Summary
Yin and Yang are two complementary forces that together describe the nature of real world elements. Yin is the dark side; Yang is the light side. We describe microRNAs having both Yin and Yang characteristics, because they can contribute to normal function (Yang) but also to autoimmunity, myeloproliferation, and cancer (Yin). We have been working on a number of microRNAs that have these dual characteristics and here we focus on two, miR-125b and miR-146a. We have concentrated on these two RNAs because we have very extensive knowledge of them, much of it from our laboratory, and also because they provide a strong contrast: the effects of overexpression of miR-125b are rapid, suggesting that it acts directly, while the effects of miR-146a are slow to develop suggesting that they arise from chronic alterations in cellular behavior.
Keywords: microRNA-125b, microRNA-146a, microRNA-155, leukemia, cancer, hematopoiesis
Introduction
MicroRNA-125b (miR-125b) is the mammalian homolog of the C. elegans heterochronic microRNA lin-4 (1), which is essential for normal temporal development of C. elegans (2). In mammalian systems, the mature sequence of miR-125b is encoded in two different genomic regions: miR-125b-1 is located in chromosomes 11 and 9, respectively, in the human and mouse genomes, and it is human chromosome 21 and mouse16 for miR-125b-2. In addition to miR-125b-1 and miR-125b-2, the mammalian genome encodes the homolog miR-125a, which shares an identical seed sequence with miR-125b. Although the functional role of lin-4 in C. elegans has been studied extensive since its discovery over three decades ago (3), the study of miR-125b’s functions in mammals is just beginning. Knockout mice models for miR-125b-1, miR-125b-2, and miR-125a are not currently available, thus limiting the ability to study the individual physiological roles of the miR-125 family. Thus, we applied a loss-of-function miR-125b ‘sponge’ decoy system, which implicated the role of miR-125b in hematopoiesis. Overexpression systems have provided the best clues to miR-125b functions. These gain-of-function systems constitutively overexpress miR-125b and thereby perturb its normal regulated expression. By observing changes in biological process upon such perturbation, these studies provide clues to the function of miR-125b. From these studies, miR-125b has been shown to be important for normal hematopoiesis by modulating cell proliferation and differentiation. In contrast, abnormal and unregulated overexpression of miR-125b leads to accelerated hematopoiesis that subsequently leads to a rapid and acute leukemia. Here, we review recent advances in understanding the functional role of miR-125b in hematopoiesis and its pathological role in leukemogenesis.
MicroRNA-146a (miR-146a) and miR-146b form a highly conserved two-member miRNA family. miR-146a and miR-146b are located on chromosome 5 and 10, respectively, in the human genome and on chromosome 11 and 19, respectively, in the mouse genome. In 2006, miR-146a first came to our attention in a systemic microarray screen to identify miRNAs that are involved in innate immune activation (4). Stimulating THP1 cells, a human monocytic cell line, with lipopolysaccharide (LPS) was found to selectively upregulate the expression of three miRNAs, miR-155, miR-146, and miR-132. Since then, we have been focused on investigating the function of miR-146 and miR-155 in the immune system. To study the physiological role of miR-146a, we generated miR-146a knockout mice to disrupt miR-146a function in these animals. By ablating miR-146a function, we and other researchers gained extensive knowledge on the role of miR-146a in the immune system. We found that miR-146a orchestrates myeloid and lymphocyte function thus impacting both innate and adaptive immunity. Whereas overexpression of miR-125b induces leukemia in mice, the deletion of miR-146a leads to hematopoietic malignancy. In contrast to miR-125b, frank cancer in miR-146a-ablated mice develops in older mice, suggesting that the effect results from chronic alterations and may require longer-term accumulation of cellular abnormalities to induce tumorigenesis. Here, we review the recent findings on role of miR-125b and miR-146a in leukemogenesis and hematopoiesis, the yin and yang of these microRNAs.
The Yang of miR-125b: function in hematopoietic development and immunity
Expression patterns of miR-125 in hematopoietic lineages
Our laboratory has been studying a variety of microRNAs, mainly ones induced by the NF-kB transcription factor. However, our attention was drawn to miR-125b in experiments designed to examine the role of miRs in the regulation of hematopoietic stem cell (HSC) development. These experiments involved performing microarray analysis to identify microRNAs enriched in hematopoietic stem-progenitor cells (HSPCs) (5). Compared to total bone marrow cells isolated from adult mice, we found 11 microRNAs to be enriched in HSPCs (defined as Lin−Sca1+cKit+). Of particular interest, four of these non-coding RNAs belonged to the group in the highly conserved microRNA cluster miR-125/miR-99/Let-7. The mouse genome encodes three separate clusters of these microRNAs: (i) miR-125a/miR-99b/Let-7e, (ii) mir-125b-2/miR-99a/Let-7c-1, and (iii) miR-125b-1/miR-100/let-7a-2. In our study, we found that miR-125b, miR-125a, miR-99a, and Let-7e were preferentially expressed in HSPCs and long-term HSCs compared to total bone marrow cells (5). The expression of miR-125b, miR-125a, and miR-99a is lower in hematopoietic progenitor cells (Lin−Sca1−cKit+ cells) compared to HSC and even lower in differentiated CD11b+ cells (5). In a separate unbiased profiling study, Ooi et al. (6) also demonstrated that the expression of miR-125b, miR-99a, and miR-100 was high in stem cells and became progressively reduced as cells become more differentiated. The expression level of these microRNAs was reported as follows: HSCs > multiple-potent progenitors (MPPs) > common myeloid progenitors (CMPs) > granulocyte-macrophage progenitors (GMPs) > megakaryocytic-eythroid progenitors (MEPs) (6). In a later study, Gerrits et al. (7) also performed microRNA expression studies and found that miR-125b-5p, miR-125a-5p, miR-99b, miR-99a, and Let-7e were expressed more highly in HSPCs compared to differentiated hematopoietic cells. Collectively, these studies indicated that some members of the miR-125/miR-99/Let-7 cluster are preferentially expressed in HSCs and their levels tend to decrease as these cells become more differentiated.
miR-125 provide a hematopoietic repopulating advantage to hematopoietic stem cells The progressive downregulation of microRNAs belonging to the miR-125/miR-99/Let-7 cluster as cells become more differentiated led us and other research groups to hypothesize that some members of this cluster might play a role in hematopoiesis at the HSC level. To address this hypothesis, we investigated whether the four microRNAs belonging to this family, which were highly expressed in HSCs (miR-125b, miR-125a, miR-99a, and Let-7e), modulate hematopoiesis (5). We performed competitive repopulation experiments by populating lethally irradiated recipient mice with combined populations of HSC-enriched CD45.1+ bone marrow cells that over-express these microRNAs and control HSC-enriched CD45.2+ cells. Whereas miR-125b overexpression conferred a competitive repopulation advantage as compared to control cells, miR-99a and Let-7e overexpression were disadvantageous to engraftment (5). These microRNAs affected hematopoiesis in multiple lineages, consistent with these molecules having an effect on stem cell homeostasis (5). A similar study of mice by Ooi et al. (6) reported that purified Lin−cKit+Sca1+CD34−Flk2− HSCs overexpressing miR-125b also exhibited enhanced repopulation potential compared to control HSCs. In addition, Guo et al. (8) and Gerritis et al. (7) showed that restricted overexpression of the miR-125a/miR-99b/Let-7e cluster or miR-125a alone in bone marrow cells also elevated the ability of these cells to repopulate the hematopoietic compartment of mice. Further analyses revealed that miR-125a provided self-renewing capacity specific to HSCs but not for CLPs, CMPs, GMPs, or MEPs, showing that the effect of miR-125a is cell specific and depends on the differentiated stage of the cells (8). In contrast to our study (5), Guo et al. (8) did not observe a disadvantage in repopulation upon Let-7e overexpression; the discrepancy may be due to differences in expression levels Let-7e. Thus, members of the miR-125/miR-99/Let-7 control hematopoiesis in part by affecting function at the HSC-level. In particular, the miR-125 family seems to provide an advantage for stem cells to repopulate (Fig. 1A). However, the role of miR-99 and Let-7 members will require further analyses.
Fig. 1. Role of miR-125b in hematopoiesis.
The figure displays a simplified diagram of the effect of miR-125b on hematopoietic development and immune cell function. (A) miR-125b enhances the repopulation of hematopoietic stem cells (HSCs). (B) miR-125b blocks the transition of common myeloid progenitors (CLPs) to pro-B cells. (C) The development of effector T cells is inhibited by miR-125b. (D) miR-125b drives the activation of macrophages. (E) Ectopic overexpression of miR-125b increases the common myeloid progenitor (CMP) pool.
The functions of miR-125b and miR-125a in all the studies described above were inferred from non-physiological over-expression systems. To provide insights into the physiological function of miR-125b and miR-125a in hematopoiesis, we utilized a ‘sponge decoy’ to competitively inhibit miR-125b and miR-125a binding to its natural targets (9). The recipient mice reconstituted with sorted miR-125-sponge transduced bone marrow cells had significantly fewer white blood cells (CD45+), myeloid cells (CD45+CD11b+), pre-erythrocytes (CD45+Ter119+), and granulocytes (CD45+Gr1+) compared to the controls one month after reconstitution (9). The miR-125 sponge loss-of-function experiments indicated an important physiological role for miR-125 in hematopoiesis and potentially stem cells since inhibiting miR-125 function affected the development of multiple lineages. Because the sponge decoy targets both miR-125b and miR-125a, further experiments will be required to determine which microRNA regulates these processes.
The role of miR-125b in development of myeloid and lymphoid cells
In addition to being able to impact stem cell repopulation, overexpression of miR-125b in the bone marrow can lead to a myeloid or lymphoid bias. Using an MSCV-based retroviral delivery system, we transduced HSPC-enriched cells from mouse adult bone marrow with a miR-125b overexpression cassette. These cells were transplanted into lethally irradiated recipient mice. The primary recipient mice that overexpressed miR-125b had a dramatic increase in all myeloid lineages, including granulocytes, macrophages, and dendritic cells (5, 9). Importantly, B-cell numbers were significantly decreased in these animals. Similarly, pre-B and pro-B-cell numbers were severely decreased, indicating that the repression of B-cell development occurred at the progenitor stage. However, the numbers of the more upstream common lymphoid progenitors (CLPs) were significantly upregulated, suggesting that miR-125b overexpression caused a developmental block between the CLP and pro-B-cell stages (Fig. 1B). Moreover, miR-125b likely can also block B-cell development at the pre-B-cell stage. Puissegur et al. (10) showed that miR-125b directly repressed the expression of Bright/Arid3a, an activator of immunoglobulin heavy-chain transcription, and blocked the differentiation of murine pre-BI cells to B cells. In addition to inhibiting the development of B cells at early progenitor stages, miR-125b can block the maturation of B cells to plasma cells. Upon overexpressing miR-125b in primary B cells, Gururajan et al. (11) found that it impaired the LPS induction of these cells into CD19loCD138hi plasma cells. It was hypothesized that miR-125b might inhibit plasma cell differentiation by directly repressing the expression of B-lymphocyte maturation protein-1 (BLIMP-1) and interferon (IFN) regulatory factor-4 (IRF4), factors important for plasma cell generation (11). In support of our data that miR-125b favors myeloid development, Klusmann et al. (12) showed that transduction of miR-125b-2 into adult murine bone marrow and human CD34+ HSPCs elevated the absolute number of myeloid colony-forming units (CFUs) in vitro. In agreement with our miR-125b studies, Gerritis et al. (7) showed that primary transplant recipients of HSPCs transduced with a miR-125a overexpression vector displayed an enrichment of myeloid cells accompanied by a decrease in B cells. Together, these studies showed that miR-125b and miR-125a are capable of expanding the myeloid compartment at the expense of B cells and can block B-cell genesis at multiple stages of development.
Ooi et al. (6) performed secondary transplant studies in which recipient mice received Lin−Sca1+cKit+CD34−Flk2− HSCs overexpressing miR-125b from primary recipients. In contrast to our primary transplant experiments, these mice exhibited increased chimerism within the preproB- and pro-B population in the spleen. The secondary transplanted mice also had higher chimerism within the CD4+ and CD8+ T-cell population in the lymph nodes. The increase in lymphoid cells in these mice could be explained by these mice having statistically significant higher percentage of Slamf1loCD34− and Slamf1−CD34− cells, which possess a lymphoid biased cell output (6). Perhaps, in the primary recipient of bone marrow cells, the lack of B cells leads to selection for a stem cell population that tries to augment the output of B cells. Whatever the case may be, enforced expression of miR-125b clearly generates a myeloid bias at the expense of B cells in primary transplants, although it also can lead to expanded populations of lymphoid cells in secondary transplants.
In addition to myeloid and B-cell development, Rossi et al. (13) reported a role of miR-125b in regulating the differentiation of naive CD4+ T cells to effector cells. In an effort to identify distinct microRNA signatures of human T-cell lymphocytes subsets, the authors found that miR-125b was highly expressed in naive CD4+ T cells compared to memory T cells. They identified targets of miR-125b in human cells and showed that it regulates a panel of genes involved in T-cell differentiation, such as IL2RB and PRDM1. Following transfection of a miR-125b-mimic, the differentiation of naive CD4+ T cells to effector T cells was blocked and, as well, there was a decreased expression of memory cell markers CXCR3 and CCR4 (Fig. 1C). Moreover, ectopic expression of miR-125b also affected the responsiveness of T cells. Overexpression of miR-125b inhibited the production of interleukin-13 (IL-13) and IFNγ upon stimulation with phorbol 12-myristate 13-acetate plus ionomycin (13). In addition to a cellular function of miR-125b in T cells, miR-125b also contributes to HIV-1 viral production and latency in these cells (14). HIV enters latency in resting CD4+ T cells in patients under antiretroviral therapy, allowing replication-competent HIV to be maintained in these cells, and thus allowing HIV to escape eradication by antiretroviral therapy (15–17). Huang et al. (14) demonstrated that inhibition of miR-125b along with miR-28, miR-150, miR-223, and miR-382 elevated HIV-1 production in resting CD4+ T cells but not in activated CD4+ T cells. miR-125b directly binds to the 3′UTR of HIV-1, thereby suppressing expression and viral production. This suggested that miR-125b might be utilized by HIV-1 to remain in latency in resting CD4+ T cells. This finding highlighted the possibility that inhibiting miR-125b along with miR-28, miR-150, miR-223, and miR-382 could be a method for eradication of the latent HIV-1 that escapes antiretroviral therapy (14). Thus, miR-125b is important for cellular T-cell function by modulating differentiation of naive T cells but also is important for controlling HIV-1 viral production and latency in these cells.
miR-125b modulates macrophage function
In an effort to further understand the role of miR-125b in hematopoiesis and immunity, we investigated the expression of miR-125b in different immune cells/tissues and found that miR-125b was much higher in macrophages compared to total splenocytes, thymocytes, splenic T cells, splenic B cells, and peritoneal macrophages from C57Bl/6 mice (18). To investigate the functional role of miR-125b in macrophages, we overexpressed miR-125b-1 (~15-fold) in bone marrow-derived macrophages (BMMs). Interestingly, miR-125b-1 overexpressing BMMs acquired a spread morphology that resembled activated macrophages. The activated status of these cells was confirmed by flow cytometric analyses, revealing an elevated expression level of macrophage activation markers, major histocompatibility complex class II, CD40, CD86, and CD80 (18). Augmented expression of miR-125b in terminally differentiated RAW264.7 macrophages gave similar results, further emphasizing that this microRNA promotes activation of macrophages. The heightened activation status may in part be due to miR-125b-1 suppressing its direct target IRF4. Knocking down IRF4 expression using RNAi recapitulated the effect of miR-125b-1 overexpression in macrophages, leading to increased surface expression of activation markers major histocompatibility complex class II, CD40, CD86, CD80, and IFNγR. Moreover, miR-125b-1 overexpressing BMMs also expressed higher levels of cell surface IFNγR, resulting in a heightened responsiveness of these cells to IFNγ stimulation. In addition to miR-125b-1 driving an elevated activation status and a hyperresponsiveness to stimulatory cues in macrophages, it also potentiated macrophage-mediated immune function. We showed that enforced augmented expression of miR-125b elevated the ability of macrophages to act as effective antigen-presenting cells for stimulating T cells in vitro (18). The effectiveness of miR-125b-stimulated macrophages at killing tumor cells was also assessed. miR-125b overexpressing macrophages were more effective at inducing apoptosis of the EL4-Fluc thymoma cell line in vitro. Macrophages with miR-125b ectopic expression were more effective in suppressing the ability of EL4 thymoma cells to expand in vivo when equal numbers of control or miR-125b overexpressing macrophages were co-injected with the cancer cells into mice (18). Our experiments thus provided evidence that miR-125b could modulate the activation status, responsiveness, and function of macrophages.
miR-125b:Lin28A axis
We are left with a reasonably clear picture of the function of miR-125a and miR-125b in the differentiation of HSCs into their downstream cell types. Clearly, miR-125 overexpression is able to potentiate the growth of HSCs and a sponge for miR-125 (and other miRs with the same seed sequence) can disadvantage HSCs, arguing that one function of miR-125 is to augment the HSC pool. As cells begin to differentiate away from the HSC stage, miR-125 appears to favor myeloid development and block B-lymphoid development, although secondary transplant gives a somewhat different picture. Not only does miR-125 stimulate commitment to the myeloid lineage, it also causes myeloid cells to adopt a more highly activated phenotype while it appears to inhibit the B-cell lineage continually at main nodes in their differentiation. The targets of miR-125 that allow it to have these disparate effects on the two lineages remain to be definitively defined. Lin28 is a direct target of miR-125b; the interaction between Lin28 and miR-125b is highly conserved from mammals to C. elegans. In bone marrow reconstitution experiments, we found that knocking down Lin28A resulted in an increased number of myeloid cells accompanied by a decrease in B cells in mice (9). However, leukemia was not observed in these mice. This might be due to insufficient knockdown of Lin28A in the experiments or that tumorigenesis induced by miR-125b is independent of Lin28A. Collectively, these data suggests that miR-125b might modulate hematopoiesis in part by directly regulating the expression of Lin28A, but the role of Lin28A in miR-125b-mediated leukemogenesis will require further experiments.
The Yin of miR-125b: its role as an oncomir in hematopoietic cells
Dysregulated expression of miR-125b in human leukemias
Because approximately 50% of microRNAs are located at fragile or cancer-associated genomic regions in the human genome (19), it is no surprise that aberrant expression of many different microRNAs has been observed in multiple cancer types. In particular, a significant number of studies have shown that dysregulated expression of miR-125b is correlated with human leukemia. Three studies separately reported a total of five patients with B-cell precursor acute lymphoblastic leukemia (BCP-ALL) harboring a genetic aberration with insertion or translocation of miR-125b-1 into the immunoglobulin heavy chain (IGH) locus (20–23). All three patients who were examined exhibited elevated miR-125b-1 expression compared to normal controls (20, 22, 23). Also, patients with myelodysplastic syndromes (MDS) who harbor t(2;11)(p21;q23) or 5q-exhibit increased expression of miR-125b-1 compared to healthy subjects (up to 70-fold) (24–26). Similarly, acute myeloid leukemic (AML) patients carrying t(2;11)(p21;q23) translocation or FLT3 mutation also have higher expression of miR-125b (24, 27). Overexpression of miR-125b appears be specific to cytogenetically distinct subsets of adult AML, because it was not found in patients harboring an NPM1 mutation (27). In pediatric AML patients, miR-125b is also expressed at higher than normal levels, with the M3 subtype being the highest (on average 200-fold) (28). Similarly, compared to normal CD34+ cells, miR-125b is also specifically upregulated in acute megakaryoblastic leukemic (AMKL) and transient leukemic (TS) patients with trisomy 21 (Down syndrome) but not AML FAB M5 patients (12). The common upregulation of miR-125b in leukemias of different cell origin suggests that it might serve as a cancer inducer or driver of cancer cells. However, some of these correlation studies are measured with tissue samples containing mixed cell types. Thus, whether the upregulation of miR-125b in leukemic patients is due to changes in the cell populations or is cell intrinsic will require more in-depth investigation.
Tumorigenic properties of miR-125b overexpression in hematopoietic cells in vitro The findings that miR-125b regulates hematopoiesis and is upregulated in leukemic samples generated a strong desire in the cancer field to determine if miR-125b plays a role in cancer development. Several in vitro studies supported the notion that miR-125b possesses tumorigenic potential. Several groups found that overexpression of miR-125b in hematopoietic cells generated cancers by inhibiting apoptosis, accelerating proliferation, and inducing abnormal self-renewal in non-stem cells. Ooi et al. (6) found that miR-125b directly repressed the anti-apoptotic genes Bmf and KLF13. The authors also found that miR-125b overexpressing cKit+ cells were quite resistant to staurosporine-induced apoptosis in vitro (6). Similarly, Puissegur et al. (10) showed that miR-125b overexpressing pre-B1 cells were also more resistant to apoptosis.
In addition to inhibiting apoptosis, constitutive miR-125b overexpression can accelerate proliferation and self-renewal of hematopoietic progenitor cells. Upon overexpressing miR-125b-2 in mouse fetal liver megakaryocytic progenitors (FL MPs), Klusmann et al. (12) found that it increases the size and numbers of megakaryocyte colony forming units (CFU-MKs) in vitro. This finding indicated that miR-125b-2 elevates the proliferative rate of MPs, which is a feature of cancer initiation. Importantly, the authors demonstrated that miR-125b elevated the re-plating efficiency of fetal liver progenitors in methylcellulose-based assays stimulated with thrombopoietin, arguing that miR-125b confers an abnormal self-renewal property on these cells, a hallmark of tumorigenesis (12). However, miR-125b-2 overexpression did not alter the differentiation potential of MP cells (12). It appears that enforced miR-125b-2 expression gives a proliferative advantage to FL MPs without interrupting their ability to differentiate. Moreover, the authors demonstrated that suppressing the activity of miR-125b in AMKL cell lines reduced cell viability. This finding suggests that some cancer cells are dependent for their viability on miR-125b expression, providing a rationale for targeting miR-125b as a form of cancer treatment. In support, downregulation of miR-125b in other types of cancer cells, e.g. HeLA, PC3, and Tera2 (embryonic carcinoma) cancer cell lines, also reduced cell growth (29). Collectively, these cell culture models reveal that miR-125b overexpression in megakaryocytic progenitors induces a growth advantage as well as a self-renewal capacity in FL MPs, providing a model in which miR-125b displays its tumorigenic potential.
Mouse models demonstrated miR-125b is an oncomir
Using murine models, we and other groups have demonstrated that restricted overexpression of miR-125b in the hematopoietic compartment induces leukemia. In bone marrow reconstitution experiments, we found that long-term enforced expression of miR-125b in mice leads to myeloproliferative disorder (MPD) or leukemia depending on the level of expression (5). Mice that received bone marrow cells overexpressing miR-125b to a modest level developed MPD characterized by increased absolute numbers of neutrophils and monocytes in the peripheral blood after two months of reconstitution (5). At this time, the bone marrow exhibited a mild expansion of myeloid cells, but they remained normal in the spleen. However, recipient mice that were transplanted with cells overexpressing miR-125b at a high level were myeloid-dominant in the bone marrow and spleens after two months of reconstitution. After 3.5 months, the peripheral blood of these mice was dominated by immature granulocytic and monocytic cells with a very high white blood cell count (5). Shortly after, the mice developed leukemia, became moribund, and subsequently succumbed to the disease. This demonstrated a dose-dependent ability of miR-125b to drive pathological myeloid expansion (5). Strikingly, we found that constitutive overexpression of miR-125a also caused mice to develop myeloid leukemia (9), further supporting the notion that the miR-125 microRNA family is an oncomir in hematopoietic cells.
Similar to our findings, Bousquet et al. (30) also demonstrated that restricted overexpression of miR-125b (~700-fold above normal) in the hematopoietic compartment caused leukemia in mice. The authors overexpressed miR-125b in fetal liver cells of mice and then performed bone-marrow reconstitution experiments. At 16 weeks after cell transplantation, the authors found that the recipient mice had higher white blood cell counts in the peripheral blood (30). The increase in white blood cell count was due to an increase in neutrophils, basophils, eosinophils, monocytes, and lymphocytes. Within 12 to 29 weeks, the recipient mice that received miR-125b overexpressing fetal liver cells developed a range of malignancies (30). Some mice developed myeloproliferative neoplasm (MPN) characterized by excess neutrophils in the blood with granulocytes dominating the bone marrow and spleen. A population of the miR-125b overexpressing mice also developed B220+ B-cell leukemia with dominance of lymphoblasts in the blood and spleen. Other mice exhibited T-cell leukemia. Interestingly, Bousquet et al. (30) revealed that mice overexpressing high levels of miR-125b (~1,500- fold) generally developed MPN whereas those that had lower over-expression (~500-fold) exhibited T-cell leukemia. Similarly, Ooi et al. (23) also showed that miR-125b overexpression in mice could cause lymphocytic leukemia. In secondary transplant experiments, Ooi et al. (6) noticed that ~11% of the recipient mice that received miR-125b overexpressing donor cells from primary transplant exhibited lymphoproliferative disease that was characterized by splenomegaly, enlarged thymi, and expansion of CD8+ T cells. Although the mechanism by which miR-125b drives leukemia remains largely un-investigated, the study by Enomoto et al. (21) supports an interpretation that miR-125b induces tumors through a cell-intrinsic mechanism at least for B-cell neoplasm. In an effort to examine the effect of the translocation of miR-125b into the IgH locus in human leukemic patient, Enomoto et al. (21) generated transgenic (TG) mice that mimicked this fusion. The authors generated Eu/miR-125b-TG mice that overexpressed miR-125b driven by the IGH enhancer and promoter. They found that mice overexpressing miR-125b driven by the IGH enhancer and promoter developed IgM− and IgM+ lethal B-cell malignancies, implying that miR-125b can drive tumorigenesis of B cells through a cell-intrinsic mechanism. Collectively, these mouse models emphasize that miR-125b is an oncomir capable of inducing tumorigenesis in different hematopoietic cells, including myeloid, T, and B cells.
To better understand the mechanism by which miR-125b induces myeloid cancer, we investigated the type of hematopoietic cells that are upregulated upon miR-125b overexpression. We focused on myeloid cancer, because we did not observe T- or B-cell malignancies in the primary transplant animals that overexpressed miR-125b in our bone marrow reconstitution experiments. At seven weeks after reconstitution, before the onset of overt leukemia, miR-125b overexpressing mice had drastic increases in all myeloid lineages, including granulocytes, macrophages, and dendritic cells. To examine the developmental stage at which miR-125b overexpression promotes myelopoiesis, we analyzed hematopoietic stem and progenitor cell (HSPC) numbers in the bone marrow. We were surprised to find that the numbers of HSCs were similar compared to control mice. However, progenitors downstream of HSCs, including multipotent progenitors (MPPs), common myeloid progenitors and granulocyte macrophage progenitors (CMP/GMPs), and myeloid-erythroid progenitors (MEPs), were all significantly increased. The CMP/GMPs were most drastically augmented compared to control mice. Thus, prior to the onset of leukemic disease, miR-125b enhances the development of the myeloid lineage likely by increasing the population of CMP/GMP (Fig. 1E). Since the number of HSCs were not increased before overt cancer development, this led us to hypothesize that miR-125b-driven leukemic development might not be through HSCs in our experimental settings. Recall that we mentioned earlier that in a competitive repopulation experiment, we found that miR-125b gave HSCs a competitive advantage. Here, we overexpressed miR-125b in most stem cells and would not have seen such an advantage. However, we did not see the HSC compartment expand, suggesting that as the rate of differentiation of the cells is such that an increased pool of HSCs is not observed. These experiments indicate that overexpression of miR-125b causes uncontrollable proliferation and/or abnormal self-renewing characteristics in early stage differentiated myeloid progenitors that subsequently leads to leukemia.
Although miR-125 appears to have an important normal role in the life of the HSC, overexpression of this microRNA can either lead to myeloid cell hyperplasia or to frank leukemia, depending on the extent of overexpression. We see tumors within a few months of transplantation, and other data suggests that the action of miR-125 is cell-intrinsic, so it is likely that miR-125 is a true oncogene for many if not all of the downstream cell types generated from the HSC.
The Yang of miR-146a: functions in immunity
Regulation of miR-146a expression
In the hematopoietic system, the expression of miR-146a is relatively low in progenitor cells and increases modestly with maturation. However, in certain specialized cell types, such as Ly-6Clo monocytes and epidermal Langerhans cells, miR-146a is constitutively expressed at a high level (31, 32, http://www.immgen.org/). In addition, miR-146a expression is upregulated upon stimulation of myeloid cells with microbial components and pro-inflammatory cytokines. miR-146a is also stimulated upon activation of T cells with T-cell receptor antigens (33–35). Viral and fungal infection also induces miR-146a expression (36–38).
Basal and induced expression of miR-146a is regulated by several important transcription factors in immune cells. Our initial characterization of miR-146a promoter locus identified two consensus NF-κB binding sites that were shown to be important in transcriptional activation of miR-146a in myeloid cells in response to inflammatory stimulation; mutations of these consensus sequences abolished the promoter activity in a luciferase reporter system (4). Subsequent studies showed that downregulation of NF-κB activity chemically or genetically inhibits basal and induced expression of miR-146a in macrophages and T cells, confirming the role of NF-κB in regulating miR-146a expression (34, 36, 37). In addition to regulation by NF-κB, expression of miR-146a is also regulated by lineage-dependent transcription factors. During myeloid differentiation, PU.1 works together with NF-κB to control the basal expression of miR-146a through dynamic occupancy at the promoter region (32, 39). In lymphocytes, the lineage-specific regulation of miR-146a may be fulfilled by other transcriptional factors, such as c-ETS (38, 40). In addition to transcriptional activation, miR-146a expression may be repressed in a lineage-specific manner. This was shown in the example of megakaryocyte differentiation, where PLZF upregulation interacts with the miR-146a promoter to inhibit its expression (41). A more extreme case involves global miRNA repression, including miR-146a, by the myc oncogenic transcriptional factor during lymphomagenesis (42). One interesting observation suggests estrogen can modestly downregulate miR-146a but not miR-146b in mouse splenic lymphocytes. However, it is not clear from the study whether the effect of estrogen on miR-146a expression is direct (43). Overall, basal miR-146a expression is regulated by a set of lineage-dependent transcription factors in a cell-lineage specific manner. This basal level can be further regulated in an activation-dependent manner by inducible transcription factors, such as NF-κB.
In comparison to miR-146a, the regulation of miR-146b expression is less well understood. Some previous reports on miR-146b expression were performed primarily with hybridization-based methods, which may have not been specific enough to separate the mature sequences of miR-146a and miR-146b because they only differ by one nucleotide. Nevertheless, miR-146b seems also to be induced by pro-inflammatory cytokines and dysregulated miR-146b expression is associated with several human non-hematologic cancers (44–49). In addition, in unpublished work, we have shown that miR-146b expression can be clearly detected by reverse transcriptase qualitative polymerase chain reaction in miR-146a-deficient bone marrow cells, indicating that it is not a pseudogene or undetectable in adult hematopoietic cells as previously suggested (33, 50).
miR-146a functions in immunity
Our initial report showed that miR-146a is highly upregulated in human monocytes in a NF-κB-dependent manner following Toll-like receptor (TLR) stimulation (4). Sequence complementation-based algorithms (e.g. TargetScan) identified tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) and IL-1 receptor-associated kinase-1 (IRAK1) as the top predicted targets of miR-146a. The regulation of these two proteins by miR-146a in monocyte/macrophages, T and B lymphocytes was then confirmed by 3′UTR luciferase reporter assays and gene expression analyses (4, 33, 34, 36, 51). Because TRAF6 and IRAK1 are known feedback regulators of the NF-κB signaling pathway, a molecular circuitry involving miR-146a, TRAF6/IRAK1, and NF-κB is proposed to be important in the regulation of both innate and adaptive immunity (52). One of the first pieces of definitive evidence of this hypothesis came from the study of miR-146a-deficient mice generated in our laboratory (33). Consistent with the proposed hypothesis, miR-146a−/− mice were found to be hyper-responsive to LPS challenge. Stimulation with sublethal level of LPS in vivo induced a higher serum level of pro-inflammatory cytokines, including TNFα and IL-6 and IL-1β, in miR-146a−/− mice than in wildtype mice. In addition, miR-146a−/− mice experienced a higher mortality rate in response to minimal lethal LPS challenge. The hyper-inflammatory response was also observed in bone marrow-derived macrophages (BMDMs) from miR-146a−/− mice in vitro. To complement the study on miR-146a−/− mice, enforced overexpression of miR-146a in THP-1 cells resulted in an attenuated inflammatory response (33). Work of others has confirmed these general observations. These studies provide further support that miR-146a is an important negative regulator of innate immune activation, potentially by targeting TRAF6 and IRAK1 and by orchestrating the silencing of TNFα gene in human monocytic cell lines in the context of endotoxin-induced tolerance (53–55).
miR-146a as effector of monocyte/macrophage function
In addition to regulating pro-inflammatory cytokine production, miR-146a negatively regulates type I IFN production in vesicular stomatitis virus-infected mouse peritoneal macrophages by targeting TRAF6, IRAK1, and IRAK2 through a RIG-I/NF-κB-dependent but TLR4/MyD88-independent pathway (36). A study in human peripheral blood mononuclear cells confirmed a role for miR-146a as a negative regulator of the type I IFN pathway (56). In a more elaborate example of cell-type-specific regulation by miR-146a in the innate immune system, miR-146a specifically controls the Ly-6Chi but not Ly-6Clo monocyte response during inflammatory challenge (31). The regulation by miR-146a in this context is cell intrinsic, apparently through directly targeting Relb, thus also connecting miR-146a to the noncanonical pathway of NF-κB activation. This study further substantiates the role of miR-146a in innate immunity by controlling monocyte functional heterogeneity during an inflammatory response. Given the hyperinflammation and the exaggerated immune response in the absence of miR-146a, we would expect miR-146a−/− mice to be more resistant to bacterial infection than wildtype animals. Indeed, miR-146a−/− mice displayed a lower bacterial load upon infection with Listeria monocytogenes (31).
Functional role of miR-146a in T cells
The role of miR-146a in adaptive immunity is particularly well studied in T cells. Aging miR-146a−/− mice develop an autoimmune disorder with lymphadenopathy, lymphocyte infiltration in various organs, and an activated T-cell phenotype (33). We carried out detailed analysis in a subsequent study to understand the physiological role of miR-146a in regulating the T-cell response to antigen stimulation (34). We showed that miR-146a-deficient CD4+ and CD8+ T cells are hyper-responsive following T-cell receptor (TCR) stimulation, as indicated by increased proliferation and survival, exaggerated activation phenotype, and enhanced effector cytokine production. The regulation of cell-intrinsic T-cell response upon TCR stimulation by miR-146a was shown to again be through the regulation of TRAF6/IRAK1 and NF-κB signaling. This study adds miR-146a to a list of well-known negative regulators of NF-κB, including IκBα and A20, acting following TCR activation and suggests that these feedback regulators function collaboratively to limit the extent and timing of activation, with each apparently fulfilling a crucial non-redundant role. These mice studies were supported by studies in human T cells showing that miR-146a overexpression impairs IL-2 production. However, contrary to our observation that TCR-induced apoptosis was reduced in miR-146a-deficient T cells and enhanced in T cells with miR-146a overexpression, activation-induced cell death seems to be inhibited by miR-146a overexpression in human Jurkat T-cell cancer line (40). The discrepancy may be due to the differences between mouse primary T cells and human cancer cell lines or the level of miR-146a overexpression achieved in the two experimental systems.
In addition to functioning in effector T cells in a cell-intrinsic manner, miR-146a also controls the regulation of T-helper 1 (Th1) responses by affecting regulatory T-cell (Treg)-function (57). Treg cells maintain immune homeostasis by suppressing the inflammatory response and preventing an overreaction. In Treg cells, expression of miR-146a, but not miR-146b, is much higher than that in naive T cells. In turn, miR-146a-deficient Treg cells are functionally defective in their ability to restrain effector T cells, leading to overproduction of IFNγ and autoimmunity. Stat1 appears to be a responsible target for miR-146a in regulating Treg cells. This study revealed an important regulatory function of miR-146a in Treg cells, suggesting upregulation of miR-146a is required for Treg cells to properly control the IFNγ-mediated Th1 response. The results imply an interesting phenomenon, which is that excessive effector cytokine signaling in Treg cells can lead to their failure to suppress the corresponding cytokine response in effector T cells. Further studies are required to elucidate the mechanistic implications behind this type of Treg cell/effector T-cell regulation.
miR-146a in B cells
Our knowledge of the function of miR-146a in B cells is limited. One study has provided evidence for miR-146a induction in a NF-κB-dependent manner by Epstein-Barr virus latent membrane protein 1 (LMP1) in human B lymphocytes (38). Study from miR-146a-deficient mice suggested that B cells in these mice are also hyperactivated and autoreactive, because we can detect anti-double stranded DNA (anti-dsDNA) antibodies in the serum and a small percentage of B-cell lymphomas in aging miR-146a−/− mice (33, 58). We have also seen de-repression of TRAF6 and IRAK1 by Western blot in splenic B cells from miR-146a−/− mice (33). Further studies will be required to understand the function of miR-146a in B cells.
Functions of miR-146a in hematopoiesis: effect of miR-146a in megakaryopoiesis
In addition to the functional role of miR-146a within immune cells, miR-146a also is important for regulating the development of hematopoietic cells. Consistent with its expression pattern, miR-146a plays a role in the development of a wide spectrum of hematopoietic cells. An early in vitro study using human leukemic cell lines and CD34+ progenitor cell culture suggested a role for miR-146a in megakaryopoiesis, during which upregulation of the transcription factor PLZF represses miR-146a expression, which in turn leads to increased expression of a direct target of miR-146a, CXCR4. In this study (41), overexpression of miR-146a led to impaired megakaryocytic proliferation, differentiation, and maturation. However, the cell-autonomous regulation of miR-146a in megakaryopoiesis is challenged by a later study, which showed no detectable changes in megakaryocyte development and platelet activation properties by overexpression of miR-146a in all hematopoietic cells in mice (59). This discrepancy may be a consequence of differences between the two experimental systems, an in vitro study with human cells versus a mouse in vivo system. Nevertheless, the study done with an overexpression system does not exclude a role for miR-146a in megakaryopoiesis, because miR-146a expression is upregulated during megakaryopoiesis and the endogenous level of miR-146a may be sufficient to downregulate the relevant target genes to ensure normal megakaryocyte development. In contrast, knocking down miR-146a and miR-145 concurrently with a ‘sponge’ in mouse hematopoietic stem and progenitor cells (HSPCs) leads to megakaryocyte expansion and thrombocytosis in vivo. However, this effect may be indirectly mediated by increased production of the positive acting cytokine IL-6 from miR-146a-deficient myeloid and lymphoid cells (51). In contrast, significant reduction in platelet counts is frequently seen in aging but not in young miR-146−/− mice (33, 58). The observed thrombocytopenia in aging miR-146a−/− is a manifestation of bone marrow failure and fibrosis or chronic dysregulation of inflammatory cytokine production. Overall, miR-146a has a regulatory function in megakaryopoiesis in a cell-intrinsic manner and/or by controlling the inflammatory environment.
miR-146a in myeloid development
The involvement of miR-146a in myeloid development is particularly interesting. Stable knockdown of both miR-145 and miR-146a concurrently in mouse HSPCs results in variable neutropenia and decreased colony-forming ability of bone marrow cells (51). In support of miR-146a as the dominant miRNA in this effect, overexpression of TRAF6, a validated target of miR-146a, also leads to mild neutropenia, with a subset of mice progressing to marrow failure or acute myeloid leukemia (51). More conclusive data came from the subsequent analysis of miR-146a−/− mice. MiR-146a−/− mice are born at the expected Mendalian frequency and show no detectable phenotype during the first 2 months of their life in the absence of an inflammatory or infectious challenge. This suggests that miR-146a is not an essential gene for the proper development of blood lineages under steady state in young mice. However, with natural aging, miR-146a−/− mice develop a progressive myeloproliferative phenotype in both spleen and bone marrow. Specifically, there is a significant increase in the percentage of CD11b+ cells in their spleens and bone marrows. It can readily be detected by FACS analysis of 6-month-old miR-146a−/− mice. The myeloid expansion becomes progressively larger by 12 months, as indicated by more than a threefold increase in the CD11b+ percentage and more than a 10-fold increase in the total number of CD11b+ cells in spleens. Bone marrows of these mice are also dominated by myeloid cells, representing close to 80% of all bone marrow cells (33, 58). Bone marrow-derived macrophages (BMDMs) developed from young knockout mice also proliferate faster in response to macrophage colony-stimulating factor (M-CSF). Consistent with this observation, increased surface expression of CSF1R, the receptor for M-CSF, is also detected in the knockout spleen, bone marrow, and peripheral blood myeloid cells (33, 58). In addition, miR-146a deficiency also confers a proliferative advantage on Ly-6Chi monocytes in bone marrow, spleen, and the peritoneal cavity in inflammatory conditions (31). Mechanistically, miR-146a−/− spleen and bone marrow cells exhibit increased transcription of NF-κB-responsive genes. Increased NF-κB p65 translocation to the nucleus is also evident in miR-146a−/− spleen cells. The increased myeloproliferation is dependent on the activation of NF-κB, because genetic ablation of an important subunit of NF-κB, p50, suppresses myeloproliferation (58). Study pursuing the mechanism of NF-κB-driven myeloproliferation will help shed further insight on the molecular link between chronic inflammation and hematopoietic malignancy.
Enforced expression of miR-146a in bone marrow transplant studies in mice has produced inconsistent results. One study reported that overexpression of miR-146a in 5-fluorouracil (5-FU) treated bone marrow cells results in a transient myeloid expansion that subsequently returns to normal (35). Another report showed no detectable change in circulating granulocytes, T cells, and B cells by overexpressing miR-146a in lineage negative bone marrow cells (59). Lastly, one other group showed that ectopic expression of miR-146a in sorted LKS (lineage−cKit+Sca1+) cells directs the selective differentiation of HSCs into peritoneal macrophages in mice. Surprisingly, other than the peritoneal cavity, this study failed to detect any transplanted miR-146a-expressing donor cells in the peripheral blood, spleen, and bone marrow, precluding comparison in these hematopoietic compartments with the other overexpression studies. The same report (39) also showed that inhibition of miR-146a impairs macrophage formation during early zebrafish development. In addition to different investigators overexpressing miR-146a in different fractions of bone marrow cells, it is also unclear whether the levels of miR-146a overexpression achieved were comparable between these studies. Despite some inconsistent findings, it appears that enforced expression of miR-146a in mouse bone marrow cells has an overall minor effect on hematopoiesis, at least in the major hematopoietic organs under steady state.
The Yin of miRNA-146a: involvement in immune system pathology
Overview of miRNA-146a in pathology
The importance of miR-146a is further exemplified by its extensive involvement in immunological pathologies, including autoimmunity and hematologic malignancy. Loss-of-function studies in mice have provided some important insight in the role of miR-146a in the pathogenesis of autoimmunity. Genetic deletion of miR-146a in all the cells in mice leads to autoimmunity, indicated by splenomegaly, lymphadenopathy, lymphocyte-infiltration in liver, kidney, and lung, and the development anti-dsDNA antibodies (33). The contribution of miR-146a to autoimmunity occurs in at least three separate ways: (i) regulation of miR-146a in effector T cells, (ii) regulation of miR-146a in Treg cells, and (iii) regulation of myeloid cell-mediated inflammation. MiR-146a-deficient T-effector cells contribute to autoimmunity in a cell-intrinsic manner. MiR-146a-deficeint T cells remain hyper-responsive to antigen stimulation when adoptively transferred into wildtype mice and they induce a spontaneous autoimmune pathology when transferred into Rag1−/− mice. Mechanistically, the enhanced proliferation, impaired activation-induced apoptosis, and exaggerated effector cytokine production seen in miR-146a-deficient CD4+ and CD8+ T cells derive from uncontrolled NF-κB activation as a result of de-repressed TRAF6 and IRAK1 (34). Whether the multi-organ autoimmune pathology is caused by a general overactivation of miR-146a-deficient T cells or by a more specific autoreactivity against a particular cell type or self-antigen remains an interesting unanswered question. In addition, miR-146a-deficient Treg cells are also functionally defective in suppressing excessive Th1 response. In the absence of miR-146a, upregulation of signal transducer and activator of transcription 1 (STAT1) in Treg cells is responsible for the failure to suppress wildtype effector T cells, leading to fatal IFNγ-mediated autoimmunity in a variety of organs (57). The chronic inflammatory environment conditioned by miR-146a-deficient myeloid cells also contributes to autoimmune pathology by further stimulating T cells and promoting tissue damage (33, 58). Lastly, anti-dsDNA antibodies are present in the serum of aging miR-146a−/− mice (33). Future studies examining of the role of miR-146a in B cells may yield additional insight into the regulation of autoantibody production. Given the widespread expression of miR-146a outside of hematopoietic cells, miR-146a may exert protective function in non-hematopoietic tissues. Evidence supporting this comes from studies on lung epithelial cells and neonate guts (60, 61). Interestingly, during the dramatic transition from a sterile environment to a bacteria-colonized surface in neonate guts, sustained miR-146a expression seems to be critical to mediate innate immune tolerance by repressing IRAK1 (60). In light of this observation, we speculate that miR-146a deficiency in intestinal epithelial cells may lead to intestinal inflammation, contributing to the overall autoimmune pathology in miR-146a−/− mice.
Corroborating the in vivo studies in mice, miR-146a has been found to be extensively dysregulated in human patients with autoimmune diseases, including systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA). In lupus patients, miR-146a expression is downregulated in peripheral blood mononuclear cells (PBMCs). In addition, the level of miR-146a expression is negatively correlated with disease severity, with lower expression correlating with more active disease and proteinuria symptoms (56). Type I IFN is suggested to be the responsible pathway upregulated in lupus, and additional targets of miR-146a, IRF5, and STAT1 have been identified based on 3′UTR luciferase reporter assay and Western blot analysis in 293T cells overexpressing miR-146a. Interestingly, enforced expression of miR-146a in PBMCs of lupus patients can downregulate a subset of IFN-responsive genes (56). Additional evidence comes from genome-wide association studies that identified SNPs (single-nucleotide polymorphisms) in the regulatory region of miR-146a that are associated with lupus disease risk (62, 63). One such SNP (rs57095329) in the promoter region of miR-146a is associated with the binding strength of transcription factor Ets-1, another lupus-susceptibility gene. The risk-associated G allele in this SNP correlates with reduced Ets-1 binding and decreased miR-146a expression (63).
In contrast to SLE where miR-146a expression is shown to be downregulated, overexpression of miR-146a is frequently detected in rheumatoid arthritis (RA) patients (64–66). miR-146a also seems to be highly expressed in osteoarthritis cartilage (67). Despite that both SLE and RA are autoimmune diseases with some shared characteristics, the underlying etiologies may be distinct. For example, chronic inflammation is a shared pathology, but the dysregulated cytokine profiles are different between SLE and RA. The IFN pathway is intimately linked to the pathogenesis of SLE, while increased TNFα, IL-1β, and IL-6 production are more prominent features of RA (64). Because pro-inflammatory cytokines can significantly upregulate miR-146a expression in various cell types, increased miR-146a expression may simply reflect the hyper-inflammatory environment and may be a useful marker for disease severity in RA. However, miR-146a deficiency may be causally related to the pathogenesis of SLE. It will be interesting to further investigate the involvement of miR-146a deficiency in the development and progression of SLE and a number of other autoimmune diseases and whether upregulation of miR-146a may have therapeutic benefit in mouse models and human clinical samples. In the addition to the role of miR-146a in Th1 cells, Treg cells, and monocyte/macrophages, understanding the function of miR-146a in Th17 cells (68), a key cell type in a number of autoimmune diseases, may provide additional insight on this miRNA’s involvement in autoimmunity.
There is also some evidence suggesting a role of miR-146a in atherosclerosis, a less studied aspect of miR-146a biology. Two reports (69, 70) suggested that upregulation of miR-146a may inhibit atherosclerosis by suppressing TLR4 and CD40L expression in low-density lipoprotein-induced macrophages and dendritic cells.
miRNA-146a in hematologic malignancy
Recent studies have also revealed an important role of miR-146a in hematologic pre-malignancy and malignancy, most notably myelodysplastic syndromes (MDS), myeloproliferative disease, myeloid cancer, and bone marrow failure. MDS represent a heterogeneous group of clonal diseases of HSC origin characterized by ineffective hematopoietic differentiation, peripheral blood cytopenias, bone marrow dysplasia, and a propensity to progress to leukemia and bone marrow failure. Recent studies have made significant advances on the molecular pathogenesis of MDS, including dysregulated expression of miRNAs (71–73). Starczynowski et al. (51) have shown that two miRNAs, miR-145 and miR-146a, are located in or near the commonly deleted region of 5q- syndrome, a common subtype of MDS. Accordingly, expression of these two miRNAs is reduced in patients of 5q- syndrome. The same study also showed that the stable knockdown of miR-145 and miR-146a concurrently in mouse HSPCs recapitulates many of the key features of human 5q- syndrome, including thrombocytosis, megakaryocytic dysplasia, and mild neutropenia. In addition, overexpression of TRAF6, a target of miR-146a, was sufficient to phenocopy many of the features of knocking down miR-145 and miR-146a, suggesting that miR-146a may be a more dominant miRNA. Studies with miR-146a knockout mouse are reaffirming. With increasing age, miR-146a−/− mice develop myeloid expansion in spleen and bone marrow, pancytopenia in the periphery, and a propensity to progress to bone marrow failure and myeloid cancer, a constellation of features reminiscent of a mixed myelodysplastic syndrome and myeloproliferative neoplasm (MDS/MPN) (33, 58). Mechanistically, increased NF-κB activation is a main driver of the myeloproliferative and bone marrow failure phenotype, because ablation of the p50 subunit greatly reduces MDS/MPN symptoms. It is now increasingly appreciated that a distinct hematopoietic developmental program, termed inflammatory hematopoiesis, is activated in bone marrow during inflammation to promote myelopoiesis at the expense of lymphopoiesis and erythropoiesis (74). We speculate that uncontrolled and persistent inflammatory hematopoiesis may promote pathological features of MDS and MPN, and over time, the pathologic hematopoietic program becomes permanent and irreversible through additional genetic mutations and/or epigenetic changes. Because chronic inflammation is a prominent feature of miR-146a−/− mice during stimulation and aging, it will be interesting to investigate whether miR-146a may serve as a molecular link between chronic inflammation and hematopoietic malignancy, such as MDS and MPN. In addition to myeloid cancers, about 15% of miR-146a−/− mice also develop B-cell or mixed B and T-cell lymphomas by 18 to 24 months (58). It is not clear whether the mechanism underlying lymphoma development in miR-146a-deficient mice is distinct from the one driving myeloid oncogenesis.
Many groups have extensively profiled miRNA expression in human leukemia in an effort to identify microRNA signatures associated with leukemia diagnosis, prognosis, and pathogenesis. Reports from different groups have identified miR-146a downregulation in CD34+ cells or total bone marrow cells from patients with 5q- syndrome (26, 51, 73). However, miR-146a downregulation is not consistently observed in leukemia studies. One expression profiling study showed that miR-146a is downregulated in bone marrow cells of AML patients compared to normal CD34+ cells (75). However, in another study that compared miRNA expression between AML and ALL samples, increased expression of miR-146a in both ALL and AML was shown to correlate with poor survival (76). miR-146a was also shown to function as a tumor suppressor in natural killer/T-cell lymphoma, and reduced expression of miR-146a was identified as a poor prognostic factor (77). It seems miR-146a downregulation is a consistent feature of 5q- syndrome or even all MDS and may be involved in the pathogenesis of MDS. The role of miR-146a in leukemia and lymphoma requires further clarification.
Relationship between miR-146a and miR-155
MicroRNA-155 (miR-155) is another miRNA identified from our original screen for miRNAs induced upon NF-κB activation (4). However, in contrast to miR-146a, miR-155 plays an almost exact opposite role in immunity and hematopoiesis. For more detailed reviews of miR-155 biology, we refer you to recent reviews published elsewhere (50, 74, 78–80). Here we focus on the aspect of miR-155 function opposing that of miR-146a. NF-κB activates an elaborate and potent transcription program central to inflammation and immunity. Because of the potency of this pathway, proper regulation of the magnitude and duration of NF-κB activation is essential (50, 81–83). NF-κB-induced miRNAs, miR-146a, and miR-155 may be two important regulators that balance the negative and positive effects driven by NF-κB activation. The opposing roles of miR-146a and miR-155 have been most clearly demonstrated in the function and development of myeloid cells. While basal expression of miR-155 is low in myeloid cells, many of the inflammatory stimuli that can induce miR-146a expression, including TLR ligands and pro-inflammatory cytokines, also upregulate miR-155 expression significantly (84, 85).
In regulating the functional capacity of myeloid cells, miR-155, upon induction, promotes expression of pro-inflammatory cytokines and the IFN response. This regulation is most likely through repression of suppressor of cytokine signaling 1 (SOCS1) and SH2 domain-containing inositol phosphatase-1 (SHIP1), both negative regulators of the pro-inflammatory pathways (86, 87). Consistent with this idea, inhibition of miR-155 in macrophages confers tolerance to endotoxin shock in mice, while mice with miR-155 overexpression become hypersensitive (87, 88). The exactly opposite effect has been observed with miR-146a inhibition and overexpression (33). In addition to pro-inflammatory cytokine production, miR-146a and miR-155 also regulate immunity in the opposite manner. In general, mice with targeted miR-155 deletion are immunocompromised, as indicated by attenuated immune response to immunization and infection (89, 90). Inhibiting miR-146a may result in a heightened immune response as shown by enhanced protection against Listeria infection (31). miR-146a and miR-155 also have an opposite effect in regulating certain aspects of T-cell function. As discussed above, miR-146a-deficient CD4+ T cells display a hyper-activated response upon TCR stimulation, while miR-155-deficient CD4+ T cells are attenuated in IL-2 and IFNγ cytokine production upon TCR stimulation (34, 89). Similarly, miR-155 and miR-146a also function in an opposite direction in autoimmunity. Contrary to miR-146a−/− mice that are prone to developing autoimmune disease, miR-155−/− mice were shown to be significantly resistant to an induced model of experimental autoimmune encephalomyelitis (EAE) because of a deficit in inflammatory T-cell development and, to a lesser extent, a defect in myeloid dendritic cell function (91). In a recent effort to directly study the opposing function of miR-146a and miR-155, mice deficient of miR-146a, miR-155, or both miRNAs were implanted with subcutaneous solid tumors and were shown to have differential ability to control tumor growth (92). This study showed again that in the context of antitumor immunity, miR-146a and miR-155 play opposing roles; miR-155 promotes while miR-146a represses antitumor immunity, correlating their functions to regulate IFNγ-producing CD4+ T-cell development.
As described above, miR-146a and miR-155 are concurrently upregulated in response to pro-inflammatory cues. Data suggest that miR-146a and miR-155 may counterbalance each other during inflammatory hematopoiesis. Sustained expression of miR-155 in mouse HSPCs results in a myeloproliferative disorder, characterized by profound myeloproliferation with dysplastic changes in the bone marrow, splenomegaly as a result of extramedullary hematopoiesis, peripheral anemia, lymphopenia, and thrombocytopenia with increased myeloid cells (84). The constellation of hematologic abnormalities seen in miR-155-overexpressing mice is strikingly similar to the one observed in aging miR-146a-deficient mice (58). miR-155-induced myeloproliferative disorder is thought to be primarily through repressing SHIP1 (86). In addition, overexpression of miR-155 in a B-cell-specific manner in a transgenic mouse model with E(mu)-promoter-driven miR-155 expression results in B-cell leukemia and lymphoma (93). Similar B-cell lymphoma pathology has also been observed in a subset of aging miR-146a−/− mice (58).
Given the above evidence, it is tempting to speculate that miR-146a and miR-155 are evolutionarily selected to function in a tug-of-war manner to properly control the magnitude and duration of NF-κB activation. Balanced level and proper timing of miR-146a and miR-155 expression represents one critical layer of regulation to ensure that NF-κB is activated in a controlled manner.
Summary remarks
Intensive study of miR-125b and miR-146a have revealed both the Yin and Yang characteristics of these microRNAs in the immune system. Whereas they regulate normal function, including hematopoiesis and immunity, abnormal and unregulated expression of these microRNAs contributes to myeloproliferative disorder and cancer.
miR-125b and miR-146a appear to share overlapping biological processes. In some cases, they have similar impacts, whereas in others, they appear to have opposing effects. Both miR-125b and miR-146a suppress T-cell activation and seemingly disfavor the B-cell axis. However, the effect of these microRNAs on myeloid development and function is strongly contrasting, with miR-125b enhancing macrophage inflammatory responses while miR-146a negatively regulates these processes. In sharp contrast, overexpression of miR-125b leads to myeloid hyperproliferative disorder and hematopoietic cancer, whereas it is ablation of miR-146a that induces similar events. The effects of miR-125b overexpression on these disorders are rapid and observed in mice shortly after bone marrow reconstitution experiments, suggesting that miR-125b may act directly on bone marrow cells to give rise to these characteristics. By contrast, induction of myeloid proliferation and cancer following ablation of miR-146a is a gradual process that has increasing impact as mice age, implying that these events arise from chronic and progressive alterations in immune function. Because of the involvement of the NF-κB pathway in this process, we speculate that continual low grade inflammatory events in miR-146a−/− mice provide repeated, slowly resolved insults to hematopoietic cells that subsequently cause irreversible cellular genetic mutations and/or epigenetic changes, of which a frequent outcome is the development of cancer.
It is become increasingly clear that microRNAs have a major impact on many physiological processes. It is thus expected that dysregulated expression of microRNAs would be closely tied to specific pathologies. Targeting microRNAs could be an invaluable methodology for the treatment of human disease, such as autoimmunity and leukemia.
Fig. 2. Expression of miR-146a is regulated by a set of transcriptional factors in a cell-lineage specific manner, involving such factors as PU.1, c-ETS, and PLZF, and in an activation-dependent manner, involving such factors as NF-κB.
Validated targets of miR-146a include Irak1, Traf6, Stat1, and RelB, which are all involved in the NF-κB and STAT pathways, highlighting the role of miR-146a as a critical negative regulator of inflammatory and interferon signaling.
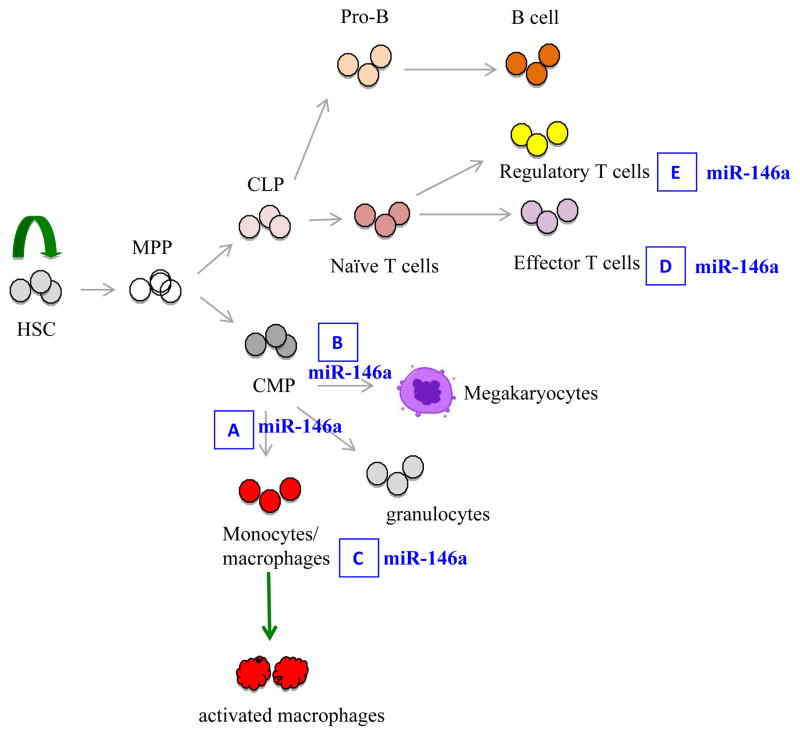
Fig. 3. A simplified schematic depiction of hematopoietic tree highlighting the role of miR-146a in hematopoiesis and immune cell function.
(A). miR-146a negatively regulates myelopoiesis and myeloproliferation during inflammation and aging. In the absence of miR-146a, increased myeloproliferation is observed. (B). miR-146a negatively regulates megakaryopoiesis. Overexpression of miR-146a inhibits megakaryocyte proliferation and differentiation, while downregulation of miR-146a promotes megakaryocyte expansion and platelet production. However, in aging miR-146a-deficient mice with bone marrow fibrosis, thrombocytopenia is observed. MiR-146a is a negative regulator of monocyte and macrophage activation (C) and effector T-cell activation (D). (E). In regulatory T cells, miR-146a positively controls Treg functional competence. In the absence of miR-146a, Treg cells are defective in suppressing effector T-cell activation.
Acknowledgments
This work was supported by NIH grants 5P01CA132681-02, 5R01AI093531-02, an Ellison Foundation award to D.B and F32 CA139883 to A.Y.S. J.L.Z. was supported by the UCLA/Caltech Joint Medical Scientist Training Program and National Research Service Award F30HL110691 from the National Institute of Health.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 4.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerrits A, et al. Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood. 2012;119:377–387. doi: 10.1182/blood-2011-01-331686. [DOI] [PubMed] [Google Scholar]
- 8.Guo S, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri AA, So AY, Mehta A, et al. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin 28A. Proc Natl Acad Sci USA. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puissegur MP, et al. B-cell regulator of immunoglobulin heavy chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia. 2012;26:2224–2232. doi: 10.1038/leu.2012.95. [DOI] [PubMed] [Google Scholar]
- 11.Gururajan M, et al. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klusmann JH, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblasticleukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi RL, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. NatMed. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 15.Chun TW, et al. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. ProcNatl Acad Sci USA. 2003;100:1908–1913. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furtado MR, et al. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, et al. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 18.Chaudhuri AA, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapiro E, et al. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–1364. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto Y, et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia. 2011;25:1849–1856. doi: 10.1038/leu.2011.166. [DOI] [PubMed] [Google Scholar]
- 22.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–2010. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 23.Tassano E, Acquila M, Tavella E, Micalizzi C, Panarello C, Morerio C. MicroRNA-125b-1 and BLID upregulation resulting from a novel IGH translocation in childhood B-Cell precursor acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:682–687. doi: 10.1002/gcc.20776. [DOI] [PubMed] [Google Scholar]
- 24.Bousquet M, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussein K, et al. Aberrant microRNA expression pattern in myelodysplastic bone marrow cells. Leukemia Res. 2010;34:1169–1174. doi: 10.1016/j.leukres.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Votavova H, et al. Differential expression of microRNAs in CD34+ cells of 5q-syndrome. J Hematol Oncol. 2011;4:1. doi: 10.1186/1756-8722-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cammarata G, et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. A, J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H, et al. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PloS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee YS, Kim HK, Chung S, Kim KS, Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem. 2005;280:16635–16641. doi: 10.1074/jbc.M412247200. [DOI] [PubMed] [Google Scholar]
- 30.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Etzrodt M, et al. Regulation of monocyte functional heterogeneity by miR-146a and Relb. Cell Rep. 2012;1:317–324. doi: 10.1016/j.celrep.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurkin J, et al. miR-146a is differentially expressed by myeloid dendritic cell subsets and desensitizes cells to TLR2-dependent activation. J Immunol. 2010;184:4955–4965. doi: 10.4049/jimmunol.0903021. [DOI] [PubMed] [Google Scholar]
- 33.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L, et al. miR-146a controls the resolution of T cell responses in mice. J Exp Med. 2012;209:1655–1670. doi: 10.1084/jem.20112218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starczynowski DT, et al. MicroRNA-146a disrupts hematopoietic differentiation and survival. Exp Hematol. 2011;39:167–178. e4. doi: 10.1016/j.exphem.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Hou J, et al. MicroRNA-146a feedback inhibits RIG-I-dependent Type IIFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 37.Monk CE, Hutvagner G, Arthur JS. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS One. 2010;5:e13669. doi: 10.1371/journal.pone.0013669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cameron JE, et al. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghani S, et al. Macrophage development from HSCs requires PU. 1-coordinated microRNA expression. Blood. 2011;118:2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Curtale G, Citarella F, Carissimi C, et al. An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood. 2010;115:265–273. doi: 10.1182/blood-2009-06-225987. [DOI] [PubMed] [Google Scholar]
- 41.Labbaye C, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- 42.Chang TC, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai R, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA. Suppression of LPS-induced Interferon-gamma and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: a novel mechanism of immune modulation. Blood. 2008;112:4591–4597. doi: 10.1182/blood-2008-04-152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia H, et al. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. doi: 10.1016/j.brainres.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 45.Chou CK, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20:489–494. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 46.Geraldo MV, Yamashita AS, Kimura ET. MicroRNA miR-146b-5p regulates signal transduction of TGF-beta by repressing SMAD4 in thyroid cancer. Oncogene. 2012;31:1910–1922. doi: 10.1038/onc.2011.381. [DOI] [PubMed] [Google Scholar]
- 47.Katakowski M, Zheng X, Jiang F, Rogers T, Szalad A, Chopp M. MiR-146b-5p suppresses EGFR expression and reduces in vitro migration and invasion of glioma. Cancer Invest. 2010;28:1024–1030. doi: 10.3109/07357907.2010.512596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Man YG, et al. Aberrant expression of chromogranin A, miR-146a, and miR-146b-5p in prostate structures with focally disrupted basal cell layers: an early sign of invasion and hormone-refractory cancer? Cancer Genomics Proteomics. 2011;8:235–244. [PubMed] [Google Scholar]
- 49.Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett. 2009;583:3349–3355. doi: 10.1016/j.febslet.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 50.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev. 2012;246:205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 51.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 52.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Nahid MA, Pauley KM, Satoh M, Chan EK. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nahid MA, Satoh M, Chan EK. Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J Immunol. 2011;186:1723–1734. doi: 10.4049/jimmunol.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.El Gazzar M, Church A, Liu T, McCall CE. MicroRNA-146a regulates both transcription silencing and translation disruption of TNF-alpha during TLR4-induced gene reprogramming. J Leukoc Biol. 2011;90:509–519. doi: 10.1189/jlb.0211074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang Y, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 57.Lu LF, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao JL, Rao DS, Boldin MP, Taganov KD, O’Connell RM, Baltimore D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. 2011 May 31;108(22):9184–9. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Opalinska JB, et al. MicroRNA expression in maturing murine megakaryocytes. Blood. 2010;116:e128–138. doi: 10.1182/blood-2010-06-292920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chassin C, et al. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe. 2010;8:358–368. doi: 10.1016/j.chom.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol. 2008;180:5689–5698. doi: 10.4049/jimmunol.180.8.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lofgren SE, et al. Genetic association of miRNA-146a with systemic lupus erythematosus in Europeans through decreased expression of the gene. Genes Immun. 2012;13:268–274. doi: 10.1038/gene.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo X, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan EK, Satoh M, Pauley KM. Contrast in aberrant microRNA expression in systemic lupus erythematosus and rheumatoid arthritis: is microRNA-146 all we need? Arthritis Rheum. 2009;60:912–915. doi: 10.1002/art.24421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abou-Zeid A, Saad M, Soliman E. MicroRNA 146a expression in rheumatoid arthritis: association with tumor necrosis factor-alpha and disease activity. Genet Test Mol Biomarkers. 2011;15:807–812. doi: 10.1089/gtmb.2011.0026. [DOI] [PubMed] [Google Scholar]
- 66.Nakasa T, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamasaki K, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 69.Chen T, et al. MicroRNA-146a regulates the maturation process and pro-inflammatory cytokine secretion by targeting CD40L in oxLDL-stimulated dendritic cells. FEBS Lett. 2011;585:567–573. doi: 10.1016/j.febslet.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Yang K, et al. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 2011;585:854–860. doi: 10.1016/j.febslet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindsley RC, Ebert BL. Molecular pathophysiology of myelodysplastic syndromes. Annu Rev Pathol. 2012 doi: 10.1146/annurev-pathol-011811-132436. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26:13–22. doi: 10.1038/leu.2011.221. [DOI] [PubMed] [Google Scholar]
- 74.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 75.Garzon R, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, et al. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paik JH, et al. MicroRNA-146a downregulates NFkappaB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin Cancer Res. 2011;17:4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 78.O’Connell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012;99:145–174. doi: 10.1016/B978-0-12-387038-4.00006-9. [DOI] [PubMed] [Google Scholar]
- 79.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 80.O’Connell RM, Zhao JL, Rao DS. MicroRNA function in myeloid biology. Blood. 2011;118:2960–2969. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baltimore D. NF-kappaB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- 82.Liew FY, Xu D, Brint EK, O’Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- 83.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 84.O’Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositolphosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Androulidaki A, et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity. 2009;31:220–231. doi: 10.1016/j.immuni.2009.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 89.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 91.O’Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huffaker T, et al. Opposing roles for microRNAs 155 and 146a during antitumor immunity. Submitted. [Google Scholar]
- 93.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]