res 14: 286–295 [PubMed] [Google Scholar]
- Kuemmerle NB, Rysman E, Lombardo PS, Flanagan AJ, Lipe BC, Wells WA, Pettus JR, Froehlich HM, Memoli VA, Morganelli PM, et al. 2011. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol Cancer Ther 10: 427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, Wongsiriroj N, Nagy HM, Ivanova PT, Scott SA, et al. 2012. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab 15: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramoto K, Okamura T, Yamaguchi T, Nakamura TY, Wakabayashi S, Morinaga H, Nomura M, Yanase T, Otsu K, Usuda N, et al. 2012. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J Biol Chem 287: 23852–23863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD 2006. Obese yeast: Triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem 281: 491–500 [DOI] [PubMed] [Google Scholar]
- Labar G, Bauvois C, Borel F, Ferrer JL, Wouters J, Lambert DM 2010. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. ChemBioChem 11: 218–227 [DOI] [PubMed] [Google Scholar]
- Lafontan M, Langin D 2009. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 48: 275–297 [DOI] [PubMed] [Google Scholar]
- Lam CW, Yuen YP, Cheng WF, Chan YW, Tong SF 2006. Missense mutation Leu72Pro located on the carboxyl terminal amphipathic helix of apolipoprotein C-II causes familial chylomicronemia syndrome. Clinica Chim Acta 364: 256–259 [DOI] [PubMed] [Google Scholar]
- Langner CA, Birkenmeier EH, Ben-Zeev O, Schotz MC, Sweet HO, Davisson MT, Gordon JI 1989. The fatty liver dystrophy (fld) mutation. A new mutant mouse with a developmental abnormality in triglyceride metabolism and associated tissue-specific defects in lipoprotein lipase and hepatic lipase activities. J Biol Chem 264: 7994–8003 [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Haemmerle G, Riederer M, Schoiswohl G, Schweiger M, Kienesberger P, Strauss JG, Gorkiewicz G, Zechner R 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab 3: 309–319 [DOI] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Oberer M, Zechner R 2011. Lipolysis—a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res 50: 14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin NN, Wang SP, Mitchell GA 2000. The hormone-sensitive lipase gene is transcribed from at least five alternative first exons in mouse adipose tissue. Mamm Genome 11: 972–978 [DOI] [PubMed] [Google Scholar]
- Lefevre C, Jobard F, Caux F, Bouadjar B, Karaduman A, Heilig R, Lakhdar H, Wollenberg A, Verret JL, Weissenbach J, et al. 2001. Mutations in CGI-58, the gene encoding a new protein of the esterase/lipase/thioesterase subfamily, in Chanarin-Dorfman syndrome. Am J Hum Genet 69: 1002–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Huang Y, Karaman R, Ivanova PT, Brown HA, Roddy T, Castro-Perez J, Cohen JC, Hobbs HH 2012. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest 122: 4130–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Ostermeyer-Fay AG, Goldberg EB, Brown WJ, Brown DA 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J Lipid Res 48: 2751–2761 [DOI] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, et al. 2009. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol 5: 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lookene A, Groot NB, Kastelein JJ, Olivecrona G, Bruin T 1997a. Mutation of tryptophan residues in lipoprotein lipase. Effects on stability, immunoreactivity, and catalytic properties. J Biol Chem 272: 766–772 [DOI] [PubMed] [Google Scholar]
- Lookene A, Savonen R, Olivecrona G 1997b. Interaction of lipoproteins with heparan sulfate proteoglycans and with lipoprotein lipase. Studies by surface plasmon resonance technique. Biochemistry 36: 5267–5275 [DOI] [PubMed] [Google Scholar]
- Lookene A, Nielsen MS, Gliemann J, Olivecrona G 2000. Contribution of the carboxy-terminal domain of lipoprotein lipase to interaction with heparin and lipoproteins. Biochem Biophys Res Commun 271: 15–21 [DOI] [PubMed] [Google Scholar]
- Ma Y, Henderson HE, Liu MS, Zhang H, Forsythe IJ, Clarke-Lewis I, Hayden MR, Brunzell JD 1994a. Mutagenesis in four candidate heparin binding regions (residues 279–282, 291–304, 390–393, and 439–448) and identification of residues affecting heparin binding of human lipoprotein lipase. J Lipid Res 35: 2049–2059 [PubMed] [Google Scholar]
- Ma Y, Ooi TC, Liu MS, Zhang H, McPherson R, Edwards AL, Forsythe IJ, Frohlich J, Brunzell JD, Hayden MR 1994b. High frequency of mutations in the human lipoprotein lipase gene in pregnancy-induced chylomicronemia: Possible association with apolipoprotein E2 isoform. J Lipid Res 35: 1066–1075 [PubMed] [Google Scholar]
- Maillard P, Walic M, Meuleman P, Roohvand F, Huby T, Le Goff W, Leroux-Roels G, Pecheur EI, Budkowska A 2011. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS ONE 6: e26637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailly F, Palmen J, Muller DP, Gibbs T, Lloyd J, Brunzell J, Durrington P, Mitropoulos K, Betteridge J, Watts G, et al. 1997. Familial lipoprotein lipase (LPL) deficiency: A catalogue of LPL gene mutations identified in 20 patients from the UK, Sweden, and Italy. Hum Mutat 10: 465–473 [DOI] [PubMed] [Google Scholar]
- Maloum K, Settegrana C, Chapiro E, Cazin B, Lepretre S, Delmer A, Leporrier M, Dreyfus B, Tournilhac O, Mahe B, et al. 2009. IGHV gene mutational status and LPL/ADAM29 gene expression as clinical outcome predictors in CLL patients in remission following treatment with oral fludarabine plus cyclophosphamide. Ann Hematol 88: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Mansouri M, Sevov M, Fahlgren E, Tobin G, Jondal M, Osorio L, Roos G, Olivecrona G, Rosenquist R 2010. Lipoprotein lipase is differentially expressed in prognostic subsets of chronic lymphocytic leukemia but displays invariably low catalytical activity. Leuk Res 34: 301–306 [DOI] [PubMed] [Google Scholar]
- Martin GA, Busch SJ, Meredith GD, Cardin AD, Blankenship DT, Mao SJT, Rechtin AE, Woods CW, Racke MM, Schafer MP, et al. 1988. Isolation and cDNA sequence of human postheparin plasma hepatic triglyceride lipase. J Biol Chem 263: 10907–10914 [PubMed] [Google Scholar]
- Mason RR, Meex RC, Lee-Young R, Canny BJ, Watt MJ 2012. Phosphorylation of adipose triglyceride lipase Ser(404) is not related to 5′-AMPK activation during moderate-intensity exercise in humans. Am J Physiol Endocrinol Metab 303: E534–E541 [DOI] [PubMed] [Google Scholar]
- Menendez JA 2009. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: Molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta 1801: 381–391 [DOI] [PubMed] [Google Scholar]
- Merkel M, Kako Y, Radner H, Cho IS, Ramasamy R, Brunzell JD, Goldberg IJ, Breslow JL 1998. Catalytically inactive lipoprotein lipase expression in muscle of transgenic mice increases very low density lipoprotein uptake: Direct evidence that lipoprotein lipase bridging occurs in vivo. Proc Natl Acad Sci 95: 13841–13846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel M, Eckel RH, Goldberg IJ 2002. Lipoprotein lipase: Genetics, lipid uptake, and regulation. J Lipid Res 43: 1997–2006 [DOI] [PubMed] [Google Scholar]
- Miesenbock G, Holzl B, Foger B, Brandstatter E, Paulweber B, Sandhofer F, Patsch JR 1993. Heterozygous lipoprotein lipase deficiency due to a missense mutation as the cause of impaired triglyceride tolerance with multiple lipoprotein abnormalities. J Clin Invest 91: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097 [DOI] [PubMed] [Google Scholar]
- Montero-Moran G, Caviglia JM, McMahon D, Rothenberg A, Subramanian V, Xu Z, Lara-Gonzalez S, Storch J, Carman GM, Brasaemle DL 2009. CGI-58/ABHD5 is a coenzyme A-dependent lysophosphatidic acid acyltransferase. J Lipid Res 51: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley N, Kuksis A 1972. Positional specificity of lipoprotein lipase. J Biol Chem 247: 6389–6393 [PubMed] [Google Scholar]
- Mottillo EP, Bloch AE, Leff T, Granneman JG 2012. Lipolytic products activate peroxisome proliferator-activated receptor (PPAR) α and δ in brown adipocytes to match fatty acid oxidation with supply. J Biol Chem 287: 25038–25048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M, Lombardi P, Jansen H, van Berkel TJ, Frants RR, Havekes LM 1993. Low density lipoprotein receptor internalizes low density and very low density lipoproteins that are bound to heparan sulfate proteoglycans via lipoprotein lipase. J Biol Chem 268: 9369–9375 [PubMed] [Google Scholar]
- Mulder H, Sorhede-Winzell M, Contreras JA, Fex M, Strom K, Ploug T, Galbo H, Arner P, Lundberg C, Sundler F, et al. 2003. Hormone-sensitive lipase null mice exhibit signs of impaired insulin sensitivity whereas insulin secretion is intact. J Biol Chem 278: 36380–36388 [DOI] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26: 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. 2010. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med 363: 2220–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, et al. 2010. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci 107: 7395–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrolles O, Hernandez-Pando R, Pietri-Rouxel F, Fornes P, Tailleux L, Barrios Payan JA, Pivert E, Bordat Y, Aguilar D, Prevost MC, et al. 2006. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS ONE 1: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF 2010. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF 2011. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol 18: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MC, Ward AM, Hahn YK, Lichtman AH, Conti B, et al. 2012. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science 334: 809–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordestgaard BG, Zilversmit DB 1988. Large lipoproteins are excluded from the arterial wall in diabetic cholesterol-fed rabbits. J Lipid Res 29: 1491–1500 [PubMed] [Google Scholar]
- Nordestgaard BG, Abildgaard S, Wittrup HH, Steffensen R, Jensen G, Tybjaerg-Hansen A 1997. Heterozygous lipoprotein lipase deficiency: Frequency in the general population, effect on plasma lipid levels, and risk of ischemic heart disease. Circulation 96: 1737–1744 [DOI] [PubMed] [Google Scholar]
- Oberer M, Boeszoermenyi A, Nagy HM, Zechner R 2011. Recent insights into the structure and function of comparative gene identification-58. Curr Opin Lipidol 22: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrowsky S, Chandak PG, Patankar JV, Pfeifer T, Povoden S, Schreiber R, Haemmerle G, Levak-Frank S, Kratky D 2012. Cholesteryl ester accumulation and accelerated cholesterol absorption in intestine-specific hormone sensitive lipase-null mice. Biochim Biophys Acta 1821: 1406–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obunike JC, Lutz EP, Li Z, Paka L, Katopodis T, Strickland DK, Kozarsky KF, Pillarisetti S, Goldberg IJ 2001. Transcytosis of lipoprotein lipase across cultured endothelial cells requires both heparan sulfate proteoglycans and the very low density lipoprotein receptor. J Biol Chem 276: 8934–8941 [DOI] [PubMed] [Google Scholar]
- O'Connell DP, LeBlanc DF, Cromley D, Billheimer J, Rader DJ, Bachovchin WW 2012. Design and synthesis of boronic acid inhibitors of endothelial lipase. Bioorg Med Chem Lett 22: 1397–1401 [DOI] [PubMed] [Google Scholar]
- Olafsen T, Young SG, Davies BS, Beigneux AP, Kenanova VE, Voss C, Young G, Wong KP, Barnes RH II, Tu Y, et al. 2010. Unexpected expression pattern for glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1) in mouse tissues revealed by positron emission tomography scanning. J Biol Chem 285: 39239–39248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivecrona G, Beisiegel U 1997. Lipid binding of apolipoprotein CII is required for stimulation of lipoprotein lipase activity against apolipoprotein CII-deficient chylomicrons. Arterioscler Thromb Vasc Biol 17: 1545–1549 [DOI] [PubMed] [Google Scholar]
- Olivecrona T, Bengtsson-Olivecrona G, Chernick SS, Scow RO 1986. Effect of combined lipase deficiency (cld/cld) on hepatic and lipoprotein lipase activities in liver and plasma of newborn mice. Biochim Biophys Acta 876: 243–248 [DOI] [PubMed] [Google Scholar]
- Olivecrona G, Ehrenborg E, Semb H, Makoveichuk E, Lindberg A, Hayden MR, Gin P, Davies BS, Weinstein MM, Fong LG, et al. 2010. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J Lipid Res 51: 1535–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong KT, Mashek MT, Bu SY, Greenberg AS, Mashek DG 2011. Adipose triglyceride lipase is a major hepatic lipase that regulates triacylglycerol turnover and fatty acid signaling and partitioning. Hepatology 53: 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuga J, Ishibashi S, Oka T, Yagyu H, Tozawa R, Fujimoto A, Shionoiri F, Yahagi N, Kraemer FB, Tsutsumi O, et al. 2000. Targeted disruption of hormone-sensitive lipase results in male sterility and adipocyte hypertrophy, but not in obesity. Proc Natl Acad Sci 97: 787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnon J, Matzaris M, Stark R, Meex RC, Macaulay SL, Brown W, O'Brien PE, Tiganis T, Watt MJ 2012. Identification and functional characterization of protein kinase A phosphorylation sites in the major lipolytic protein, adipose triglyceride lipase. Endocrinology 153: 4278–4289 [DOI] [PubMed] [Google Scholar]
- Pappan KL, Pan Z, Kwon G, Marshall CA, Coleman T, Goldberg IJ, McDaniel ML, Semenkovich CF 2005. Pancreatic β-cell lipoprotein lipase independently regulates islet glucose metabolism and normal insulin secretion. J Biol Chem 280: 9023–9029 [DOI] [PubMed] [Google Scholar]
- Park SY, Kim HJ, Wang S, Higashimori T, Dong J, Kim YJ, Cline G, Li H, Prentki M, Shulman GI, et al. 2005. Hormone-sensitive lipase knockout mice have increased hepatic insulin sensitivity and are protected from short-term diet-induced insulin resistance in skeletal muscle and heart. Am J Physiol Endocrinol Metab 289: E30–E39 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart J-C, Krauss RM, Rubin EM 2001. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294: 169–173 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC 2002. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet 11: 3031–3038 [DOI] [PubMed] [Google Scholar]
- Peterfy M, Ben-Zeev O, Mao HZ, Weissglas-Volkov D, Aouizerat BE, Pullinger CR, Frost PH, Kane JP, Malloy MJ, Reue K, et al. 2007. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet 39: 1483–1487 [DOI] [PubMed] [Google Scholar]
- Peyot ML, Guay C, Latour MG, Lamontagne J, Lussier R, Pineda M, Ruderman NB, Haemmerle G, Zechner R, Joly E, et al. 2009. Adipose triglyceride lipase is implicated in fuel- and non-fuel-stimulated insulin secretion. J Biol Chem 284: 16848–16859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA, et al. 2008. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science 322: 1702–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previato L, Guardamagna O, Brunzell JD, Santamarina-Fojo S, Brewer HB Jr 1992. A novel missense mutation in the C-terminal domain of lipoprotein lipase leads to enzyme inactivation. Circulation 86: 1–608 [PubMed] [Google Scholar]
- Quiroga AD, Lehner R 2012. Liver triacylglycerol lipases. Biochim Biophys Acta 1821: 762–769 [DOI] [PubMed] [Google Scholar]
- Radner FP, Streith IE, Schoiswohl G, Schweiger M, Kumari M, Eichmann TO, Rechberger G, Koefeler HC, Eder S, Schauer S, et al. 2010. Growth retardation, impaired triacylglycerol catabolism, hepatic steatosis, and lethal skin barrier defect in mice lacking comparative gene identification-58 (CGI-58). J Biol Chem 285: 7300–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Daum G 2010. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell 21: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BN, Ables GP, Otlivanchik OA, Schoiswohl G, Zechner R, Blaner WS, Goldberg IJ, Schwabe RF, Chua SC Jr, Huang LS 2008. Hepatic overexpression of hormone-sensitive lipase and adipose triglyceride lipase promotes fatty acid oxidation, stimulates direct release of free fatty acids, and ameliorates steatosis. J Biol Chem 283: 13087–13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina M, Brunzell JD, Deeb SS 1992. Molecular basis of familial chylomicronemia: Mutations in the lipoprotein lipase and apolipoprotein C-II genes. J Lipid Res 33: 1823–1832 [PubMed] [Google Scholar]
- Rios JJ, Shastry S, Jasso J, Hauser N, Garg A, Bensadoun A, Cohen JC, Hobbs HH 2011. Deletion of GPIHBP1 causing severe chylomicronemia. J Inherit Metab Dis 35: 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Ben Ali Y, Abdelkafi S, Mendoza LD, Leclaire J, Fotiadu F, Buono G, Carriere F, Abousalham A 2010. In vitro stereoselective hydrolysis of diacylglycerols by hormone-sensitive lipase. Biochim Biophys Acta 1801: 77–83 [DOI] [PubMed] [Google Scholar]
- Roduit R, Masiello P, Wang SP, Li H, Mitchell GA, Prentki M 2001. A role for hormone-sensitive lipase in glucose-stimulated insulin secretion: A study in hormone-sensitive lipase-deficient mice. Diabetes 50: 1970–1975 [DOI] [PubMed] [Google Scholar]
- Rogalska E, Cudrey C, Ferrato F, Verger R 1993. Stereoselective hydrolysis of triglycerides by animal and microbial lipases. Chirality 5: 24–30 [DOI] [PubMed] [Google Scholar]
- Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC 2007. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet 39: 513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH 2008. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40: 1461–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Yin W, Kozlitina J, Pennacchio LA, Boerwinkle E, Hobbs HH, Cohen JC 2009. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest 119: 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, Forsdyke DR 1991. A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell Biol 10: 581–591 [DOI] [PubMed] [Google Scholar]
- Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser–Asp catalytic dyad. Biochemistry 42: 6696–6708 [DOI] [PubMed] [Google Scholar]
- Ryden M, Agustsson T, Laurencikiene J, Britton T, Sjolin E, Isaksson B, Permert J, Arner P 2008. Lipolysis—not inflammation, cell death, or lipogenesis—is involved in adipose tissue loss in cancer cachexia. Cancer 113: 1695–1704 [DOI] [PubMed] [Google Scholar]
- Sanan DA, Fan J, Bensadoun A, Taylor JM 1997. Hepatic lipase is abundant on both hepatocyte and endothelial cell surfaces in the liver. J Lipid Res 38: 1002–1013 [PubMed] [Google Scholar]
- Sanchez-Pinera P, Micol V, Corbalan-Garcia S, Gomez-Fernandez JC 1999. A comparative study of the activation of protein kinase C α by different diacylglycerol isomers. Biochem J 337: 387–395 [PMC free article] [PubMed] [Google Scholar]
- Santamarina-Fojo S, Gonzalez-Navarro H, Freeman L, Wagner E, Nong Z 2004. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler Thromb Vasc Biol 24: 1750–1754 [DOI] [PubMed] [Google Scholar]
- Sapiro JM, Mashek MT, Greenberg AS, Mashek DG 2009. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor α activity. J Lipid Res 50: 1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, et al. 2010. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci 13: 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, Tornqvist H, Zechner R, Zimmermann R 2006. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem 281: 40236–40241 [DOI] [PubMed] [Google Scholar]
- Schweiger M, Schoiswohl G, Lass A, Radner FP, Haemmerle G, Malli R, Graier W, Cornaciu I, Oberer M, Salvayre R, et al. 2008. The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J Biol Chem 283: 17211–17220 [DOI] [PubMed] [Google Scholar]
- Schweiger M, Lass A, Zimmermann R, Eichmann TO, Zechner R 2009. Neutral lipid storage disease: Genetic disorders caused by mutations in adipose triglyceride lipase/PNPLA2 or CGI-58/ABHD5. Am J Physiol Endocrinol Metab 297: E289–E296 [DOI] [PubMed] [Google Scholar]
- Schweiger M, Paar M, Eder C, Brandis J, Moser E, Gorkiewicz G, Grond S, Radner FP, Cerk I, Cornaciu I, et al. 2012. G0/G1 switch gene-2 regulates human adipocyte lipolysis by affecting activity and localization of adipose triglyceride lipase. J Lipid Res 53: 2307–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya M, Osuga J, Yahagi N, Okazaki H, Tamura Y, Igarashi M, Takase S, Harada K, Okazaki S, Iizuka Y, et al. 2008. Hormone-sensitive lipase is involved in hepatic cholesteryl ester hydrolysis. J Lipid Res 49: 1829–1838 [DOI] [PubMed] [Google Scholar]
- Semenkovich CF, Luo CC, Nakanishi MK, Chen SH, Smith LC, Chan L 1990. In vitro expression and site-specific mutagenesis of the cloned human lipoprotein lipase gene. Potential N-linked glycosylation site asparagine 43 is important for both enzyme activity and secretion. J Biol Chem 265: 5429–5433 [PubMed] [Google Scholar]
- Sendak RA, Melford K, Kao A, Bensadoun A 1998. Identification of the epitope of a monoclonal antibody that inhibits heparin binding of lipoprotein lipase: New evidence for a carboxyl-terminal heparin-binding domain. J Lipid Res 39: 633–646 [PubMed] [Google Scholar]
- Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, Santini L, Biolchi A, Seib KL, Giuliani MM, et al. 2010. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci 107: 3770–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lookene A, Zhang L, Olivecrona G 2010. Site-directed mutagenesis of apolipoprotein CII to probe the role of its secondary structure for activation of lipoprotein lipase. J Biol Chem 285: 7484–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WJ, Yu Z, Patel S, Jue D, Liu LF, Kraemer FB 2011. Hormone-sensitive lipase modulates adipose metabolism through PPARγ. Biochim Biophys Acta 1811: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Gill PJ, Silbert JE, Douglas WH, Fanburg BL 1981. Involvement of cell surface heparin sulfate in the binding of lipoprotein lipase to cultured bovine endothelial cells. J Clin Invest 68: 995–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Hishiki T, Sugiyama K, Ogawa K, Funami K, Kato A, Ohsaki Y, Fujimoto T, Takaku H, Shimotohno K 2010. Lipoprotein lipase and hepatic triglyceride lipase reduce the infectivity of hepatitis C virus (HCV) through their catalytic activities on HCV-associated lipoproteins. Virology 407: 152–159 [DOI] [PubMed] [Google Scholar]
- Soni KG, Mardones GA, Sougrat R, Smirnova E, Jackson CL, Bonifacino JS 2009. Coatomer-dependent protein delivery to lipid droplets. J Cell Sci 122: 1834–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler KR, Welton AR, Lim ES, Duvvuru S, Althaus IW, Imperiale JE, Daoud AI, Chesler EJ 2010. The major locus for mouse adenovirus susceptibility maps to genes of the hematopoietic cell surface-expressed LY6 family. J Immunol 184: 3055–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner G, Myher JJ, Kuksis A 1985. Milk and plasma lipid composition in a lactating patient with type I hyperlipoproteinemia. Am J Clin Nutr 41: 121–128 [DOI] [PubMed] [Google Scholar]
- Strauss JG, Frank S, Kratky D, Hammerle G, Hrzenjak A, Knipping G, von Eckardstein A, Kostner GM, Zechner R 2001. Adenovirus-mediated rescue of lipoprotein lipase-deficient mice. Lipolysis of triglyceride-rich lipoproteins is essential for high density lipoprotein maturation in mice. J Biol Chem 276: 36083–36090 [DOI] [PubMed] [Google Scholar]
- Strom K, Hansson O, Lucas S, Nevsten P, Fernandez C, Klint C, Moverare-Skrtic S, Sundler F, Ohlsson C, Holm C 2008. Attainment of brown adipocyte features in white adipocytes of hormone-sensitive lipase null mice. PLoS ONE 3: e1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom K, Gundersen TE, Hansson O, Lucas S, Fernandez C, Blomhoff R, Holm C 2009. Hormone-sensitive lipase (HSL) is also a retinyl ester hydrolase: Evidence from mice lacking HSL. FASEB J 23: 2307–2316 [DOI] [PubMed] [Google Scholar]
- Sukonina V, Lookene A, Olivecrona T, Olivecrona G 2006. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci 103: 17450–17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Lazar MA 2013. Dissociating fatty liver and diabetes. Trends Endocrinol Metab 24: 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Jiang L, Luo X, Jin W, He Q, An J, Lui K, Shi J, Rong R, Su W, et al. 2013. Potential tumor-suppressive role of monoglyceride lipase in human colorectal cancer. Oncogene 32: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran RP, Visser ME, Heemelaar S, Wang J, Peter J, Defesche JC, Kuivenhoven JA, Hosseini M, Peterfy M, Kastelein JJ, et al. 2012. Mutations in LPL, APOC2, APOA5, GPIHBP1 and LMF1 in patients with severe hypertriglyceridaemia. J Intern Med 272: 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschler U, Radner FP, Heier C, Schreiber R, Schweiger M, Schoiswohl G, Preiss-Landl K, Jaeger D, Reiter B, Koefeler HC, et al. 2011. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J Biol Chem 286: 17467–17477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomssen R, Bonk S 2002. Virolytic action of lipoprotein lipase on hepatitis C virus in human sera. Med Microbiol Immunol (Berl) 191: 17–24 [DOI] [PubMed] [Google Scholar]
- Tornqvist H, Belfrage P 1976. Purification and some properties of a monoacylglycerol-hydrolyzing enzyme of rat adipose tissue. J Biol Chem 251: 813–819 [PubMed] [Google Scholar]
- Turpin SM, Hoy AJ, Brown RD, Rudaz CG, Honeyman J, Matzaris M, Watt MJ 2011. Adipose triacylglycerol lipase is a major regulator of hepatic lipid metabolism but not insulin sensitivity in mice. Diabetologia 54: 146–156 [DOI] [PubMed] [Google Scholar]
- Tuzgol S, Bijvoet SM, Bruin T, Kastelein JJ, Hayden MR 1994. Apolipoprotein CII-Padova (Tyr37 → stop) as a cause of chylomicronaemia in an Italian kindred from Siculiana. J Med Genet 31: 622–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Cho Y, Moradian S, Kim J, Nakajima K, Crumrine D, Park K, Ujihara M, Akiyama M, Shimizu H, et al. 2010. Neutral lipid storage leads to acylceramide deficiency, likely contributing to the pathogenesis of Dorfman-Chanarin syndrome. J Invest Dermatol 130: 2497–2499 [DOI] [PubMed] [Google Scholar]
- Ullrich NF, Purnell JQ, Brunzell JD 2001. Adipose tissue fatty acid composition in humans with lipoprotein lipase deficiency. J Investig Med 49: 273–275 [PubMed] [Google Scholar]
- Vallet-Erdtmann V, Tavernier G, Contreras JA, Mairal A, Rieu C, Touzalin AM, Holm C, Jegou B, Langin D 2004. The testicular form of hormone-sensitive lipase HSLtes confers rescue of male infertility in HSL-deficient mice. J Biol Chem 279: 42875–42880 [DOI] [PubMed] [Google Scholar]
- Van Bockstaele F, Pede V, Janssens A, Callewaert F, Offner F, Verhasselt B, Philippe J 2007. Lipoprotein lipase mRNA expression in whole blood is a prognostic marker in B cell chronic lymphocytic leukemia. Clin Chem 53: 204–212 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ 2009. Cold-activated brown adipose tissue in healthy men. N Engl J Med 360: 1500–1508 [DOI] [PubMed] [Google Scholar]
- Vatannejad A, Khodadadi I, Amiri I, Vaisi-Raygani A, Ghorbani M, Tavilani H 2011. Genetic variation of hormone sensitive lipase and male infertility. Syst Biol Reprod Med 57: 288–291 [DOI] [PubMed] [Google Scholar]
- Vaughan M, Berger JE, Steinberg D 1964. Hormone-sensitive lipase and monoglyceride lipase activities in adipose tissue. J Biol Chem 239: 401–409 [PubMed] [Google Scholar]
- Vega GL, Clark LT, Tang A, Marcovina S, Grundy SM, Cohen JC 1998. Hepatic lipase activity is lower in African American men than in white American men: Effects of 5′ flanking polymorphism in the hepatic lipase gene (LIPC). J Lipid Res 39: 228–232 [PubMed] [Google Scholar]
- Vilaro S, Camps L, Reina M, Perez-Clausell J, Llobera M, Olivecrona T 1990. Localization of lipoprotein lipase to discrete areas of the guinea pig brain. Brain Res 506: 249–253 [DOI] [PubMed] [Google Scholar]
- Villena JA, Roy S, Sarkadi-Nagy E, Kim KH, Sul HS 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: Ectopic expression of desnutrin increases triglyceride hydrolysis. J Biol Chem 279: 47066–47075 [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. 2009. Functional brown adipose tissue in healthy adults. N Engl J Med 360: 1518–1525 [DOI] [PubMed] [Google Scholar]
- Vogt AM, Barragan A, Chen Q, Kironde F, Spillmann D, Wahlgren M 2003. Heparan sulfate on endothelial cells mediates the binding of Plasmodium falciparum-infected erythrocytes via the DBL1α domain of PfEMP1. Blood 101: 2405–2411 [DOI] [PubMed] [Google Scholar]
- Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Holm H, Ding EL, Johnson T, et al. 2012. Plasma HDL cholesterol and risk of myocardial infarction: A Mendelian randomisation study. Lancet 380: 572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voshol PJ, Haemmerle G, Ouwens DM, Zimmermann R, Zechner R, Teusink B, Maassen JA, Havekes LM, Romijn JA 2003. Increased hepatic insulin sensitivity together with decreased hepatic triglyceride stores in hormone-sensitive lipase-deficient mice. Endocrinology 144: 3456–3462 [DOI] [PubMed] [Google Scholar]
- Voss CV, Davies BS, Tat S, Gin P, Fong LG, Pelletier C, Mottler CD, Bensadoun A, Beigneux AP, Young SG 2011. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci 108: 7980–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SP, Laurin N, Himms-Hagen J, Rudnicki MA, Levy E, Robert MF, Pan L, Oligny L, Mitchell GA 2001. The adipose tissue phenotype of hormone-sensitive lipase deficiency in mice. Obes Res 9: 119–128 [DOI] [PubMed] [Google Scholar]
- Wang S, Soni KG, Semache M, Casavant S, Fortier M, Pan L, Mitchell GA 2008. Lipolysis and the integrated physiology of lipid energy metabolism. Mol Genet Metab 95: 117–126 [DOI] [PubMed] [Google Scholar]
- Wang H, Astarita G, Taussig MD, Bharadwaj KG, DiPatrizio NV, Nave KA, Piomelli D, Goldberg IJ, Eckel RH 2011a. Deficiency of lipoprotein lipase in neurons modifies the regulation of energy balance and leads to obesity. Cell Metab 13: 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Bell M, Sreenevasan U, Hu H, Liu J, Dalen K, Londos C, Yamaguchi T, Rizzo MA, Coleman R, et al. 2011b. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem 286: 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein MM, Yin L, Beigneux AP, Davies BS, Gin P, Estrada K, Melford K, Bishop JR, Esko JD, Dallinga-Thie GM, et al. 2008. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem 283: 34511–34518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein MM, Yin L, Tu Y, Wang X, Wu X, Castellani LW, Walzem RL, Lusis AJ, Fong LG, Beigneux AP, et al. 2010. Chylomicronemia elicits atherosclerosis in mice–brief report. Arterioscler Thromb Vasc Biol 30: 20–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein MM, Goulbourne CN, Davies BS, Tu Y, Barnes RH II, Watkins SM, Davis R, Reue K, Tontonoz P, Beigneux AP, et al. 2012. Reciprocal metabolic perturbations in the adipose tissue and liver of GPIHBP1-deficient mice. Arterioscler Thromb Vasc Biol 32: 230–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock PH, Bisgaier CL, Aalto-Setälä K, Radner H, Ramakrishnan R, Levak-Frank S, Essenburg AD, Zechner R, Breslow JL 1995. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired low density lipoprotein clearance in heterozygotes. J Clin Invest 96: 2555–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead RH 1909. A note on the absorption of fat. Am J Physiol 24: 294–296 [Google Scholar]
- Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ 2006. Characterization of the human patatin-like phospholipase family. J Lipid Res 47: 1940–1949 [DOI] [PubMed] [Google Scholar]
- Wion KL, Kirchgessner TG, Lusis AJ, Schotz MC, Lawn RM 1987. Human lipoprotein lipase complementary DNA sequence. Science 235: 1638–1641 [DOI] [PubMed] [Google Scholar]
- Wong H, Davis RC, Thuren T, Goers JW, Nikazy J, Waite M, Schotz MC 1994. Lipoprotein lipase domain function. J Biol Chem 269: 10319–10323 [PubMed] [Google Scholar]
- Wong H, Yang D, Hill JS, Davis RC, Nikazy J, Schotz MC 1997. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci 94: 5594–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Wang SP, Alvarez F, Casavant S, Gauthier N, Abed L, Soni KG, Yang G, Mitchell GA 2011. Deficiency of liver adipose triglyceride lipase in mice causes progressive hepatic steatosis. Hepatology 54: 122–132 [DOI] [PubMed] [Google Scholar]
- Yang X, Lu X, Liu J 2010a. Identification of a novel splicing isoform of murine CGI-58. FEBS Lett 584: 903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lu X, Lombes M, Rha GB, Chi YI, Guerin TM, Smart EJ, Liu J 2010b. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab 11: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Hirata K, Ishida T, Kojima Y, Tanaka H, Okada T, Quertermous T, Yokoyama M 2007. Endothelial lipase is increased by inflammation and promotes LDL uptake in macrophages. J Atheroscler Thromb 14: 192–201 [DOI] [PubMed] [Google Scholar]
- Yasuda T, Ishida T, Rader DJ 2010. Update on the role of endothelial lipase in high-density lipoprotein metabolism, reverse cholesterol transport, and atherosclerosis. Circ J 74: 2263–2270 [DOI] [PubMed] [Google Scholar]
- Ye L, Zhang B, Seviour EG, Tao KX, Liu XH, Ling Y, Chen JY, Wang GB 2011. Monoacylglycerol lipase (MAGL) knockdown inhibits tumor cells growth in colorectal cancer. Cancer Lett 307: 6–17 [DOI] [PubMed] [Google Scholar]
- Yin W, Romeo S, Chang S, Grishin NV, Hobbs HH, Cohen JC 2009. Genetic variation in ANGPTL4 provides insights into protein processing and function. J Biol Chem 284: 13213–13222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Shimizugawa T, Ono M, Furukawa H 2002. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res 43: 1770–1772 [DOI] [PubMed] [Google Scholar]
- Young SG, Davies BS, Voss CV, Gin P, Weinstein MM, Tontonoz P, Reue K, Bensadoun A, Fong LG, Beigneux AP 2011. GPIHBP1, an endothelial cell transporter for lipoprotein lipase. J Lipid Res 52: 1869–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Rieke CJ, Rimon G, Wingerd BA, Smith WL 2006. Partnering between monomers of cyclooxygenase-2 homodimers. Proc Natl Acad Sci 103: 6142–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C, Sidhu RS, Kuklev DV, Kado Y, Wada M, Song I, Smith WL 2009. Cyclooxygenase allosterism, fatty acid-mediated cross-talk between monomers of cyclooxygenase homodimers. J Biol Chem 284: 10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res 50: 3–21 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Repa JJ, Gauthier K, Mangelsdorf DJ 2001. Regulation of lipoprotein lipase by the oxysterol receptors, LXRα and LXRβ. J Biol Chem 276: 43018–43024 [DOI] [PubMed] [Google Scholar]
- Zhang X, Qi R, Xian X, Yang F, Blackstein M, Deng X, Fan J, Ross C, Karasinska J, Hayden MR, et al. 2008. Spontaneous atherosclerosis in aged lipoproteinl lipase deficient mice with wevere hypertriglyceridemia on a normal chow diet. Circ Res 102: 250–256 [DOI] [PubMed] [Google Scholar]
- Zhu H, Cong JP, Yu D, Bresnahan WA, Shenk TE 2002. Inhibition of cyclooxygenase 2 blocks human cytomegalovirus replication. Proc Natl Acad Sci 99: 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann R, Haemmerle G, Wagner EM, Strauss JG, Kratky D, Zechner R 2003. Decreased fatty acid esterification compensates for the reduced lipolytic activity in hormone-sensitive lipase-deficient white adipose tissue. J Lipid Res 44: 2089–2099 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, Lass A, Neuberger G, Eisenhaber F, Hermetter A, et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306: 1383–1386 [DOI] [PubMed] [Google Scholar]
- Ziouzenkova O, Perrey S, Asatryan L, Hwang J, MacNaul KL, Moller DE, Rader DJ, Sevanian A, Zechner R, Hoefler G, et al. 2003. Lipolysis of triglyceride-rich lipoproteins generates PPAR ligands: Evidence for an antiinflammatory role for lipoprotein lipase. Proc Natl Acad Sci 100: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]