Abstract
Methionine residues in proteins are susceptible to oxidation by reactive oxygen species, but can be repaired via reduction of the resulting methionine sulfoxides by methionine-S-sulfoxide reductase (MsrA) and methionine-R-sulfoxide reductase (MsrB). However, the identity of all methionine sulfoxide reductases involved, their cellular locations and relative contributions to the overall pathway are poorly understood. Here, we describe a methionine-R-sulfoxide reduction system in mammals, in which two MsrB homologues were previously described. We found that human and mouse genomes possess three MsrB genes and characterized their protein products, designated MsrB1, MsrB2, and MsrB3. MsrB1 (Selenoprotein R) was present in the cytosol and nucleus and exhibited the highest methionine-R-sulfoxide reductase activity because of the presence of selenocysteine (Sec) in its active site. Other mammalian MsrBs contained cysteine in place of Sec and were less catalytically efficient. MsrB2 (CBS-1) resided in mitochondria. It had high affinity for methionine-R-sulfoxide, but was inhibited by higher concentrations of the substrate. The human MsrB3 gene gave rise to two protein forms, MsrB3A and MsrB3B. These were generated by alternative splicing that introduced contrasting N-terminal and C-terminal signals, such that MsrB3A was targeted to the endoplasmic reticulum and MsrB3B to mitochondria. We found that only mitochondrial forms of mammalian MsrBs (MsrB2 and MsrB3B) could compensate for MsrA and MsrB deficiency in yeast. All mammalian MsrBs belonged to a group of zinc-containing proteins. The multiplicity of MsrBs contrasted with the presence of a single mammalian MsrA gene as well as with the occurrence of single MsrA and MsrB genes in yeast, fruit flies, and nematodes. The data suggested that different cellular compartments in mammals maintain a system for repair of oxidized methionine residues and that this function is tuned in enzyme- and stereo-specific manner.
INTRODUCTION
Methionine sulfoxide reductases catalyze reduction of free and protein-bound methionine sulfoxides to corresponding methionines (Brot et al., 1981; Weissbach et al., 2002). The oxidation of methionine by reactive oxygen species (ROS) generates a diastereomeric mixture of methionine-S-sulfoxide (Met-S-SO) and methionine-R-sulfoxide (Met-R-SO). Two distinct enzyme families evolved for reduction of these sulfoxides, with methionine-S-sulfoxide reductase (MsrA) being stereospecific for Met-S-SO and methionine-R-sulfoxide reductase (MsrB) for Met-R-SO. Previously proposed functions of these enzymes include repair of oxidatively damaged proteins, regulation of protein function and elimination of oxidants through reversible formation of methionine sulfoxides (Levine et al., 2000).
Recently, MsrB proteins have been identified and characterized in various organisms including bacteria (Grimaud et al., 2001; Olry et al., 2002; Etienne et al., 2003), yeast (Kryukov et al., 2002; Koc and Gladyshev, unpublished data), fruit fly (Kumar et al., 2002), and mammals (Jung et al., 2002; Kryukov et al., 2002; Moskovitz and Stadtman, 2003). To date, two mammalian MsrB proteins have been identified: selenocysteine (Sec)-containing protein, designated selenoprotein R (SelR; Kryukov et al., 1999, 2002) and its homolog, designated CBS-1, in which Cys is present in place of Sec (Jung et al., 2002). The Sec-containing MsrB has only been described in mammals. Members of the MsrB family have been characterized mechanistically (Kumar et al., 2002; Olry et al., 2002) and structurally (Lowther et al., 2002). However, what is the number of methionine sulfoxide reductases that are present in various organisms and what are their cellular locations and relative contributions to the overall pathway are not known.
For recoding UGA as a Sec codon, several trans-acting elements are used, including a unique tRNASec and a Sec-specific elongation factor SelB. In eukaryotes, the SelB function is served by two proteins, elongation factor EFSec and a SECIS-binding protein SBP2 (Atkins and Gesteland, 2000). In addition to these factors, a cis-acting stem-loop structure within selenoprotein mRNA, designated selenocysteine insertion sequence (SECIS) element, is required for Sec insertion. Eukaryotic SECIS elements are located in 3′ untranslated regions of selenoprotein genes (Hatfield and Gladyshev, 2002), whereas bacterial SECIS elements are present within selenoprotein ORFs, immediately downstream of the Sec-encoding UGA codon (Zinoni et al., 1990; Sandman and Noren, 2000). Recent studies on the Escherichia coli SelB and formate dehydrogenase H SECIS element revealed that 1) only the upper stem-loop structure with a proper distance from UGA is required for Sec incorporation, and 2) a GpU sequence at the tip of the apical loop and a bulged uracil that is located five Watson-Crick base pairs upstream of the GpU motif are essential for Sec insertion in E. coli (Liu et al., 1998; Fourmy et al., 2002). However, the overall bacterial consensus SECIS element is not known.
In the present study, we identified a new MsrB gene in mammals and characterized products of three mammalian MsrBs genes with respect to catalytic functions, cellular locations, and alternative splicing variants. The data suggest that various cellular compartments in mammals maintain systems for repair of oxidized methionines and that this function is tuned in enzyme- and stereo-specific manner.
MATERIALS AND METHODS
Preparation of Recombinant Mammalian MsrB Proteins
The cDNA for Cys mutant of mouse MsrB1 (mSelR; Kryukov et al., 2002) was cloned into pET28a expression vector (Novagen, Madison, WI) to generate a construct coding for a protein with an N-terminal His tag. The mouse MsrB2 (mCBS-1) cDNA was amplified from a mouse EST clone (ID 5150285, Open Biosystems, Huntsville, AL) and cloned into pET28a to code for a protein with a C-terminal His tag. Human MsrB3A (from an EST clone, ID 5109534, Research Genetics, Huntsville, AL) and MsrB3B (from an EST clone, ID 6025598; Open Biosystems) cDNAs were cloned into pET21a to generate constructs with C-terminal His tags. The MsrB3Δ(1-31) construct, which codes for amino acids 32-192 of human MsrB3 (a protein without the signal peptide, numbering follows MsrB3A), was made by inserting the corresponding cDNA into pET21a. This construct coded for a protein with a C-terminal His tag. All constructs were verified by DNA sequencing. MsrB proteins were expressed in E. coli, purified using TALON metal affinity resin (Clontech, Palo Alto, CA) and analyzed for purity by SDS-PAGE. Protein concentration was determined by the Bradford method using bovine serum albumin as a standard.
Preparation of Sec-containing MsrB1
To express a recombinant Sec-containing MsrB1 in E. coli (Sec in the amino acid position 95 is encoded by UGA), we mutated sequences within the ORF downstream of UGA to generate a bacterial SECIS element. The SECIS element was designed to introduce as few mutations as possible in the protein sequence, yet to build a functional SECIS element. To avoid the contamination of the full-length Sec-containing MsrB1 with the truncated form, in which the UGA codon designates termination of translation, we also introduced a His tag at the C-terminus of MsrB1. The ORF of the wild-type MsrB1 was subcloned into the NdeI and XhoI sites of pET21a. Through site-directed mutagenesis using QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), the plasmid pMR1-SECIS4 was constructed, which resulted in high MsrB catalytic activity when expressed in E. coli. The nucleotide sequence downstream of the Sec-coding UGA (underlined) in the plasmid pMR1-SECIS4 was as follows: UGAATATTTAGCAGGCTACTGGGTCCAGTGCCTAAAGGCAAAGAAGCTGCTGCCTCCCAGGGGCAC (mutated nucleotides are shown in bold). The resulting protein was designated as MsrB1-Sec/SECIS. This protein differed from the wild-type MsrB1 by four amino acid replacements: Ser99Arg, Ser100Leu, Lys102Gly, and Phe103Pro. To generate a Sec95Cys mutant of MsrB1-Sec/SECIS, site-directed mutagenesis was carried out using pMR1-SECIS4 as a template. The resulting protein was designated as MsrB1-Cys/SECIS.
To prepare the recombinant MsrB1-Sec/SECIS, E. coli BL21(DE3) cells harboring pMR1-SECIS4 were grown in 18 liters of LB medium supplemented with 2 μM Na2SeO3 and 100 μg/ml ampicillin at 37°C to A600 = 0.6, induced with 0.1 mM IPTG at 30°C for 6 h, and harvested by centrifugation. The harvested cells were resuspended in an extraction buffer (50 mM sodium phosphate, pH 7.0, 300 mM NaCl, 15 mM imidazole, 1 mM PMSF) and treated by sonication. After removal of insoluble material by centrifugation, the supernatant was applied to a TALON metal affinity column (Clontech). The protein was eluted with an elution buffer containing 100 mM imidazole, 50 mM sodium phosphate, pH 7.5, 50 mM NaCl. Because of small amounts of the affinity-purified Sec-containing protein, protein concentration was determined by Western blotting using MsrB1-Cys as an internal standard, followed by analysis of the Western signals with a densitometer.
Determination of Methionine Sulfoxide Reductase Activity and Analysis of Enzyme Kinetics
Methionine sulfoxide reductase activity was assayed using dabsylated L-Met-SO or free L-Met-SO as a substrate. The assay for the reduction of dabsyl-Met-SO to dabsyl-Met was performed as described previously (Kumar et al., 2002). Briefly, the reaction mixture (100 μl) contained 50 mM sodium phosphate (pH 7.5), 20 mM DTT, 200 μM dabsyl-Met-R or S-SO, and 0.1-25 μg of purified proteins. The reaction was carried out at 37°C for 30 min and stopped by adding 200 μl of acetonitrile. The reaction product, dabsyl-Met, was analyzed by HPLC as described previously (Kumar et al., 2002). A typical reaction mixture (100 μl) for reduction of free Met-SO to Met contained 50 mM sodium phosphate (pH 7.5), 20 mM DTT, 1 mM free Met-R-SO, and 5-50 μg of purified proteins. The reaction was carried out at 37°C for 2 h. TLC (n-butanol:acetic acid:water 60:15:25) was used to identify Met as the reaction product. The Met was detected using ninhydrin spray.
All kinetic constants in this study were determined using dabsyl-Met-R-SO as a substrate. Km and kcat values were determined using Lineweaver-Burk plots.
Different diastereomers of Met-SO were prepared as described (Lavine, 1947). Dabsyl derivatives of Met, Met-R-SO, and Met-S-SO were also prepared as described (Minetti et al., 1994).
Determination of Zinc Content
Zinc content of MsrB proteins was analyzed using inductively coupled argon plasma (ICAP) at the Chemical Analysis Laboratory of University of Georgia. Simultaneously, 19 other trace elements were analyzed, but these did not show significant levels. In addition, buffers against which MsrBs were dialyzed for metal analysis were assayed in parallel, and these showed no significant zinc concentrations.
Expression of MsrB Proteins in Saccharomyces cerevisiae
The constructs for expression of MsrB proteins in yeast were made using a yeast expression vector p423 GPD (Mumberg et al., 1995). MsrB1-Cys, MsrB2, MsrB2Δ(1-23), MsrB3A, MsrB3Δ (1-31), and MsrB3B were cloned into the EcoRI/XhoI sites of p423. The constructs were transformed into yeast GY5 cells (MATa his3 leu2 met15 ura3 ΔmsrB::KAN ΔmsrA::URA3), a methionine auxotroph lacking both MsrA and MsrB genes (Kryukov et al., 2002), and transformants were selected for histidine prototrophy. Cells containing p423 only or plasmids with MsrB genes were grown in yeast nitrogen base (YNB) minimal medium at 30°C.
Constructs for Subcellular Localization
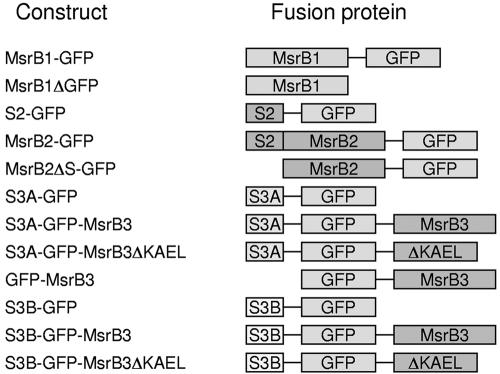
Green fluorescent protein (GFP) expression constructs described in this work were prepared using pEGFP-N1 (Clontech). The coding region of MsrB1-Cys was PCR-amplified and cloned into the XhoI/EcoRI sites of pEGFP-N1, resulting in an MsrB1-GFP construct. To create an MsrB1 only expression construct (MsrB1ΔGFP), the amplified PCR fragment with introduced XhoI and BsrGI sites was cloned into pEGFP-N1 that was pretreated with XhoI/ BsrGI and lacked GFP coding sequences. To characterize the subcellular location of MsrB2, we generated MsrB2-GFP, MsrB2ΔS-GFP (an MsrB2-GFP fusion protein without the first 23 amino acids that corresponded to the predicted mitochondrial signal peptide), and S2-GFP (only the first 23 amino acids of MsrB2 were fused to GFP) constructs using the XhoI/EcoRI sites of pEGFP-N1. To characterize the subcellular location of MsrB3, the following constructs were prepared. The sequences coding for the initial 31 amino acids of the wild-type MsrB3A (predicted N-terminal signal peptide) were cloned into the XhoI/EcoRI sites of pEGFP-N1, resulting in an S3A-GFP construct. The S3A-GFP-MsrB3 (GFP inserted in between the predicted signal peptide and the rest of MsrB3) and the S3A-GFP-MsrB3ΔKAEL (the corresponding construct that lacked the C-terminal KAEL sequence) were obtained by cloning the corresponding PCR-amplified fragments into the BsrGI/NotI sites of the S3A-GFP construct. The GFP-MsrB3 (32-192 amino acids of MsrB3A) construct was made by cloning the corresponding PCR-amplified fragment into the BsrGI/NotI site of pEGFP-N1. Finally, the sequences coding for the N-terminal 23 amino acids of MsrB3B were PCR-amplified and cloned into the XhoI/EcoRI sites of pEGFP-N1 and the same sites of pGFP-MsrB3. The resulting constructs were designated S3B-GFP and S3B-GFP-MsrB3, respectively.
Fluorescence Confocal Microscopy
Transfections into CV-1 cells with appropriate constructs were performed using Lipofectamine (Invitrogen, Carlsbad, CA). MsrB1 was stained with anti-MsrB1 antibodies, followed by secondary anti-rabbit Cy5-conjugated antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Mitochondria were stained with MitoTracker Red (Molecular Probes, Eugene, OR). ER was stained with Calregulin (C-17; Santa Cruz Biotechnology, Santa Cruz, CA) followed by secondary anti-goat Cy5-conjugated antibodies (Jackson ImmunoResearch Laboratories). Images were collected using a Bio-Rad MRC1024ES laser scanning microscope (Richmond, CA).
RESULTS
Identification of a New Mammalian MsrB Gene
Two mammalian MsrB protein forms have been recently identified, including selenoprotein R (also designated as selenoprotein X and most recently as MsrB) and its homolog designated CBS-1 (Huang et al., 1999; Kryukov et al., 1999, 2002; Lescure et al., 1999; Jung et al., 2002). By homology searches of NCBI nonredundant and EST databases using these known MsrB sequences, we identified a new mammalian MsrB gene. The new MsrB sequences were identified in human, mouse, and rat genomes. No additional MsrB homologues were detected, suggesting that mammalian genomes code for three MsrB genes. Here, we designate selenoprotein R as MsrB1, CBS-1 as MsrB2, and the newly identified MsrB homolog as MsrB3.
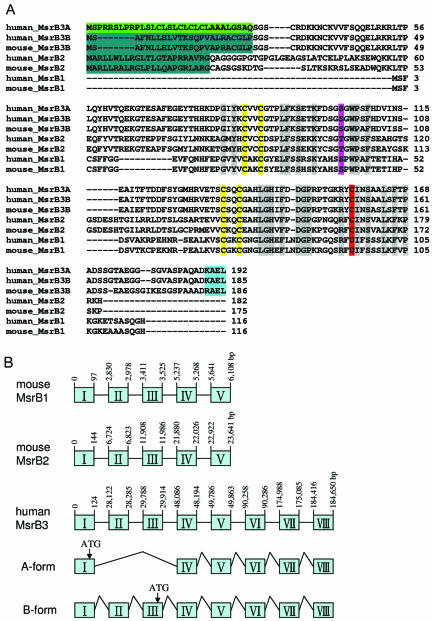
Analysis of human EST sequences corresponding to MsrB3 revealed two cDNA forms that differed in their 5′ sequences. One form coded for an ORF of 192 residues and was designated as MsrB3A; the second coded for an ORF of 185 residues and was designated as MsrB3B (Figure 1A). Both coding sequences were identical except in their N-terminal regions. Analysis of the human MsrB3 gene structure revealed that these two forms were generated by alternative splicing (Figure 1B). The 184-kb human MsrB3 gene was located on chromosome 12 and contained 8 exons. Human MsrB1 and MsrB2 genes had 5 exons each, were of considerably smaller size, and were located on chromosomes 16 and 10, respectively.
Figure 1.
Mammalian methionine-R-sulfoxide reductases. (A) Alignment of mammalian MsrBs. The conserved catalytic Cys and Sec (shown as U) residues are shown in red. The violet shows Ser and Thr residues that replaced the cysteine that forms a disulfide bond with the catalytic Cys in many MsrBs. Zinc-coordinating Cys residues are high-lighted in yellow. Other identical residues are indicated in gray. The predicted signal peptides are indicated as follows: green, N-terminal ER or secretory signal peptide of MsrB3A; bluish green, mitochondrial target signal of MsrB3B or MsrB2; turquoise, ER retention signal at C-terminus of MsrB3. GenBank accession numbers are as follows: human MsrB3A, BI257829; human MsrB3B, BC040053; mouse MsrB3B, NM_177092; human MsrB2, AF122004; mouse MsrB2, BC021619; human MsrB1, NM_016332; mouse MsrB1, NP_038787. (B) Genomic structures of MsrBs. Boxes indicate exons. Mouse MsrB1, MsrB2, and human MsrB3 are located on chromosomes 17, 2, and 12, respectively. MsrB3A consists of six exons, whereas MsrB3B is composed of eight exons. Translation start sites are shown by arrows.
MsrB3 showed 22% identity to MsrB1 and 39% identity to MsrB2. Like MsrB2, it had a conserved catalytic Cys residue (Cys158, numbering follows MsrB3A sequence), which corresponded to the catalytic Sec95 in MsrB1. In addition, the protein had conserved Asp148 and Arg156, the residues that were predicted to activate the nucleophilic group at the catalytic Cys (or Sec) and together with this thiolate (or selenolate) to form a catalytic triad (Lowther et al., 2002). A Cys residue that was shown to function in thiol-disulfide exchange through formation of a disulfide bond with the active site Cys residue (this residue is conserved in at least 50% of known MsrBs) was replaced in MsrB3 with Ser (Ser102). MsrB1 and MsrB2 sequences also lacked this Cys residue.
Predicted Signal Peptides and Retention Signals in MsrB2, MsrB3B, and MsrB3A Sequences
Examination of the MsrB2 sequences with SignalP, MitoProt II, and PSORT programs revealed the presence of a typical mitochondrial signal peptide (a hydrophobic sequence with a high proportion of lysine or arginine residues that forms an amphiphilic α-helix). In addition, we found that MsrB3A and MsrB3B contained a putative ER retention sequence, KAEL, at the C-terminus. The above programs also revealed that MsrB3A had an N-terminal ER or secretory signal peptide, whereas MsrB3B contained a different N-terminal signal peptide that was most similar to mitochondrial signals. No signal peptides/sequences were detected in MsrB1. This analysis suggested different subcellular locations for mammalian MsrBs.
MsrB3 Stereospecifically Reduces Met-R-SO
To test whether MsrB3, like previously characterized MsrBs, reduces specifically the Met-R-SO stereoisomer to Met, we prepared a full-length recombinant MsrB3A, MsrB3Δ(1-31) that lacked the first 31 residues (this protein corresponded to signal-less MsrB3A or MsrB3B) and a full-length MsrB3B using the pET21a expression vector. We found that the growth of E. coli BL21(DE3) cells expressing MsrB3A (after induction with 0.1 mM IPTG at 37°C) was strongly inhibited, but low-density cells expressed the recombinant protein. The purified MsrB3A indeed reduced the dabsylated Met-R-SO (but not Met-S-SO) stereoisomer to the corresponding methionine derivative in the presence of DTT, demonstrating that the protein was methionine-R-sulfoxide reductase. The cells expressing MsrB3Δ(1-31) or MsrB3B showed a normal growth after induction with IPTG. These proteins also catalyzed the stereospecific reduction of dabsylated Met-R-SO.
Preparation of Sec-containing MsrB1
To address how the Sec residue in MsrB1 affects its enzyme activity and to compare enzymatic characteristics of mammalian MsrBs, we expressed the recombinant Sec-containing MsrB1 (MsrB1-Sec/SECIS) in E. coli. The MsrB1 gene has a mammalian SECIS element in its 3′-UTR; however, a bacterial Sec insertion system is different from that of mammals. Specifically, 1) the essential sequences within the bacterial SECIS elements are different from those present in mammals; 2) SECIS structures are different in bacteria and mammals; and 3) bacterial SECIS elements are located within ORFs, immediately downstream of Sec-coding UGA codons.
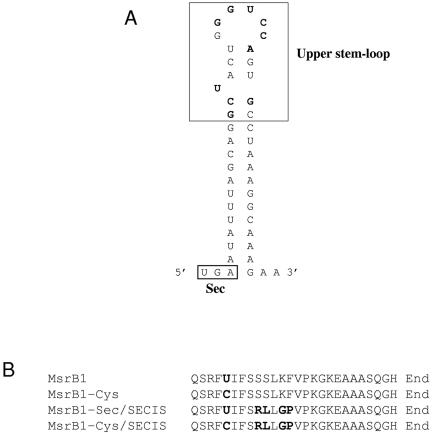
We generated various versions of a bacterial SECIS element downstream of the UGA codon (codon position 95) by site-directed mutagenesis (Figure 2) such that the downstream coding sequences had 2, 3, or 4 mutated residues. In each construct, the protein was designed to contain a C-terminal His tag. This allowed us to separate, using affinity chromatography, the full-length protein from the truncated form, in which the UGA terminated protein synthesis. Previous studies indicated that Sec insertion into recombinant mammalian selenoproteins in E. coli was inefficient and the major protein products were truncated proteins (Arner et al., 1999). We found that most mutations downstream of the UGA resulted in either low efficiency of Sec insertion or in completely insoluble proteins. The truncated form of MsrB1, in which the UGA codon was used as a stop signal, was also insoluble.
Figure 2.
SECIS element in MsrB1-Sec/SECIS construct and amino acid sequences of the Sec-containing region. (A) Secondary structure of SECIS element for incorporating Sec in MsrB1. The mutated nucleotides are indicated as bold letters. (B) Alignment of regions downstream of Sec in MsrB1 sequences.
However, we were able to obtain a soluble and functional Sec-containing protein (MsrB-Sec/SECIS) by introducing a SECIS element with concomitant changes in four residues in MsrB1: Ser99Arg, Ser100Leu, Lys102Gly, and Phe103Pro. We also prepared a Cys mutant of MsrB-Sec/SECIS (MsrB1-Cys/SECIS) as well as a Cys mutant of the wild-type MsrB1 (MsrB1-Cys), which served as controls in determining whether mutations of the four residues affected the catalytic activity of the enzyme.
Catalytic Properties of Mammalian MsrBs: Key Role of Sec in MsrB1
Table 1 summarizes the specific activities and enzyme kinetics of mammalian MsrBs using dabsyl-Met-R-SO as substrate. MsrB1-Sec/SECIS exhibited the highest specific activity. The specific activity of MsrB1-Cys/SECIS was similar to that of MsrB1-Cys (Cys mutant of the wild-type enzyme), suggesting that mutation of the four residues to generate the SECIS element did not alter the enzyme activity. The specific activity of MsrB1-Sec/SECIS was ∼800-fold higher than those of the two Cys mutants, MsrB1-Cys/SECIS and MsrB1-Cys, indicating that high activity of MsrB was due to the presence of Sec. Furthermore, MsrB1-Sec/SECIS showed ∼4-fold higher specific activity than MsrB2 and MsrB3. Bar-Noy and Moskovitz recently reported that a recombinant Sec-containing MsrB1, in which six residues were mutated downstream of Sec, had 100–200-fold higher specific activity than the Cys mutant (Bar-Noy and Moskovitz, 2002).
Table 1.
Specific activity and kinetic constants of mammalian MsrBs
| Protein | Specific activity (nmol/min/mg protein) | Km (mM) | kcat (s-1) | kcat/Km (M-1 s-1) |
|---|---|---|---|---|
| MsrB1-Sec/SECIS | 1560 | 1.0 | 2.28 | 2280 |
| MsrB1-Cys/SECIS | 1.6 | 0.9 | 0.02 | 22 |
| MsrB1-Cys | 2.1 | 1.1 | 0.03 | 27 |
| MsrB2 | 353 | 0.17 | 0.23 | 1353 |
| MsrB3 | 423 | 2.9 | 2.29 | 790 |
Enzyme activity was determined using dabsyl-Met-R-SO as described in MATERIALS AND METHODS. The purified MsrB3Δ(1-31) lacking the N-terminal signal peptide was used as MsrB3. The specific activity was determined at 200 μM dabsyl-Met-R-SO. For determination of Km the range of concentration from 50 to 800 μM of dabsyl-Met-R-SO was used except for MsrB2, where the substrate concentration varied from 25 to 200 μM. All values are the average of three measurements and the standard deviations for all values are ≤20%.
Further kinetic characterization of mammalian MsrBs revealed that MsrB2 was inhibited by concentrations of dabsyl-Met-R-SO >300 μM (Figure 3). In contrast, no substrate inhibition was observed for MsrB1 and MsrB3 up to the highest tested substrate concentration (3.2 mM).
Figure 3.
Substrate inhibition of MsrB2. One microgram of purified MsrB2 was used in the enzyme reaction. The assay was carried out at various concentrations of dabsyl-Met-R-SO as described in MATERIALS AND METHODS.
Km values for dabsyl-Met-R-SO were 1.0 mM for MsrB1-Sec/SECIS, 0.9 mM for MsrB1-Cys/SECIS, 1.1 mM for MsrB1-Cys, 0.17 mM for MsrB2, and 2.9 mM for MsrB3. These data, along with the finding of substrate inhibition, suggested that MsrB2 is adapted to work with low concentrations of methionine sulfoxide, whereas MsrB1 and MsrB3 are most active at higher concentrations of the substrate. The Km values of MsrB1 and MsrB3 were similar to that of Drosophila MsrB (Km value of 2.1 mM; Kumar et al., 2002).
The kcat value of MsrB1-Sec/SECIS (2.28 s-1) was similar to that of MsrB3 (2.29 s-1), but 80- and 10-fold higher than that of the corresponding Cys mutant and MsrB2, respectively. Moreover, the catalytic efficiency, kcat/Km, of MsrB1-Sec/SECIS was the highest of all tested MsrBs: ∼80-, 2-, and 3-fold higher than those of MsrB1-Cys, MsrB2, and MsrB3, respectively. These data revealed that the high catalytic efficiency of MsrB1-Sec/SECIS was due to Sec, which increased catalytic activity without significantly affecting substrate binding. Thus, it appears that selenium provides a key contribution to the MsrB activity in mammalian cells.
Mammalian MsrBs Reduce Free Met-R-SO
Although MsrB and MsrA isolated from various sources catalyze reduction of protein methionine sulfoxides, they are also known to have low activity with free methionine sulfoxides. Therefore, we also assayed for free Met-R-SO reductase activity of mammalian MsrBs. As shown in Figure 4, all three MsrBs have the ability to reduce free Met-R-SO to Met.
Figure 4.
Free Met-R-SO reductase activity of mammalian MsrBs. The assay was carried out using 1 mM free Met-R-SO as described in MATERIALS AND METHODS. Fifty micrograms of MsrB1-Cys and 10 μg of MsrB2 and MsrB3, respectively, were used in the enzyme reaction. Equal amounts of the reaction mixture were spotted on to TLC plate. Lane 1, reaction mixture without enzymes (control); lane 2, MsrB1-Cys; lane 3, MsrB2; lane 4, MsrB3; lane 5, Met as a standard.
Mammalian MsrBs Are Zinc-containing Enzymes
MsrB1 is a zinc-containing enzyme (Kryukov et al., 2002). Recent study demonstrated that zinc was essential for the catalytic function of Drosophila MsrB and that it was coordinated in the enzyme through four cysteine residues organized in two CxxC motifs (Kumar et al., 2002). However, these residues were absent in a number of bacterial MsrBs, apparently because they were lost during evolution. It was proposed that zinc serves a structural role and is not involved directly in catalytic reaction (Kumar et al., 2002). Primary sequences of MsrB2 and MsrB3 conserved the two CxxC motifs, suggesting that these enzymes also contained zinc. We analyzed the content of 20 trace elements in dialyzed preparations of MsrB2 and MsrB3. Only zinc was detected in significant amounts: 0.36 equivalents in MsrB2 and 0.61 equivalents in MsrB3. Thus, all three mammalian MsrBs are zinc-containing proteins.
In an attempt to remove zinc from the enzymes, MsrB1-Cys, MsrB2, or MsrB3 were incubated with 5 mM EDTA or 5 mM CDTA (a stronger chelating agent) at 4°C for 1 h and then dialyzed against a buffer containing 50 mM sodium phosphate (pH 7.5) and 50 mM NaCl. However, the treatment with either EDTA or CDTA did not affect the enzyme activity of any of three MsrBs, suggesting that zinc was strongly bound in these enzymes. A similar strong binding of the metal was previously observed in Drosophila MsrB (Kumar et al., 2002). We further examined whether in the presence of zinc, the enzyme activity of MsrBs may be increased and found that it was not affected by the addition of 0.1 mM ZnCl2. However, at higher concentration (1.0 mM ZnCl2), we observed 25-50% inhibition of MsrB1-Cys and MsrB2 activities.
Expression of Mammalian MsrB Proteins in Yeast
To test if mammalian MsrBs can significantly contribute to methionine sulfoxide reduction in vivo, we used a yeast model system. A yeast strain that lacked MsrA and MsrB genes cannot grow in the presence of Met-S-SO, primarily because of the lack of MsrA. However, these double mutant cells can grow in the presence of Met-R-SO, suggesting that the yeast might have an additional Met-R-SO activity (Koc and Gladyshev, unpublished data).
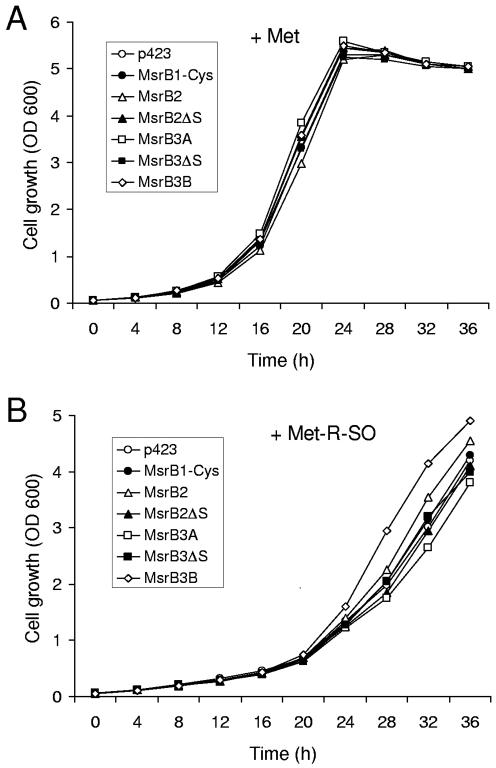
We made various mammalian MsrB1, MsrB2, and MsrB3 constructs with or without relevant signal peptides using a yeast expression vector p423 and expressed the proteins in the yeast double mutant strain. Growth characteristics of cells containing various MsrB proteins were similar to those of control cells containing p423, when cells were grown in the presence of methionine (Figure 5A). However, in the presence of Met-R-SO, the cells expressing the full-length MsrB3B grew better than the control cells containing p423 (Figure 5B), suggesting that MsrB3B compensated for methionine sulfoxide reductase deficiency. Interestingly, both MsrB3B and yeast MsrB had predicted mitochondrial signal peptides. In addition, a slight growth complementation effect was observed in cells expressing the full-length MsrB2, also a predicted mitochondrial protein. In contrast, MsrB1-Cys, MsrB2Δ(1-23) (MsrB2 without the signal peptide), MsrB3A, and MsrB3Δ(1-31) (MsrB3 without the signal peptides of the A- or B-forms) did not compensate for yeast MsrA/MsrB deficiency. Moreover, the growth of cells expressing MsrB3A was slightly inhibited. Thus, growth compensation in yeast was observed only when expressed proteins had predicted mitochondrial signals.
Figure 5.
Expression of mitochondrial targeted mammalian MsrBs compensates for growth of yeast cells lacking MsrA and mitochondrial MsrB genes. The yeast cells expressing various mammalian MsrBs were grown at 30°C in YNB media containing 2% dextrose in the presence of 0.14 mM L-Met (A) or 0.14 mM L-Met-R-SO (B). The constructs used in this experiment were as follows: MsrB1-Cys, full-length mouse MsrB1-Cys mutant; MsrB2, full-length mouse MsrB2; MsrB2ΔS, MsrB2 lacking N-terminal 23 aa signal peptide; MsrB3A, full-length human MsrB3A; MsrB3ΔS, MsrB3 without signal peptide of either A- or B-form; MsrB3B, full-length human MsrB3B.
MsrB1 Is a Nuclear and Cytosolic Protein
To investigate subcellular distribution of mammalian MsrBs, we generated various GFP-fusion constructs using a pEGFP-N1 vector (Figure 6). In the case of MsrB1, its cysteine mutant was used. The constructs were transfected into monkey kidney CV-1 cells. The MsrB1-GFP protein was localized in the cytoplasm and also in the nucleus (Figure 7A). However, nuclear location of MsrB1-GFP could have been due to GFP, which is partially enriched in the nucleus when expressed by itself. To examine whether MsrB1 is present in the nucleus, we generated an MsrB1ΔGFP construct, which encoded only the cysteine mutant of MsrB1. As shown in Figure 7B, MsrB1 was present in both nucleus and cytoplasm. Although MsrB1 does not have a pronounced nuclear localization signal, it is rich in positively charged residues, which could serve to transfer the protein in the nucleus.
Figure 6.
Schematic representation of GFP fusion constructs. The GFP fusion constructs used in this study were as follows: MsrB1-GFP, full-length mouse MsrB1-Cys fused to the N-terminus of GFP; MsrB1ΔGFP, only full-length MsrB1-Cys expressing construct without GFP; S2-GFP, N-terminal signal peptide of mouse MsrB2 fused to the N-terminus of GFP; MsrB2-GFP, full-length MsrB2 fused to the N-terminus of GFP; MsrB2ΔS-GFP, MsrB2 without signal peptide fused to the N-terminus of GFP; S3A-GFP, N-terminal signal peptide of human MsrB3A fused to the N-terminus of GFP; S3A-GFP-MsrB3, signal peptide of MsrB3A and MsrB3 without signal peptide fused to the N-terminus and the C-terminus of GFP, respectively; S3A-GFP-MsrB3ΔKAEL, the same construct as S3A-GFP-MsrB3 except for deletion of KAEL sequence at the C-terminus; GFP-MsrB3, MsrB3 without signal peptide fused to the C-terminus of GFP; S3B-GFP, N-terminal signal peptide of MsrB3B fused to the N-terminus of GFP; S3B-GFP-MsrB3, signal peptide of MsrB3B and MsrB3 without signal peptide fused to the N-terminus and the C-terminus of GFP, respectively; S3B-GFP-MsrB3ΔKAEL, the same construct as S3B-GFP-MsrB3 except for deletion of KAEL sequence.
Figure 7.
MsrB1 is located in both cytoplasm and nucleus. (A) Green fluorescence images of CV-1 cells expressing GFP-fused MsrB1 and control proteins at 20 h posttransfection. (B) Confocal images of MsrB1ΔGFP-expressing cells. The transfected cells at 18 h posttransfection were stained with anti-MsrB1 antibodies followed by secondary anti-rabbit Cy5. A set of two images is shown: left panel, immunofluorescence; right panel, phase contrast.
MsrB2 Is Targeted to Mitochondria
MsrB2 contained a typical predicted mitochondrial signal at the N-terminus (MARLLRALRGLPLLQAPGRLARG). As shown in Figure 8, both S2-GFP (the N-terminal signal peptide of MsrB2 fused to GFP) and MsrB2-GFP (the full-length MsrB2 fused to GFP) colocalized with mitochondria, whereas MsrB2ΔS-GFP (MsrB2 lacking the signal peptide and fused to GFP) exhibited cytoplasmic and nuclear location. Interestingly, the fluorescence in some cells transfected with S2-GFP or MsrB2-GFP constructs was detected in the cytoplasm and the nucleus. However, Western blots with anti-GFP antibodies revealed that a 26-kDa band corresponding to the GFP protein was detected besides the MsrB2-GFP fusion protein in MsrB2-GFP transfected cells (unpublished data). It appears that this GFP contributed to the nonmitochondrial location of the GFP fluorescence. The data suggested that the GFP was either produced by cotranslation (even though the ∼600-nucleotide MsrB2 fragment was present upstream of the possible ATG initiation codon of GFP) or the expressed MsrB2-GFP fusion protein was partially cleaved to generate GFP.
Figure 8.
MsrB2 is targeted to mitochondria. Confocal images of CV-1 cells expressing various GFP-tagged MsrB2 and control proteins at 12 h posttransfection. A set of three images is shown for each construct. Left panels: green fluorescence corresponding to transiently expressed fusion proteins. Center panels; fluorescence of the mitochondrial marker, Mito-Tracker Red. Right panels: images obtained by merging left and center panels. The GFP fusion constructs used in this experiment are shown on the left.
MsrB3A Resides in the ER and MsrB3B in Mitochondria
The primary sequence of human MsrB3 contained a KAEL tetrapeptide at the C-terminus, an analog of the classical ER retention signal, the KDEL sequence. Furthermore, MsrB3A contained a typical secretory signal peptide at the N-terminus, whereas MsrB3B had a different signal peptide. To determine the subcellular distribution of MsrB3A and MsrB3B, we used a set of constructs that encoded various fusion proteins between GFP and MsrB3 (Figure 6). We first tested whether MsrB3A or MsrB3B are located in the ER (Figure 9). S3A-GFP-MsrB3 (GFP inserted between the predicted ER signal and the rest of MsrB3) colocalized with the ER marker. However, GFP-MsrB3 (a fusion protein that lacked the signal peptide) was not detected in the ER and instead was seen in the cytoplasm. Likewise, S3A-GFP (a protein with the N-terminal signal peptide fused to GFP) was not present in the ER. To test whether the tetrapeptide KAEL functioned as an ER retention signal, we developed a construct that encoded S3A-GFP-MsrB3ΔKAEL (a fusion protein with the deletion of KAEL). This protein was not localized in the ER. These data demonstrated that the ER location of MsrB3A was dependent on the tetrapeptide KAEL sequence.
Figure 9.
MsrB3A is located in ER. Images of CV-1 cells expressing various GFP-tagged MsrB3 proteins at 20 h posttransfection. A set of three images is shown for each construct. Left panels: green fluorescence corresponding to transiently expressed fusion proteins. Center panels: fluorescence of the ER marker. ER was stained with Calregulin (C-17), followed by secondary antigoat Cy5-conjugated antibody. Right panels: images obtained by merging left and center panels. The GFP fusion constructs are shown on the left.
In contrast to the ER location of MsrB3A, MsrB3B was not seen in this compartment when the corresponding fusion proteins were expressed in CV-1 cells, even though MsrB3B also had the KAEL retention signal. As shown in Figure 9, S3B-GFP-MsrB3 and S3B-GFP proteins did not colocalize with the ER standard. Similar data were obtained with S3B-GFP-MsrB3ΔKAEL. Instead, the fluorescence from these fusion proteins colocalized with MitoTracker, a mitochondrial standard. As shown in Figure 10, S3B-GFP and S3B-GFP-MsrB3 were observed in mitochondria, whereas GFP-MsrB3 did not show mitochondrial location. Thus, MsrB3A was present in the ER because of its N-terminal and C-terminal signals. Although MsrB3B also had the C-terminal ER retention signal, its N-terminal signal peptide targeted this protein to mitochondria.
Figure 10.
MsrB3B is a mitochondrial protein. Images of CV-1 cells expressing various GFP-tagged MsrB3B proteins at 20 h posttransfection are shown. A set of three images is shown for each construct: left panel, green fluorescence; center panel, mitochondrial marker (MitoTracker Red); right panel, merged image. The GFP fusion constructs are shown on the left.
DISCUSSION
We identified a third MsrB gene in mammals and characterized kinetic properties, subcellular distribution, and alternative splicing forms of three methionine-R-sulfoxide reductases. Our data revealed that methionine sulfoxide reduction in mammals occurs in different cellular compartments and is fine-tuned in an enzyme- and stereo-specific manner.
The newly identified human MsrB3 is unusual in that it occurs in two protein forms, MsrB3A and MsrB3B. Genomic analyses revealed that these forms are generated by alternative first exon splicing and differ in their N-terminal sequences.
The first mammalian MsrB identified was selenoprotein R, in which the position of the active site Cys residue is occupied by Sec. Substitution of Sec with Cys decreased the enzyme activity ∼800-fold, indicating the essential role of Sec in this enzyme. On the other hand, the specific activity of Cys mutants of MsrB1 was ∼170- and ∼210-fold lower than that of MsrB2 and MsrB3, respectively, suggesting that the other mammalian MsrBs were tuned for the use of Cys. The Sec-containing MsrB1 had ∼fourfold higher specific activity than MsrB2 and MsrB3. Thus, MsrB1 appears to be the major MsrB in mammalian cells and, together with MsrA, it contributes significantly to the reduction of methionine sulfoxides.
Our study also revealed that mammalian cells have at least four different MsrB protein products that reside in different compartments: MsrB1 in the cytoplasm and nucleus, MsrB2 in mitochondria, MsrB3A in the ER, and MsrB3B in mitochondria. Therefore, it is clear that oxidized methionine in proteins can be repaired in different cellular compartments in mammals.
Both MsrB2 and MsrB3B were targeted to mitochondria. Although MsrB2 could easily be predicted as a mitochondrial protein, the location of MsrB3B was not immediately clear because it had contrasting N- and C-terminal signals. However, our data clearly showed that the human MsrB3B was a mitochondrial protein. The MsrB3B form was not unique to humans because we also found a mouse homolog of this protein (GenBank accession number NM_177092). The mouse homolog of MsrB3A form has not yet been found in the EST database. The occurrence of two different mitochondrial MsrBs raises the question of why mammalian cells use two enzymes in this organelle. Mitochondria are thought to be the major source of ROS, and therefore, mitochondrial proteins may be particularly susceptible to damage induced by ROS. In addition, MsrB2 was most active at lower concentrations of methionine sulfoxide but was inhibited by high concentrations of the substrate, whereas MsrB3B was most active at concentrations of methionine sulfoxide > 1 mM. A further difference in MsrB2 and MsrB3B functions may be due to differential tissue expression of these proteins. For example, MsrB2 is mainly expressed in muscle tissues, whereas in the brain the expression of this enzyme is weak (Jung et al., 2002).
The fact that only mammalian mitochondrial MsrBs compensated for deficiency in methionine sulfoxide reductase function in the yeast mutant strain lacking both MsrA and MsrB genes provided independent demonstration of the importance of methionine sulfoxide reduction in this organelle. The exact location of MsrA in mammals is not fully understood. Mammals contain only one known MsrA gene, and our BLAST analyses revealed no additional MsrA genes. Whether a single MsrA gene can generate multiple protein products that are targeted to different cellular compartments is not known. Recent reports provided evidence that MsrA was present in mitochondria and cytosol (Hansel et al., 2002; Vougier et al., 2003). However, this protein has neither an ER signal peptide nor a C-terminal ER retention signal, and whether it resides in the nucleus is also not known. If there are no nuclear and ER-resident MsrA forms, mammalian cells might have an epimerase activity that interconverts Met-S-SO to Met-R-SO and prevents the accumulation of Met-S-SO in these cellular compartments.
The abundance of mammalian MsrB forms also contrasts with the presence of single MsrA and MsrB genes in most organisms, including yeast, fruit flies, and nematodes. It is not clear how Met-S-SO and Met-R-SO are reduced in these organisms in different cellular compartments.
Acknowledgments
We thank You Zhou for help with localization experiments, Ahmet Koc with yeast expression, and Sergey Novoselov with mammalian cell culture. We also thank Abhilash Kumar for initial attempts to express MsrB1. This study was supported by National Institutes of Health Grant AG021518 (to V.N.G.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-08-0629. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0629.
References
- Arner, E.S., Sarioglu, H., Lottspeich, F., Holmgren, A., and Bock, A. (1999). High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J. Mol. Biol. 292, 1003-1016. [DOI] [PubMed] [Google Scholar]
- Atkins, J.F., and Gesteland, R.F. (2000). The twenty-first amino acid. Nature 407, 463-465. [DOI] [PubMed] [Google Scholar]
- Bar-Noy, S., and Moskovitz, J. (2002). Mouse methionine sulfoxide reductase B: effect of selenocysteine incorporation on its activity and expression of the seleno-containing enzyme in bacterial and mammalian cells. Biochem. Biophys. Res. Commun. 297, 956-961. [DOI] [PubMed] [Google Scholar]
- Brot, N., Weissbach, L., Werth, J., and Weissbach, H. (1981). Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl. Acad. Sci. USA 78, 2155-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne, F., Spector, D., Brot, N., and Weissbach, H. (2003). A methionine sulfoxide reductase in Escherichia coli that reduces the R enantiomer of methionine sulfoxide. Biochem. Biophys. Res. Commun. 300, 378-382. [DOI] [PubMed] [Google Scholar]
- Fourmy, D., Guittet, E., and Yoshizawa, S. (2002). Structure of prokaryotic SECIS mRNA hairpin and its interaction with elongation factor SelB. J. Mol. Biol. 324, 137-150. [DOI] [PubMed] [Google Scholar]
- Grimaud, R., Ezraty, B., Mitchell, J.K., Lafitte, D., Briand, C., Derrick, P.J., and Barras, F. (2001). Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276, 48915-48920. [DOI] [PubMed] [Google Scholar]
- Hansel, A., Kuschel, L., Hehl, S., Lemke, C., Agricola, H.J., Hoshi, T., and Heinemann, S.H. (2002). Mitochondrial targeting of the human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 16, 911-913. [DOI] [PubMed] [Google Scholar]
- Hatfield, D.L., and Gladyshev, V.N. (2002). How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22, 3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Escribano, J., Sarfarazi, M., and Coca-Prados, M. (1999). Identification, expression and chromosome localization of a human gene encoding a novel protein with similarity to the pilB family of transcriptional factors (pilin) and to bacterial peptide methionine sulfoxide reductases. Gene 233, 233-240. [DOI] [PubMed] [Google Scholar]
- Jung, S., Hansel, A., Kasperczyk, H., Hoshi, T., and Heinemann, S.H. (2002). Activity, tissue distribution and site-directed mutagenesis of a human peptide methionine sulfoxide reductase of type B: hCBS1. FEBS Lett. 527, 91-94. [DOI] [PubMed] [Google Scholar]
- Kryukov, G.V., Kryukov, V.M., and Gladyshev, V.N. (1999). New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 274, 33888-33897. [DOI] [PubMed] [Google Scholar]
- Kryukov, G.V., Kumar, R.A., Koc, A., Sun, Z., and Gladyshev, V.N. (2002). Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc. Natl. Acad. Sci. USA 99, 4245-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R.A., Koc, A., Cerny, R.L., and Gladyshev, V.N. (2002). Reaction mechanism, evolutionary analysis, and role of zinc in Drosophila methionine-R-sulfoxide reductase. J. Biol. Chem. 277, 37527-37535. [DOI] [PubMed] [Google Scholar]
- Lavine, F.T. (1947). The formation, resolution and optical properties of the diastereoisometric sulfoxides derived from L-methionine. J. Biol. Chem. 169, 477-491. [PubMed] [Google Scholar]
- Lescure, A., Gautheret, D., Carbon, P., and Krol, A. (1999). Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 274, 38147-38154. [DOI] [PubMed] [Google Scholar]
- Levine, R.L., Moskovitz, J., and Stadtman, E.R. (2000). Oxidation of methionine in proteins: roles in antioxidant defense and cellular regulation. IUBMB Life 50, 301-307. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Reches, M., Groisman, I., and Engelberg-Kulka, H. (1998). The nature of the minimal 'selenocysteine insertion sequence'(SECIS) in Escherichia coli. Nucleic Acids Res. 26, 896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther, W.T., Weissbach, H., Etienne, F., Brot, N., and Matthews, B.W. (2002). The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat. Struct. Biol. 9, 348-352. [DOI] [PubMed] [Google Scholar]
- Minetti, G., Balduini, C., and Brovelli, A. (1994). Reduction of DABS-L-methionine-dl-sulfoxide by protein methionine sulfoxide reductase from polymorpho-nuclear leukocytes: stereospecificity towards the l-sulfoxide. Ital. J. Biochem. 43, 273-283. [PubMed] [Google Scholar]
- Moskovitz, J., and Stadtman, E.R. (2003). Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc. Natl. Acad. Sci. USA 100, 7486-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg, D., Muller, R., and Funk, M. (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- Olry, A., Boschi-Muller, S., Marraud, M., Sanglier-Cianferani, S., Van Dorsselear, A., and Branlant, G. (2002). Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J. Biol. Chem. 277, 12016-12022. [DOI] [PubMed] [Google Scholar]
- Sandman, K.E., and Noren, C.J. (2000). The efficiency of Escherichia coli selenocysteine insertion is influenced by the immediate downstream nucleotide. Nucleic Acids Res. 28, 755-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vougier, S., Mary, J., and Friguet, B. (2003). Subcellular localization of methionine sulfoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem. J. 373, 531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach, H., Etienne, F., Hoshi, T., Heinemann, S.H., Lowther, W.T., Matthews, B., St. John, G., Nathan, C., and Brot, N. (2002). Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 397, 172-178. [DOI] [PubMed] [Google Scholar]
- Zinoni, F., Heider, J., and Bock, A. (1990). Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl. Acad. Sci. USA 87, 4660-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]