Abstract
G protein-coupled receptors (GPCRs) mediate transmembrane signaling. Before ligand binding, GPCRs exist in a basal state. Crystal structures of several GPCRs bound with antagonists or agonists have been solved. However, the crystal structure of the ligand-free basal state of a GPCR, the starting point of GPCR activation and function, has not been determined. Here we report the X-ray crystal structure of the first ligand-free basal state of a GPCR in a lipid membrane-like environment. Oligomeric turkey β1-adrenergic receptors display two alternating dimer interfaces. One interface involves the transmembrane domain (TM) 1, TM2, the C-terminal H8, and the extracellular loop 1. The other interface engages residues from TM4, TM5, the intracellular loop 2 and the extracellular loop 2. Structural comparisons show that this ligand-free state is in an inactive conformation. This provides the structural information regarding GPCR dimerization and oligomerization.
INTRODUCTION
G protein-coupled receptors (GPCRs) are transmembrane proteins that act as key gatekeepers between external signals and cellular responses1,2. These receptors are activated by a diverse array of ligands, including photons, odorants, chemokines, hormones, growth factors and neurotransmitters. GPCRs play critical roles in regulating many physiological functions of eukaryotic cells3. They constitute the largest group of cell surface receptors involved in signal transduction, and have been one of the best pharmaceutical drug targets4,5. Both endogenous and exogenous substances can modulate the activity of GPCRs. An agonist increases the activity of its GPCR above the basal level, presumably through shifting GPCRs into an active state capable of interacting with downstream signaling G proteins. An inverse agonist decreases the GPCR activity below its basal level, likely by stabilizing GPCRs in an inactive state uncoupled from G proteins. A neutral antagonist itself has no effect on the receptor activity but can prevent the interaction of agonists or inverse agonists with GPCRs, while they do not affect the equilibrium of different GPCR conformations6.
Crystal structures of several GPCRs have been determined7–24. Most of these GPCRs were bound with antagonists or agonists. No crystal structures of the ligand-free basal states of GPCRs have been determined, except in the unusual case of rhodopsin7. Rhodopsin is a special case among GPCRs because, in its basal state, rhodopsin is covalently bound with its inverse agonist cis-retinal. It is activated by photon absorption that isomerizes cis-retinal to the agonist all-trans-retinal25. Structural studies have shown that, while the basal (inverse agonist-bound) rhodopsin adopts an inactive state, the ligand-free opsin has an active state conformation7,12. We set out to determine the crystal structure of a GPCR without ligands. Here we report the X-ray crystal structure of the β1-adrenerigc receptor (β1-AR) in a ligand-free basal state. In this structure, β1-AR forms oligomers in a lipid membrane-like environment, and adopts an inactive state conformation.
RESULTS
Structure determination
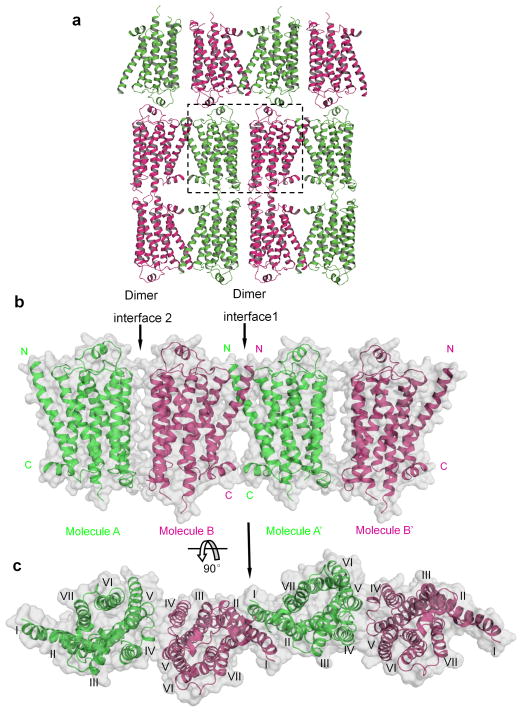
We determined the X-ray crystal structure of turkey β1-AR without ligands in the presence of synthetic lipids (Fig. 1, Table 1, and Supplementary Fig. 1, 2). Several complete data sets from individual crystals were recorded and they gave similar structures. Only one representative data set from a single crystal was described here (Table 1). Diffraction of the crystal was highly anisotropic, and the structure was solved and refined at 3.5 Å resolution. The overall quality of the electron density map was high. The structure of the ligand-free state of β1-AR was determined by molecular replacement using the partial agonist (salbutamol)-bound β1-AR as a search model18. Crystal packing indicated a membrane-like environment and was largely impacted by lipids in one direction (Fig. 1, Supplementary Fig. 2a and b). β1-ARs parallely packed as oligomers with two distinct dimer interfaces (Fig. 1a–c). Each crystallographic asymmetric unit had twoβ1-AR molecules (Supplementary Fig. 2c). These two β1-AR molecules had similar configurations; the root mean square deviation was ~0.02 Å for all equivalent Cα atoms (Supplementary Fig. 2d). Although most of the experiments (including screening for crystallization conditions) described in this paper were done with the construct β1-AR(H0), the presented ligand-free structure was from the construct β1-AR(m23) since it gave a slightly higher resolution (Supplementary Fig. 1).β1-AR(H0) is with deletions of amino acid sequences of 3–32, of 249–283 and of 366–483 and with point mutations of Cys116Leu and Cys358Ala (Supplementary Figure 1).β1-AR(m23) is a thermostabilized β1-AR mutant that was used in previous crystal structural studies with antagonists or agonists 6,7.
Figure 1.
Structure of the ligand-free basal state β1-AR. a, β1-AR crystallographic packing. The dashed box indicates one crystallographic asymmetric unit. Chain A: green; chain B: magenta. b and c, Molecular surface representation of oligomers of β1-AR. Within the same layer, β1-ARs form oligomers with two dimer interfaces. The N- and C-termini are indicated. c, Top view (from the extracellular surface) of the β1-AR oligomers. The TMs are labeled as I to VII.
Table 1.
Data collection and refinement statistics
| Structure of ligand-free β1-ARa | |
|---|---|
| Data collection b | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 229.66, 79.59, 69.04 |
| αβγ(°) | 90, 101.83, 90 |
| Resolution (Å) | 67.57-3.35 (3.44-3.35) c |
| Rmerge | 0.141 (>1.0) |
| I/σ(I) | 6.3 (1.7) |
| Completeness (%) | 98.2 (97.0) |
| Redundancy | 4.3 (4.2) |
| Refinement | |
| Resolution (Å) | 29.78-3.50 |
| No. reflections (test set) | 13006 (642) |
| Rwork/Rfree (%) | 30.99/35.46 |
| No. atoms | |
| Protein | 4442 |
| Overall B factor (Å2) d | 79.3 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.136 |
One crystal was used for data collection and refinement.
The data set was anisotropically truncated to 3.3 × 3.3 × 4.3 Å after merging and scaling.
Values in parentheses are for highest-resolution shell.
An additional isotropic B factor of − 54.13 was applied to the scaled data for map sharpening.
Receptor oligomerization: the TM1-TM2-H8 dimer interface
Within the same lipid bilayer, oligomers of β1-ARs were parallely packed with two alternating dimer interfaces (Figs. 1b, 1c, 2 and 3). This oligomeric architecture was remarkably similar to the proposed models for oligomeric GPCRs based on a large body of functional data26–34. GPCRs can exist and function as dimers or oligomers 26–29,35. Dimerization and oligomerization modulate various GPCR functions such as cell surface targeting, cooperativity, activation, G-protein coupling, signaling and internalization28,29,36. In previous crystallographic studies with β1-ARs bound with antagonists or agonists, β1-ARs were observed as anti-parallel dimers 10,18. The lack of oligomeric arrangement in those studies might be due to the exclusion of phospholipids in the crystallization conditions. Here, in one dimer interface (dimer interface 1), the interaction was mainly through TM1 as well as some residues from the C-terminal helical domain H8, TM2, and the extracellular loop 1 (Figs. 1b, 1c, and 2). The total buried contact surface (from both protomers) was ~1700 Å2 (Fig. 2a and b). Interacting residues were mainly from TM1 (including Gln38, Gln39, Ala42, Leu46, Ala49, Leu50, Val52, Leu53, and Leu54) (Fig. 2c). Residues from other parts of the receptor also contributed to this dimer interface, including residues from TM2 (Pro96, Ala99, Thr100 and Val103) (Fig. 2c), the extracellular loop 1 (Thr106, Leu108, and Trp109) (Fig. 2c and d), and the C-terminal H8 (Arg351, Lys354, Arg355 and Leu356) (Fig. 2e). In addition to these hydrophobic and van der Waals interactions, Ser45 in one TM1 formed a hydrogen bond with Ser45 from another TM1 (Fig. 2c). Glu41 in TM1 from one monomer formed a salt bridge with Arg104 in TM2 from the second monomer (Fig. 2c). This dimer interface is similar to the one observed in the dimer of rhodopsin in the active state which uses TM1 and H8 as an interface12,13.
Figure 2.
Dimer interface 1 of β1-AR oligomers. a and b, The surface involved in dimer interface 1 is highlighted in green (chain A) and in magenta (chain B). The helix 8 is labeled as VIII and the extracellular loop 1 as ECL1. c and d, Residues in TM1, TM2 and ECL1 are involved in the dimer formation. e, Residues in H8 are involved in the dimer formation.
Figure 3.
Dimer interface 2 of β1-AR oligomers. a and b, The surface involved in dimer interface 2 is highlighted in green (chain A) and in magenta (chain B). The intracellular loop 2 is labeled as ICL2 and the extracellular loop 2 as ECL2. c, Residues in TM5 and ECL2 are involved in dimer formation. d, Residues in TM4 and ICL2 are involved in dimer formation. e. Disulfide crosslinking experiments with Cys mutants of β1-AR with copper phenanthroline. One representative experiment of three is shown (left panel). The dimer fraction is quantified as dimer/(monomer + dimer). Results are means and s.d. (n = 3; *, p < 0.05; **, p < 0.001, Student’s t-test) (right panel).
Receptor oligomerization: the TM4-TM5-ICL2 dimer interface
In the second dimer interface, the interacting regions involved residues from TM4, TM5, the intracellular loop 2 (ICL2) and the extracellular loop 2 (Figs. 1b, 1c, and 3). The total buried surface (from both receptors) was ~900 Å2 (Fig. 3a and b). Residues from both TM4 (including Leu171) and TM5 (including Arg205, Ala206, Ala210, Ile218 and Arg229) contributed to the hydrophobic interaction (Fig. 3c and d). The intracellular loop 2 also played a critical role in this dimer interaction (including residues Tyr140, Leu141, Thr144, Ser145, Phe147, Arg148, Ser151, and Leu152) (Fig. 3d). Two residues (Trp181 and Arg183) from extracellular loop 2 also participated in this interaction (Fig. 3c). Previous studies with human β1-ARs using the bioluminescence resonance energy transfer method had shown that human β1-ARs formed dimers, and that TM4 was involved in human β1-AR dimerization 37,38.
Disulfide trapping of β1-AR dimers
Next we used disulphide-trapping experiments to biochemically test some representative residues identified from our structural studies for their involvement in β1-AR dimerization in cells. Cysteine replacement of an appropriately disposed pair of residues at the dimer interfaces is expected to generate a disulphide bridge 30,33,39–41. Based on our structural model, we selected a few residues from the two interfaces and mutated these residues into cysteines on the background of β1-AR(H0). In dimer interface 1, Lys354 from one protomer interacted with Lys354 from the second protomer (Fig. 2e). In dimer interface 2, Arg148 from one protomer interacted with Arg148 from the other protomer (Fig. 3d). As a negative control, we also mutated Phe112 to Cys. Phe112 is on the extracellular side of TM3 and was not involved in the dimer interfaces based on our crystal structure. We transfected these mutants individually into CHO cells and stable cell lines were selected. After exposing the membrane preparations to the hyperoxidizing environment of Cu-phenanthroline (CuP), dimer formation was assessed by Western blot analysis of detergent-solubilized protein samples with β1-AR antibodies (Fig. 3e). Wild-type β1-AR (no cysteine residues in the two dimer interfaces) and Phe112Cys mutant showed only monomers with or without CuP treatment (Fig. 3e). Mutants Arg148Cys and Lys354Cys showed increased formation of dimers after CuP treatment (Fig. 3e). Arg148Cys mutants formed some dimers without CuP treatment, likely the result of air oxidation as shown for some Cys mutants of serotonin 5HT2c receptors 41. These data confirm that the two dimer interfaces identified in our structure are used under physiological conditions. Hence, our crystal structure provides a structural context for the dimerization and oligomerization of GPCRs, and the co-existence of the two types of dimers within the oligomers.
Ligand-free basal state of β1-AR in an inactive conformation
One of the characteristics of the inactive state of class A GPCRs is the presence of the ionic-lock salt bridge between the highly conserved D(E)R3.50Y motif in TM3 and an E/D6.30 residue in TM6 (Ballesteros-Weinstein numbering system is in superscripts)7,14,16,42. This ionic-lock salt bridge between Arg1393.50 and Glu2856.30 was present in the ligand-free state of β1-AR (Fig. 4a and b). Hence, our data are consistent with the ligand-free basal state β1-AR being in an inactive state.
Figure 4.
The ligand-free basal state of β1-AR in an inactive conformation and with a contracted ligand-binding pocket. a and b, 2Fo-Fc map (blue mesh) of the cytoplasmic ends of TM3 and TM6 showing the ionic-lock salt bridge between Arg1393.50 and Glu2856.30. The electron density is contoured at 1.0 σ level and the dashed line shows the distance between Arg1393.50 and Glu2856.30. c, Comparison of the ligand-free state of β1-AR (in cyan, molecule B) and the cyanopindolol-bound β1-AR with TM6 in the bent conformation (in magenta, PDB code 2YCX, molecule A). The ionic-lock is present in both structures. d, Comparison of the ligand-free state of β1-AR (in cyan, molecule B) and the cyanopindolol-bound β1-AR with TM6 in the straight conformation (in gold, PDB code 2YCY, molecule B). The ionic-lock is present in the ligand-free state, but not in the cyanopindolol-bound β1-AR with TM6 in the straight conformation. Structures were aligned using all seven TM segments. Parts of the helices in front are removed for clarity. e, Representative regions of 2Fo-Fc map (blue mesh) around the ligand-binding pocket of β1-AR (molecule B, cyan), showing the empty pocket. The electron density is contoured at 1.2 σ level. f, Comparison of the ligand-free state of β1-AR (in cyan, molecule B) and the antagonist cyanopindolol-bound β1-AR (in yellow, PDB code 2VT4, molecule B). g, Comparison of the ligand-free state of β1-AR (in cyan, molecule B) and the agonist isoprenaline-bound β1-AR (in magenta, PDB code 2Y03, molecule A). The ligand-binding pockets are viewed from the extracellular surface and ECL2 is hidden for clarity. The dash lines represent the key hydrogen bonds involved in ligand binding. h, Comparison of the ligand-binding pockets for the empty ligand-free state β1-AR structure (molecule B, cyan), β1-AR with the antagonist cyanopindolol-bound (molecule B, yellow), and β1-AR with the agonist isoprenaline-bound (molecule A, magenta). The ligands and ECL2 are removed for clarity. The distances between the Cα atoms of Ser211 and Asn329 are represented as dashes and labeled.
In the first report of the crystal structure of β1-AR bound with the antagonist cyanopindolol, the ionic-lock was absent10. In a subsequent report of the crystal structures of β1-AR with cyanopindolol, the ionic-lock was present in some structures, but absent in others43. In the structure of cyanopindolol-bound β1-AR with the ionic-lock, the cytoplasmic end of TM6 (the G protein-interacting region) was in a bent conformation (Fig. 4c)43. In the cyanopindolol-bound β1-AR without the ionic lock, the cytoplasmic end of TM6 was in a straight conformation (Fig. 4d)43. Thus, it was proposed that the presence of the ionic-lock was associated with the bent conformation of the cytoplasmic end of TM6 43. However, in the ligand-free basal state structure of β1-AR described here, the ionic-lock existed concomitantly with the straight conformation of TM6 (Fig. 4c and d).
The basal state with a contracted ligand-binding pocket
Based on the comparisons of the crystal structures of several GPCRs in inactive and active states, it has been proposed that, while the overall GPCR structures did not change significantly, an outward movement of the cytoplasmic end of TM6 (to a lesser degree TM5 as well) relative to the receptor helix bundle core is a hallmark of the active state13,17,22–24. The ligand-free basal state of β1-AR did not display this characteristic outward movement of TM6 and TM5, consistent with its inactive conformation. Furthermore, agonist binding to β1-AR induces the contraction of the ligand-binding pocket by ~1 Å (as measured between the Cα atoms of Ser211 and Asn329)18. The ligand-binding pocket in the ligand-free state of β1-AR was empty (Fig. 4e and Supplementary Fig. 3). Moreover, the ligand-binding pocket of the ligand-free state of β1-AR was narrower than those of the antagonist-bound and similar to the agonist-bound structures of β1-AR (Fig. 4f–h). Thus, the contraction of the ligand-binding pocket may not be an essential feature of the binding of full agonists to β1-AR.
DISCUSSION
The ligand-free basal state of GPCRs
Before ligand binding, GPCRs are in a basal state. Since agonists or inverse agonists could shift the ligand-free state to an activated state or an inactive state, respectively, the ligand-free state is likely conformationally flexible. This may partly explain the difficulty in crystallizing the ligand-free GPCRs. On the other hand, many GPCRs including β1-AR have a low basal activity in the absence of ligands, suggesting that, in the ligand-free state, a large fraction of the receptor population is in the inactive state. For the ligand-free GPCRs, while opsin is in an active state, the ligand-free β1-AR is in an inactive state. These differences might reflect the different crystallization conditions such as the presence of membrane-like environment in the β1-AR structure, or the different crystal packings leading to different stabilized conformations. The crystal structures only provide a snapshot of the lowest energy conformations that these receptors could adopt under the specific crystallization conditions.
It might also be argued that the ligand-free β1-AR observed here in the inactive state was stabilized by the use of thermostabilizing mutations. However, that is unlikely since these thermostabilizing mutations, although making β1-AR proteins more stable at higher temperatures, do not stabilize β1-AR(m23) in an inactive conformation. As recently reported, this mutated β1-AR(m23) is still a functional receptor capable of binding agonists and antagonists and activating intracellular agonist responses (Supplementary Fig. 4) 44. Furthermore, most crystal structures of this thermostabilized β1-AR(m23) with agonists or antagonists displayed intermediate conformations without the ionic-lock and without the outward movement of the cytoplasmic end of TM6 10,18,43. The structure presented here was determined from β1-AR proteins purified with alprenolol affinity purification (eluted with cyanopindolol) as the second purification step. Although the protein samples were dialyzed with buffers without cyanopindolol and the crystallization condition was at ~pH 4 which reduced antagonist binding to β1-ARs (Supplementary Fig. 4), we could not completely exclude the possibility of a very low occupancy of cyanopindolol in the presented structure. However, use of β1-AR proteins purified with two-rounds of nickel affinity purifications (without the alprenolol affinity purification step) resulted in similar structures although at lower resolutions.
It is known that some GPCRs display varying levels of constitutive activity (i.e. the basal activity in the absence of any ligands) which are critical for their physiological functions. Structural determinations of the ligand-free states of these GPCRs should provide molecular insights into the activation processes of GPCRs, the basal activity, and development of agents for therapeutic applications since the ligand-free state is the starting state and offers a point of comparison.
Dimer interfaces and G-protein interaction
In our crystal structure of β1-AR oligomers, there are two dimer interfaces: one involves TM1-TM2-H8 and the other engages TM4-TM5-ICL2 (Fig. 1). Among the published crystal structures of GPCRs, there are four other GPCRs showing parallel dimers. In rhodopsin and κ-opioid receptor, the dimer interface involves residues from TM1-TM2-H8 12,13,45,46 (Supplementary Fig. 5a and b). This dimer interface is similar to dimer interface 1 in our β1-AR structure. In the CXCR4 structure, there is a dimer interface involving residues from TM5 and TM6 15 (Supplementary Fig. 5c). However, dimer interface 2 of β1-ARs involves TM4 and TM5. Compared to β1-AR, one monomer of CXCR4 rotates ~40° towards another monomer (Supplementary Fig. 5c). In a recent crystal structure of oligomeric μ-opioid receptor, two dimer interfaces were observed: one involves TM1-TM2-H8, and the other involves TM5-TM6 47. Hence, the TM1-TM2-H8 interface is rather conserved in various GPCRs. On the other hand, the TM5 interface sometime functions with TM4 and other times with TM6. Remarkably, the crystal structures of GPCRs so far only displayed these two types of dimer interfaces which are in agreement with a large body of experimental data, indicating that these two dimer interfaces are likely physiological relevant.
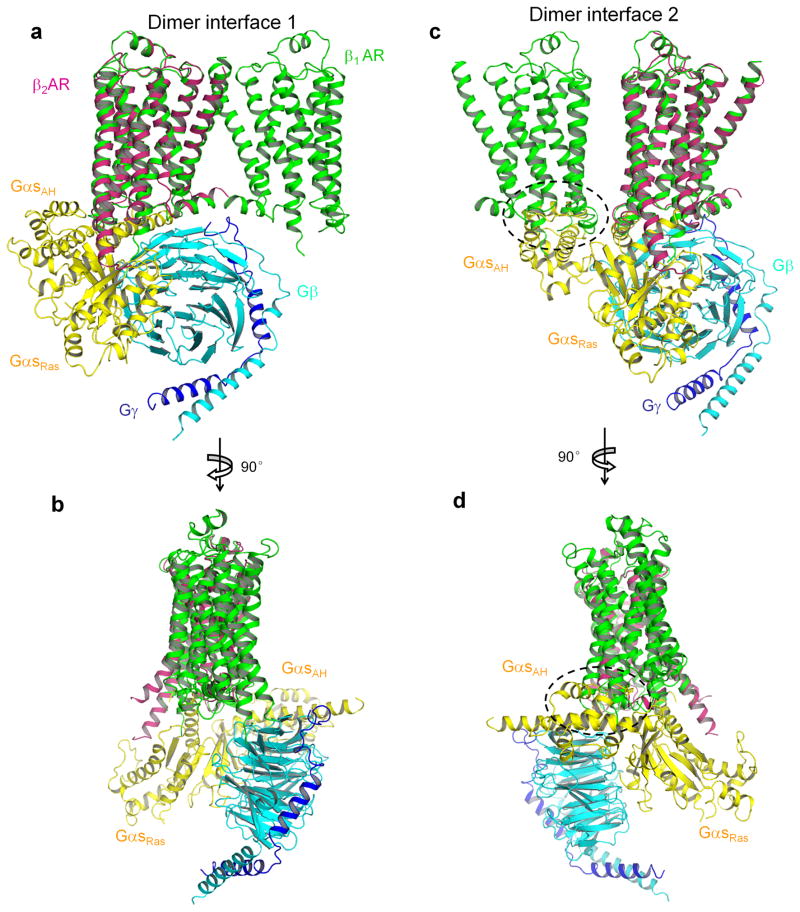
In addition to the TMs, intracellular regions contribute significantly to the dimer interfaces. There are four residues from H8 involved in the TM1-TM2-H8 dimer interface. Eight residues from ICL2 contribute to the TM4-TM5-ILC2 dimer interface. ICL2 is critical for interacting with G proteins based on the structural model of the complex of β2-AR and Gs 24. A Gs trimer could be docked onto a β1-AR dimer formed through the TM1-TM2-H8 dimer interface (Fig. 5a and b). On the other hand, it was not possible to dock a Gs trimer onto the β1-AR dimer formed via the TM4-TM5-ICL2 dimer interface without steric collisions (Fig. 5c and d). Participation of ICL2 in this dimer interface may prevent G protein coupling to the dimer formed through TM4-TM5-ICL2 interface, or G protein binding may disrupt this dimer interface. Therefore, we propose that, if the signaling unit is a pentamer (two GPCRs and one trimeric G protein), the GPCR dimer interface in this signaling unit is TM1-TM2-H8 (Fig. 5a and b). In this model, only one β1-AR contacts with the G protein trimer, and the other β1-AR is “spared” or could function through trans-protomer allosteric regulation (see below).
Figure 5.
Docking of Gs onto β1-AR dimer. a and b, The complex of β2-AR and Gs (PDB code 3SN6) was aligned with molecule B of the β1-AR dimer with the TM1-TM2-H8 interface. β1-AR is in green and β2-AR is in magenta. Gs α-subunit (the Ras-like and the α-helical (AH) domains) is in yellow. Gβ subunit is in cyan. Gγ subunit is in blue. c and d, The complex of β2-AR and Gs was aligned with molecule B of the β1-AR dimer with the TM4-TM5 interface. The steric collision is indicated by dashed circles.
GPCR oligomerization and receptor activation
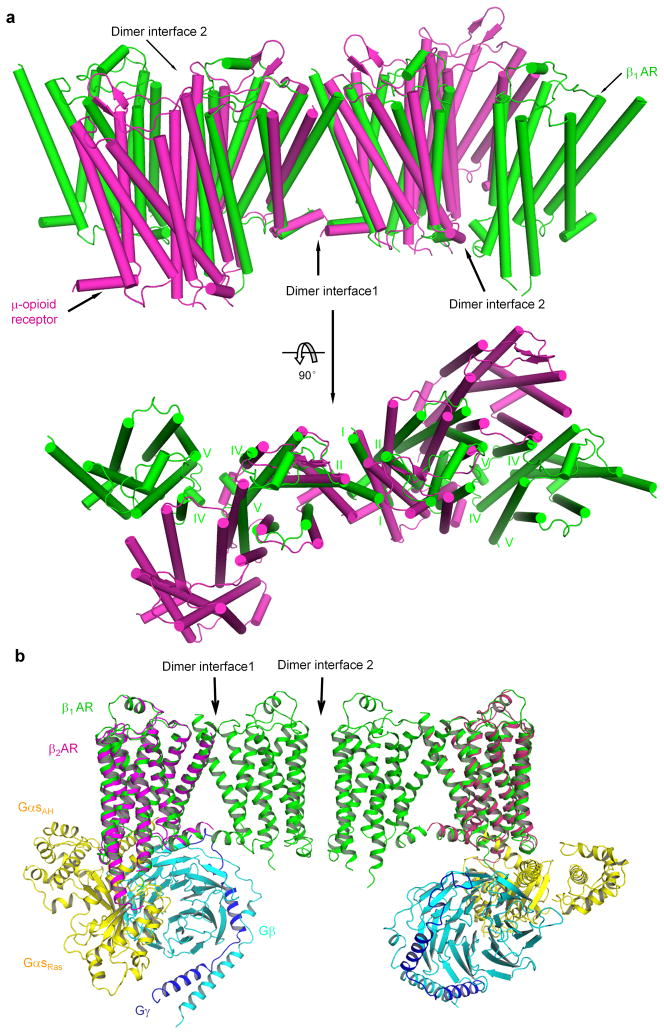
In our crystal structure, β1-ARs form oligomers in a membrane-like environment. Although the physiological functions are not clear, studies using various approaches have indicated that GPCRs could form oligomers in cells 28,29,34,48,49. While our manuscript was under review, a recent crystal structure of μ-opioid receptor also showed oligomers 47. The β1-AR oligomers show some similarities and some differences from the oligomers of μ-opioid receptors (Fig. 6a). Both oligomers have two dimer interfaces and share the same TM1-TM2-H8 dimer interface (Fig. 6a). However, in the second dimer interface, TM5 works together with TM4 in β1-ARs while TM5 acts together with TM6 in μ-opioid receptors (Fig. 6a). Furthermore, while β1-AR oligomers form a linear array in one direction, μ-opioid receptors are in a sine wave arrangement (Fig. 6a).
Figure 6.
Comparison of the β1-AR oligomer with the μ-opioid receptor oligomer. a, β1-AR is in green and μ-opioid receptor (PDB code 4DKL) is in magenta. Top panel, side view of the oligomers. Bottom panel, top view (from the extracellular surface) of the oligomers. The alignment was performed between molecule A ofβ1-AR and one molecule in μ-opioid receptor using all seven TM segments. b, Docking of Gs ontoβ1-AR tetramer. The complex of β2-AR and Gs (PDB code 3SN6) was aligned with molecule B of the β1-AR dimer with the TM1-TM2-H8 interface.
A docking exercise revealed that, with the oligomeric arrangement, trimeric G proteins could not be fitted in without steric hindrance (Supplementary Fig. 6). Even though this docking was speculative, it suggests that either G protein binding may disrupt the oligomers or change the oligomeric arrangement, or the oligomeric architecture would prevent the dramatic sideway-rotation and upward-translation of the helical-domain of the Gαs subunit, relative to the Ras-like domain of the Gαs subunit, as observed in the crystal structure of the complex of β2-AR and Gs 24. Indeed, the extent of membrane-driven oligomerization of a GPCR (such as D2 receptors and 5HT2c receptors) in the inverse agonist-bound state may be larger than in the agonist-bound state 40,41. Moreover, inverse agonists stabilize β2-AR oligomers, while Gs reduced the extent of oligomerization of β2-ARs 50. Hence oligomerization of GPCRs is sensitive to ligand binding. That is to say, agonist binding may disrupt the oligomerization of GPCRs into dimers and/or tetramers.
It is possible to dock two trimeric G proteins into a β1-AR tetramer (Fig. 6b). Whether this model is physiologically relevant requires further experimental testing. In this model, the two protomers via the TM4-TM5-ICL2 dimer interface were “spared” (not interacting with G proteins) (Fig. 6b). An asymmetric functioning for GPCR dimers has been proposed 34,51. Previously a dimer interface involving TM1 has been shown to be insensitive to ligand binding and the receptor activation state as shown for dopamine D2 receptors and serotonin 5HT2c receptors 31,41. The similarity of dimer interface 1 (involving TM1) in the inactive β1-AR and the active rhodopsin is consistent with the notion that this dimer interface involving TM1 does not undergo significant conformational changes from inactive to active states of GPCRs (Supplementary Fig. 5a). On the other hand, the dimer interface involving TM4 makes structural rearrangement during the GPCR activation process, at least in the cases of dopamine D2 receptors and serotonin 5HT2c receptors 40,41. These imply a possible role for the TM4-TM5 dimer interface in GPCR transactivation even though the two promoters do not directly interact with G proteins in the proposed model (Fig. 6b). Based on the structures of active GPCRs, the intracellular end of TM5 moves away from the TM bundle core 24. Therefore it is possible that, upon the agonist binding, the configuration change at the TM4-TM5 dimer interface is part of the receptor activation process.
ONLINE METHODS
β1-AR constructs and purification of β1-AR proteins
A cDNA plasmid for the turkey β1-AR was obtained from Dr. E. Ross (Dallas, Texas)52. For pre-crystallization screening of β1-AR constructs for structural studies, we used the fluorescence-detection size-exclusion chromatography (FSEC) method for integral membrane proteins developed by Dr. E. Gouaux and colleagues53. In this screening method, the target proteins are covalently fused to GFP. The resultant fusion proteins are monitored first for expression level and pattern in whole cells by epifluorescence microscopy. After solubilization of whole cells or crude membranes, the resulting unpurified protein is analyzed by fluorescence-detection size-exclusion chromatography. A monodisperse and folded protein would generally yield a single symmetrical Gaussian peak, while a polydisperse, unstable, or unfolded protein would typically yield multiple asymmetric peaks53. We PCR-subcloned different β1-AR constructs into the pCGFP vector (from Drs. O. Boudker and E. Gouaux) and transfected them into HEK 293 cells. Based on previous studies with many different GPCRs, we focused on deletions on the N-terminus, the C-terminus, and the intracellular loop 3 region of β1-AR. Two days after transfection, the subcellular localizations of the β1-AR receptors were checked by fluorescence microscopy. All tested constructs expressed proteins at the plasma membrane. Membrane preparations were solubilized in a buffer containing the nonionic detergent n-dodecyl-β-D-maltoside (DDM), and the resulting supernatant was analyzed by fluorescence-detection size-exclusion chromatography. Among the ~40 β1-AR constructs, several β1-AR constructs displayed a nearly symmetric fluorescence peak with an apparent molecular weight of a monomer of β1-AR in DDM (the protein-detergent complex) (Supplementary Fig. 7a). We purified the recombinant β1-AR proteins from High5 insect cells. The stability of these β1-AR proteins in different detergents was tested at 18°C. Most of the studies were with a β1-AR construct [β1-AR(H0)] with deletions of amino acid sequences of 3–32, of 249–283 and of 366–483 and with point mutations of Cys116Leu and Cys358Ala (Supplementary Figure 1). β1-AR(H0) generated similar cAMP responses as wild-type β1-AR when expressed in β1-AR−/−/β2-AR−/− MEF cells 54 (Supplementary Fig. 7b).
β1-AR mutants (with a C-terminal His6 tag) were subcloned into a baculoviral expression vector pVL1393. Recombinant baculoviruses were picked and amplified. High5 insect cells were grown suspension in High5 Express Medium (Invitrogen) at 27°C with shaking at 110 rpm. Cells were infected at a multiplicity of infection of 5–10. After shaking for one hour, an equal volume of fresh medium was added. Cells were harvested by centrifugation 48 h after infection. Infected cells from cultures were harvested by centrifugation and the resulting pellet was resuspended in 20 mM Tris–HCl pH 8, 1 mM EDTA. Cells were flash frozen in liquid nitrogen and stored at −80°C. Cells were broken by sonication. After centrifugation at 2,000 rpm for 10 min, the supernatant was collected and centrifuged for 1 h at 45,000 rpm at 4°C in a Beckman Ti45 rotor. Membrane pellets were resuspended in the same volume of buffer and the centrifugation was repeated. The final pellet was resuspended in a buffer with a reduced EDTA concentration (0.2 mM) at 10–20 mg protein/ml and frozen in liquid nitrogen and stored at −80°C. Membranes containing 2 g of total proteins were thawed and diluted to 10 mg/ml protein in ice-cold 20 mM Tris–HCl pH 8 with 0.35 M NaCl, 2% DDM and protease inhibitors, and then stirred at 4°C for 1 hour. After centrifugation for 1 h at 45,000 rpm in a Ti45 rotor (4°C), solubilized membrane proteins were mixed with Ni-NTA beads pre-equilibrated with buffer A (20 mM Tris–HCl pH 8, 0.35 M NaCl, 10 mM imidazole, protease inhibitors and 0.025% DDM). The mixture was rolled at 4°C for 6 hours. The protein-loaded resin was washed with buffer B (20 mM Tris–HCl (pH 8.0), 500 mM NaCl, 0.025% DDM), and the bound protein was eluted by using 3 X bed volume buffer C (20 mM Tris–HCl (pH 8.0), 50 mM NaCl, 250 mM imidazole, 0.025% DDM). In some preparations, β1-AR proteins were purified again with a second-round Ni-NTA affinity purification and used for crystallization. In other preparations, β1-AR proteins were purified by alprenolol affinity purification. For alprenolol affinity purification, after dilute the protein sample with four volumes of buffer D (Buffer C without imidazole), the sample was incubated with alprenolol-Affi-Gel beads overnight at 4°C. Alprenolol-NH2 was synthesized at Cornell’s chemistry core facility following a published protocol55. Alprenolol was crosslinked to Aff-Gel-15. After washing the alprenolol beads with buffer D, β1-AR was eluted with buffer D containing 50 μM cyanopindolol, and then dialyzed, concentrated and changed buffer to 10 mM Tris pH 7.7, 50 mM NaCl, 0.02% DDM and 0.1 mM EDTA with centricon (100 KDa cutoff)(Millipore)56,57. SDS-PAGE showed that β1-AR protein was >90% pure. The yield was ~ 2 mg of purified β1-AR proteins from 1L of insect cells.
Crystallization
β1-AR(H0) proteins were initially used for screening crystallization conditions. β1-AR at a final concentration of ~8 mg/ml in 20 mM Tris-HCl pH 7.7, 50 mM NaCl, 0.1 mM EDTA, 0.02% DDM, 0.1 mg/ml lipid (3:1:1:1 = POPC:POPE:POPG: Cholesterol) was incubated on ice for 1 hour prior to set up the tray. Crystals were obtained in several crystallization conditions. To further make sure that there were no ligands in the final crystals, we selected conditions with low pH which decreased the binding of ligands from β1-AR (ref.58) (Supplementary Fig. 4). Crystallization was performed by the vapour diffusion hanging drop method at 18°C. 1 μl β1-AR protein sample was mixed with 1 μl crystallization buffer (0.1–0.3 M (NH4)2SO4, 0.02 M NaAc, pH 3.6~4, 26~30% PEG200). With β1-AR(H0) construct, the screened crystals yielded diffraction to ~8 Å. We then introduced the six point mutations (Arg68Ser, Met90Val, Tyr227Ala, Ala282Leu, Phe327Ala and Phe338Met) and generated a construct same as the thermostabilized β1-AR(m23) (Supplementary Figure 1)18. Thus the ligand-free structure described here was for β1-AR(m23) and this thermostabilized β1-AR mutant was used in previous crystal structural studies with antagonists or agonists 10,18. This mutant β1-AR is able to adopt different conformations, to bind antagonists, partial agonists and agonists, as well as to activate G proteins and increase cAMP levels in cells in response to agonists10.18,44. Crystals were formed within one week and were directly frozen in liquid nitrogen. Crystals were screened and diffraction data were collected at the National Synchrotron Light Source (beamlines X6A and X25) of the Brookhaven National Laboratory or the Advanced Photon Source beamline NE-CAT 24E at Argonne National Laboratory.
Data collection, structure determination and refinement
Diffraction data presented in this paper were collected at 100 K using synchrotron radiation (λ = 1.1000 Å) at the beamline X25, National Synchrotron Light Source (Brookhaven, USA), using a PILATUS 6M CCD detector. The crystal diffracted to ~ 3.3 Å and diffraction data were indexed and integrated with XDS, followed by merging and scaling with XSCALE59. The crystal belongs to space group C2 and the corresponding data-collection statistics are shown in Table 1. Analysis of the final data set by the UCLA diffraction anisotropy server indicated that the diffraction was highly anisotropic, strong in two directions while weak in the third direction along the reciprocal space axis c* 60. As guided by an <F>/<σF> cutoff of 3.0 along each reciprocal space axis, reflections were anisotropically truncated to 3.3 × 3.3 × 4.3 Å along a*, b*, c* and sharpened by application of a negative isotropic B factor of − 54.13 before use in refinement.
The structure of the ligand-free state β1-AR was solved by molecular replacement with PHASER of the CCP4 suite using a monomer of the salbutamol-bound β1AR-m23 structure (PDB ID Code: 2Y04, chain A) as the search model61. A total of two copies of the monomer were observed per asymmetric unit (ASU). Model refinements were performed with REFMAC5 and PHENIX followed by employing the program COOT for iterative cycles of rebuilding based on sigma-A weighted 2Fo-Fc and Fo-Fc maps, as well as non-crystallographic symmetry (NCS) averaged and unaveraged maps62–64. During refinement, reflections within the resolution range 30-3.5 Å were selected and tight 2-fold NCS restraints were applied to chains A and B, with a notable reduction in Rfree with good geometry. Refinement statistics were also presented in Table 1, and the stereochemical quality of the refined structure was validated using MolProbity65. The Ramachandran plot distribution for residues in the structure was 95.6% in the favored region and 4.4% in allowed region. The high R factors might be partially attributed by the unmodeled discontinuous density maps in the gaps between protein molecule layers, which could not be fitted into any ordered lipid molecules or solvents due to the resolution limit of 3.5 Å. The interfaces of two β1AR dimers were analyzed using the EBI PDBe PISA web server66. Global alignment of various structural models of β1-ARs was performed using PyMOL (super_align) (DeLano Scientific LLC) and all structural model figures were created with PyMOL as well.
Cysteine crosslinking
Disulfide trapping experiments were performed as described 30,41.
Supplementary Material
Acknowledgments
We thank O. Andersen, O. Boudker, J. McCoy, C. Steegborn, G. Verdon, and W. Xu for advice, discussions and help. We thank E. Ross (UT Southwestern Medical Center, Dallas, Texas, USA) for the turkey β1-AR plasmid, E. Gouaux (Vollum Institute, Portland, Oregon, USA) and O. Boudker (Weill Cornell Medical College, New York, New York, USA) for the pCGFP-EU plasmid, and Cornell’s chemistry core facility for the synthesis of alprenolol-NH2. We thank I. Kourinov at the Advanced Photon Source beamline 24-ID-E and J. Jakoncic at the Brookhaven National Laboratory NSLS beamlines X6A and X25 for their assistance with X-ray data collection. We are grateful to Olga Boudker, Harel Weinstein, and members of our laboratory for critically reading the manuscript. This work was supported by an NIH grant HL 91525 (XYH).
Footnotes
Accession code. Atomic coordinates and structure factor files for the oligomeric turkey β1-ARs have been deposited in the Protein Data Bank under ID code 4GPO.
AUTHOR CONTRIBUTIONS: J.H. performed the protein purification, set up crystallization trials, grew crystals for data collection and participated in data collection. S.C. processed the diffraction data, solved and refined the structures. J.J.Z. participated in project strategy and manuscript preparation. X.Y.H. was responsible for the overall project strategy and management, participated in data collection, and wrote the manuscript.
References
- 1.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 2.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 3.Vassart G, Costagliola S. G protein-coupled receptors: mutations and endocrine diseases. Nat Rev Endocrinol. 2011;7:362–72. doi: 10.1038/nrendo.2011.20. [DOI] [PubMed] [Google Scholar]
- 4.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 6.Brunton, Goodman L, Gilman’s . The pharmacological basis of therapeutics. 12. McGraw-Hill Professional; 2010. [Google Scholar]
- 7.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 8.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 10.Warne T, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–91. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–7. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 13.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 14.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–7. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 15.Wu B, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chien EY, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–5. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warne T, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–4. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimamura T, et al. Structure of the human histamine H(1) receptor complex with doxepin. Nature. 2011 doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322–7. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebon G, et al. Agonist-bound adenosine A(2A) receptor structures reveal common features of GPCR activation. Nature. 2011 doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choe HW, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–5. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 23.Standfuss J, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002;14:189–95. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- 26.Angers S, Salahpour A, Bouvier M. Dimerization: an emerging concept for G protein-coupled receptor ontogeny and function. Annu Rev Pharmacol Toxicol. 2002;42:409–35. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 27.Filizola M, Weinstein H. The study of G-protein coupled receptor oligomerization with computational modeling and bioinformatics. FEBS J. 2005;272:2926–38. doi: 10.1111/j.1742-4658.2005.04730.x. [DOI] [PubMed] [Google Scholar]
- 28.Milligan G. The role of dimerisation in the cellular trafficking of G-protein-coupled receptors. Curr Opin Pharmacol. 2010;10:23–9. doi: 10.1016/j.coph.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 29.Palczewski K. Oligomeric forms of G protein-coupled receptors (GPCRs) Trends Biochem Sci. 2010;35:595–600. doi: 10.1016/j.tibs.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klco JM, Lassere TB, Baranski TJ. C5a receptor oligomerization. I. Disulfide trapping reveals oligomers and potential contact surfaces in a G protein-coupled receptor. J Biol Chem. 2003;278:35345–53. doi: 10.1074/jbc.M305606200. [DOI] [PubMed] [Google Scholar]
- 31.Guo W, et al. Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 2008;27:2293–304. doi: 10.1038/emboj.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y, et al. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–62. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kota P, Reeves PJ, Rajbhandary UL, Khorana HG. Opsin is present as dimers in COS1 cells: identification of amino acids at the dimeric interface. Proc Natl Acad Sci U S A. 2006;103:3054–9. doi: 10.1073/pnas.0510982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688–95. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274–86. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 36.Lohse MJ. Dimerization in GPCR mobility and signaling. Curr Opin Pharmacol. 2010;10:53–8. doi: 10.1016/j.coph.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Mercier JF, Salahpour A, Angers S, Breit A, Bouvier M. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo-and heterodimerization by bioluminescence resonance energy transfer. J Biol Chem. 2002;277:44925–31. doi: 10.1074/jbc.M205767200. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi H, Ogawa K, Yao R, Lichtarge O, Bouvier M. Functional rescue of beta-adrenoceptor dimerization and trafficking by pharmacological chaperones. Traffic. 2009;10:1019–33. doi: 10.1111/j.1600-0854.2009.00932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo W, Shi L, Javitch JA. The fourth transmembrane segment forms the interface of the dopamine D2 receptor homodimer. J Biol Chem. 2003;278:4385–8. doi: 10.1074/jbc.C200679200. [DOI] [PubMed] [Google Scholar]
- 40.Guo W, Shi L, Filizola M, Weinstein H, Javitch JA. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc Natl Acad Sci U S A. 2005;102:17495–500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–9. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 43.Moukhametzianov R, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta1-adrenergic receptor. Proc Natl Acad Sci U S A. 2011;108:8228–32. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker JG, Proudman RG, Tate CG. The pharmacological effects of the thermostabilising (m23) mutations and intra and extracellular (beta36) deletions essential for crystallisation of the turkey beta-adrenoceptor. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:71–91. doi: 10.1007/s00210-011-0648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salom D, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–8. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–32. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith NJ, Milligan G. Allostery at G protein-coupled receptor homo-and heteromers: uncharted pharmacological landscapes. Pharmacol Rev. 2010;62:701–25. doi: 10.1124/pr.110.002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hebert TE, et al. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–92. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 50.Fung JJ, et al. Ligand-regulated oligomerization of beta(2)-adrenoceptors in a model lipid bilayer. EMBO J. 2009;28:3315–28. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damian M, Martin A, Mesnier D, Pin JP, Baneres JL. Asymmetric conformational changes in a GPCR dimer controlled by G-proteins. EMBO J. 2006;25:5693–702. doi: 10.1038/sj.emboj.7601449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parker EM, Ross EM. Truncation of the extended carboxyl-terminal domain increases the expression and regulatory activity of the avian beta-adrenergic receptor. J Biol Chem. 1991;266:9987–96. [PubMed] [Google Scholar]
- 53.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–81. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, Huang J, Xiang Y, Bastepe M, Juppner H, Kobilka BK, Zhang JJ, Huang XY. Dosage-dependent switch from G protein-coupled to G protein-independent signaling by a GPCR. EMBO J. 2007;26:53–64. doi: 10.1038/sj.emboj.7601502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henis YI, Hekman M, Elson EL, Helmreich EJ. Lateral motion of beta receptors in membranes of cultured liver cells. Proc Natl Acad Sci U S A. 1982;79:2907–11. doi: 10.1073/pnas.79.9.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caron MG, Srinivasan Y, Pitha J, Kociolek K, Lefkowitz RJ. Affinity chromatography of the beta-adrenergic receptor. J Biol Chem. 1979;254:2923–7. [PubMed] [Google Scholar]
- 57.Warne T, Chirnside J, Schertler GF. Expression and purification of truncated, non-glycosylated turkey beta-adrenergic receptors for crystallization. Biochim Biophys Acta. 2003;1610:133–40. doi: 10.1016/s0005-2736(02)00716-2. [DOI] [PubMed] [Google Scholar]
- 58.Modest VE, Butterworth JF. Effect of pH and lidocaine on beta-adrenergic receptor binding. Interaction during resuscitation? Chest. 1995;108:1373–9. doi: 10.1378/chest.108.5.1373. [DOI] [PubMed] [Google Scholar]
- 59.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–5. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658– 674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 65.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.