SUMMARY
DNA sequence is a major determinant of the binding specificity of transcription factors (TFs) for their genomic targets. However, eukaryotic cells often express, at the same time, TFs with highly similar DNA binding motifs but distinct in vivo targets. Currently, it is not well understood how TFs with seemingly identical DNA motifs achieve unique specificities in vivo. Here, we used custom protein binding microarrays to analyze TF specificity for putative binding sites in their genomic sequence context. Using yeast TFs Cbf1 and Tye7 as our case study, we found that binding sites of these bHLH TFs (i.e., E-boxes) are bound differently in vitro and in vivo, depending on their genomic context. Computational analyses suggest that nucleotides outside E-box binding sites contribute to specificity by influencing the 3D structure of DNA binding sites. Thus, local shape of target sites might play a widespread role in achieving regulatory specificity within TF families.
Keywords: transcription factors, bHLH, DNA binding sites, protein binding microarrays, DNA shape, support vector regression
INTRODUCTION
Transcriptional regulation is effected primarily by sequence-specific transcription factors (TFs) that recognize short DNA sequences (5–15 base pairs long) in the promoters or enhancers of the genes whose expression they regulate (Bulyk, 2003). Determination of the DNA recognition properties of TFs is essential for understanding how these proteins achieve their unique regulatory roles in the cell.
TFs are typically annotated according to the structural class of their DNA binding domains. Members of a particular class (i.e., paralogous TFs) often have similar DNA binding preferences (Badis et al., 2009). However, despite apparently shared binding specificities, individual TF family members often exhibit non-redundant functions. In some cases, differences in the core DNA binding site motifs have been shown to contribute to differential in vivo binding by closely related TFs (Busser et al., 2012; Fong et al., 2012; Grove et al., 2009; Wei et al., 2010). However, in many cases the DNA motifs of paralogous TFs are virtually identical, and still the proteins select different genomic targets in vivo. In these cases, interactions with protein cofactors are thought to be responsible for differential in vivo DNA binding of paralogous TFs. However, such cofactors can be difficult to identify and only a few conclusive examples are known (e.g., (Hollenhorst et al., 2009; Mann and Chan, 1996; Slattery et al., 2011)). Another factor that determines in vivo TF binding is the local chromatin environment (Arvey et al., 2012; Lelli et al., 2012; Thurman et al., 2012; Zhou and O’Shea, 2011). Nevertheless, protein cofactors and chromatin context are unlikely to completely explain differential binding specificity of paralogous TFs.
Here, we investigate a potential mechanism through which TFs with highly similar DNA binding motifs can achieve differential binding in vivo. Several studies have indicated that nucleotides flanking TF binding sites (i.e., nucleotides outside the core DNA binding site motif) can affect binding specificity (Leonard et al., 1997; Morin et al., 2006; Nagaoka et al., 2001; Rajaram and Kerppola, 1997). Thus, we investigated whether the genomic context of putative TF binding sites differentially affects binding of paralogous TFs.
In this case study, we examined S. cerevisiae basic helix-loop-helix (bHLH) TFs Cbf1 and Tye7. These factors have highly similar DNA binding motifs (MacIsaac et al., 2006; Zhu et al., 2009) but interact with different sets of genomic regions in vivo (Harbison et al., 2004) (Figure 1). Importantly, these differences in in vivo DNA binding are not due to the TFs being active under different conditions, in which the accessibility of potential DNA binding sites might be different (as has been observed for other bHLH factors (Fong et al., 2012)). Instead, the Cbf1 and Tye7 ChIP-chip data (Harbison et al., 2004) were both collected from the same culture conditions (YPD), in which the two proteins had access to the same E-box (CAnnTG) binding sites. Thus, mechanisms other than chromatin accessibility contribute to differential in vivo DNA binding by these two TFs.
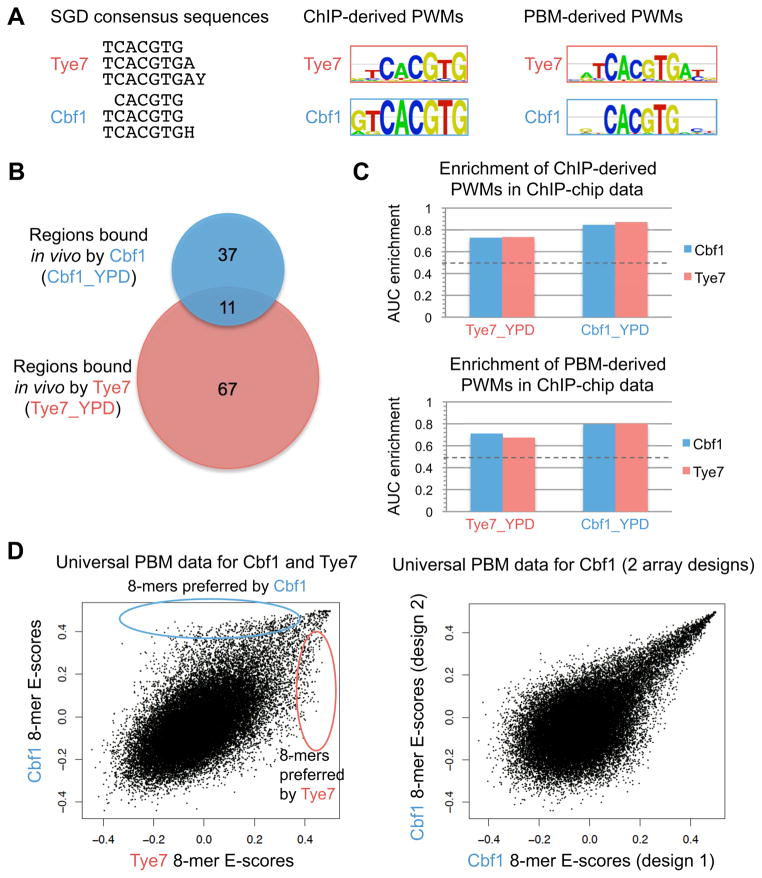
Figure 1.
DNA binding specificities of S. cerevisiae Cbf1 and Tye7. (A) Cbf1 and Tye7 have highly similar DNA binding specificities according to consensus sequences in SGD, PWMs from ChIP-chip data (Harbison et al., 2004), or PWMs from universal PBM data (Zhu et al., 2009)). (B) Cbf1 and Tye7 have little overlap in genomic regions bound in rich medium (YPD) (ChIP-chip P > 0.005 (Harbison et al., 2004)). (C) PWMs of Cbf1 and Tye7 are enriched both in genomic regions bound in Cbf1_YPD and Tye7_YPD ChIP-chip data. Dotted line shows expected enrichment for a random PWM. (D) Universal PBM data for Cbf1 and Tye7 show differences not seen in replicate PBM experiments for the same TF (not shown) nor in PBM experiments for the same factor on two different universal array designs (right plot). See also Figure S2.
Using custom-designed ”genomic context protein binding microarrays” (gcPBMs), we analyzed binding of Cbf1 and Tye7 to their putative E-box binding sites centered within native genomic sequences. Our gcPBM data show that when placed within genomic flanking sequences, E-box sites are bound with different preferences by these two proteins. Importantly, these differences in binding are observed not just in vivo, but also in vitro, where cofactors or histones are not present. Thus, the DNA sequence itself is responsible for differential binding by these two TFs.
Notably, the identified differences in DNA binding preferences between Cbf1 and Tye7 are not apparent from these proteins’ binding site motifs (Figure 1). Therefore, to further investigate the source of the binding differences, we used the gcPBM data in a regression analysis to build computational models of the DNA binding specificities of Cbf1 and Tye7. Compared to traditional DNA motif models (i.e., position weight matrices, PWMs), these new models are more accurate in predicting in vitro DNA binding. Examination of the sequence features that are important for our regression models revealed that features located in the genomic sequences flanking the E-boxes contribute to DNA binding specificity. Our results show that differences in the intrinsic sequence preferences of related TFs, even when they occur outside the core DNA binding site motif can contribute to differential TF-DNA binding. Importantly, these differences in intrinsic sequence preferences, as identified through our in vitro studies, can partly explain differential DNA binding in vivo.
DNA sequences flanking the E-box motif, which were found to affect binding of Cbf1 and Tye7, do not typically form base-specific contacts with bHLH proteins (De Masi et al., 2011). Thus, we hypothesized that these sequences contribute to binding specificity indirectly by influencing the three-dimensional structure of the DNA binding sites. A role of DNA shape in achieving binding specificity of TFs has been suggested for Drosophila Hox proteins (Joshi et al., 2007; Slattery et al., 2011) and other protein families (Rohs et al., 2010; Rohs et al., 2009). However, for these examples DNA shape was a result of the nucleotide sequence within the TF binding site. Here, we found that nucleotides flanking Cbf1 and Tye7 binding sites alter structural properties of their DNA targets, and thus contribute to their differential binding preferences. This finding, for the first time, reveals a mechanistic explanation for the role of nucleotides that are located outside of a binding motif to TF binding specificity. Moreover, this finding suggests why TFs bind in vivo to only a subset of available target sites with identical core motifs. Future studies will investigate the generality of our findings within the bHLH family as well as other TF families. Our results suggest that the local shape of DNA binding sites might play a critical role in achieving regulatory specificity within TF families.
RESULTS
S. cerevisiae TFs Tye7 and Cbf1 recognize highly similar DNA sequence motifs despite binding different target sites in vivo
TFs from the bHLH protein family recognize DNA binding sites containing the E-box motif (CAnnTG) (Atchley and Fitch, 1997), with different family members sometimes having different preferences for the two central base pairs of the E-box (De Masi et al., 2011; Fong et al., 2012; Grove et al., 2009). In S. cerevisiae, the bHLH family comprises 8 TFs that have diverse functions. Among these TFs, Cbf1 and Tye7 are most similar in terms of their DNA binding specificities (Figure 1A) (Cherry et al., 2012; MacIsaac et al., 2006; Zhu et al., 2009), with both having a strong preference for the E-box CACGTG. However, the sets of in vivo targets bound by Cbf1 and Tye7, as determined by ChIP-chip (Harbison et al., 2004), barely overlap (Figure 1B), and the two TFs regulate different processes: Cbf1 is involved in methionine biosynthesis and chromatin remodeling (Cai and Davis, 1990; Kent et al., 2004), while Tye7 plays a major role in the regulation of glycolytic genes (Nishi et al., 1995). It is currently unclear how two TFs with highly similar DNA binding motifs attain their regulatory specificities.
The Cbf1 and Tye7 DNA binding motifs, although very similar, are not identical. For this reason, we first asked whether the small differences in these motifs (Figure 1A) can explain, at least in part, their differential binding in vivo. Using DNA motifs derived from in vivo (ChIP-chip) and in vitro (PBM) data, we computed an AUC-based enrichment score (see Experimental Procedures) (Gordân et al., 2009) for the enrichment of Cbf1 and Tye7 motifs in in vivo DNA binding data (Harbison et al., 2004), where a value of 1.0 corresponds to perfect enrichment and a value of 0.5 corresponds to the enrichment of a random motif. If the DNA motifs can explain, even in part, the differential in vivo binding, then we would expect the Cbf1 motif to be significantly more enriched in the Cbf1 ChIP-chip data, and the Tye7 motif to be significantly more enriched in the Tye7 ChIP-chip. However, we find that the motifs of both of these TFs are equally well enriched in both the Cbf1 and Tye7 ChIP-chip data sets (Figure 1C), which indicates that the information in the existing PWMs does not explain why these TFs bind different sites in vivo. A similar enrichment analysis that included S. cerevisiae bHLH protein Pho4, which also has a strong preference for the E-box CACGTG, revealed that the Pho4 PWM was not significantly enriched in the Cbf1 or Tye7 ChIP-chip data (Gordân et al., 2009). The same study showed that the Tye7 PWM was not significantly enriched in the Pho4 ChIP-chip data, and the Cbf1 PWM was only marginally enriched (in agreement with previous studies of Pho4 and Cbf1 (Zhou and O’Shea, 2011)). Thus, difference in the PWMs of Pho4 versus Cbf1/Tye7 can explain, at least in part, the differences in their in vivo DNA binding. However, the Cbf1 and Tye7 PWMs are too similar to explain why these two TFs interact with distinct sets of E-box sites in vivo.
An alternative way to represent the DNA binding specificities of TFs utilizes data generated by universal PBMs. PBM experiments performed on universal arrays (Berger et al., 2006) provide measurements of TF binding to all possible 8-bp sequences (8-mers), as well as a measure of the PBM enrichment score (E-score) for each 8-mer. E-scores range from −0.5 to +0.5, with higher values corresponding to higher sequence preference; typically, E-scores > 0.35 correspond to specific TF-DNA binding (Berger et al., 2006; Gordan et al., 2011). We compared previously published 8-mer E-scores for Cbf1 and Tye7 (Zhu et al., 2009), and found that, although they are correlated, the binding specificities of the two TFs are not identical (Figure 1D); there are many 8-mers that are strongly preferred by only one of these two TFs. We did not observe such differences between universal PBM experiments performed for the same factor (Cbf1) on two different universal array designs (Figure 1D). This suggests that Cbf1 and Tye7 have slightly different specificities in vitro.
Tye7 and Cbf1 bind with different specificities to putative DNA binding sites in their genomic context
To further investigate the differences in the in vitro DNA binding specificities between Cbf1 and Tye7, we designed a custom PBM containing putative Cbf1 and Tye7 DNA binding sites in their native genomic context (Figure 2A, B, C). In this novel array design, termed ”genomic context PBM” (gcPBM), we initially focused on genomic regions bound in vivo by either of the two TFs, defined as regions with P < 0.005 in Cbf1 or Tye7 ChIP-chip data (Harbison et al., 2004). To identify putative TF binding sites in the S. cerevisiae genome, we used universal PBM data for Cbf1 and Tye7 (Zhu et al., 2009) to search for DNA sites containing two consecutive, overlapping 8-mers with E-score > 0.35 (Busser et al., 2012). Next, we selected 30-bp genomic regions centered at the putative binding sites to create a set of “ChIP-chip bound” probes for our gcPBM. Similarly, we created a set of “ChIP-chip unbound” probes by searching for putative Cbf1 and Tye7 binding sites in the genomic regions not bound in the ChIP-chip experiments (ChIP-chip P > 0.5).
Figure 2.
Design of genomic context PBM to compare Cbf1 and Tye7 DNA binding preferences. Arrays included (A) “ChIP-chip bound” probes and (B) “ChIP-chip unbound” probes, representing 30-bp genomic regions; see Extended Experimental Procedures for details. Cbf1 and Tye7 show significant differences in binding in vitro to (D) “ChIP-chip bound” (E) and “ChIP-chip unbound” probes. Both proteins were tested at 200 nM in PBMs. The plots show the natural logarithm of the normalized PBM signal intensities, with higher numbers corresponding to higher affinity binding. See also Figure S1.
For two proteins with identical specificities, we expect their in vitro DNA binding signals (here, the natural logarithm of the PBM fluorescence signal intensity) to be highly correlated. However, comparison of the in vitro DNA binding specificities of Cbf1 and Tye7 for their putative “ChIP-chip bound” sites (Figure 2D) clearly shows that the two TFs interact differently with these genomic sites. Importantly, even when we extend the comparison to include the “ChIP-chip unbound” probes, we observe the same trend (Figure 2E). Finally, although Cbf1 and Tye7 were tested at the same concentration (200 nM) on the array, Cbf1 bound with higher affinity to a larger number of probes than did Tye7. To ensure that the generally higher-affinity binding by Cbf1 is not the reason for the low correlation between in vitro DNA binding by these two TFs, we repeated the PBM experiment at a lower concentration of Cbf1 (100 nM). As expected, we saw lower overall PBM signal for Cbf1, but the differences in DNA binding specificity between Cbf1 and Tye7 were maintained (Figure S1). In conclusion, our gcPBM data show that, despite having highly similar DNA binding motifs, the two TFs exhibit different binding preferences for their putative genomic binding sites.
Base pairs flanking the E-box binding site contribute to DNA binding specificity in vitro
The DNA binding signal observed in our gcPBM experiments reflects the specificities of Cbf1 and Tye7 for E-box binding sites and their genomic flanks. Henceforth we will refer to the two base pairs immediately upstream and downstream of the E-box as the “proximal flanks”, and the base pairs more than two positions away from the E-box as the “distal flanks” (Figure 3A). Previous studies of bHLH DNA binding specificity focused either on the core E-box or the 2-bp proximal flanks (e.g., (De Masi et al., 2011; Fong et al., 2012; Grove et al., 2009; Maerkl and Quake, 2007; Wang et al., 2012)). Our analyses of the gcPBM data revealed that in addition to the E-box site and the proximal flanks, the distal flanks also contribute to the differential DNA binding specificities of Cbf1 and Tye7.
Figure 3.
Flanking sequences contribute to Cbf1 and Tye7 DNA binding specificity. (A) Proximal or distal flanks surrounding the E-box result in (B) variation in Tye7 DNA binding signal for probes that contain the preferred E-box CACGTG, or any of the possible 8-mers centered at this E-box. Numbers in parentheses indicate number of probes containing each 6-mer or 8-mer. (C) Wide variation in DNA binding signal is observed even when we restrict the analysis to probes containing specific 10-mers. See also Figure S3.
We first investigated whether the central two base pairs in the E-box binding sites are responsible for the different binding preferences. Analysis of the binding of these two TFs for all possible E-boxes revealed that the 2-bp central spacer does not appear to be the cause of the binding specificity differences, and, as expected, both proteins have a strong preference for the E-box CACGTG (Figure S2). Thus, in our analyses of the gcPBM data we focused primarily on genomic regions containing this E-box.
Our gcPBM data indicate that not all CACGTG sites across the genome are bound equally well by Tye7: depending on the flanking genomic regions, this E-box is bound in vitro with a wide range of affinities, ranging from highly preferential to non-specific binding (Figure 3B). We observed a similar trend for Cfb1 (Figure S3). Even when we expanded the binding sites to include the 1-bp or 2-bp proximal flanks, we still observed wide variation in Cbf1 and Tye7 binding signal (Figure 3B, C, Figure S3), which indicates that the distal flanks contribute significantly to DNA binding specificity. Importantly, the wide range of binding affinities is not due to probes containing different numbers of binding sites, as the probes contain a single binding site located in the center of the probe (see Experimental Procedures). Thus, the differences in TF-DNA binding observed for probes that contain identical E-boxes and proximal flanks (e.g., ATCACGTGAA in Figure 3C) are due to contributions from the distal flanks.
Regression-based models can accurately predict in vitro DNA binding of Cbf1 and Tye7
To understand what features in the genomic flanks contribute to the DNA binding specificities of Cbf1 and Tye7, we performed a regression analysis of the gcPBM data. We used Support Vector Regression (SVR) (Drucker et al., 1997) to train linear models that use sequence features derived from the proximal and distal flanks to predict the DNA binding signal observed with gcPBMs (Figure 4A, B). Because both Cbf1 and Tye7 bind DNA as homodimers and their E-box binding sites are palindromic, we combined the two flanking regions 5′ and 3′ of the E-box motif (Figure 4A) and derived the sequence features from the combined flanks. Next, we derived features that reflect the number of occurrences of each possible 1-mer, 2-mer, and 3-mer at each position in the combined flanks. Thus, each feature derived from the combined flanks can take one of three values: 0, 1, or 2 (see example shown in Figure 4A).
Figure 4.
Regression analysis of gcPBM data. (A) For each 30-bp probe, we combined the two flanking regions and we generated 1-mer, 2-mer, and 3-mer features. We used ε-SVR to train linear models that predict the PBM log signal intensity of each probe based on its sequence features. Positions are numbered starting from the center of the CACGTG core. (B) Leave-one-out cross-validation analysis indicates that regression models for Cbf1 and Tye7 accurately predict PBM signal intensity. (C) Analysis of the sequence features with the largest positive and negative weights in SVR models shows that base pairs in both the proximal and distal flanks are important for predicting DNA binding specificity. Bar plots show the top 20 positive and negative weights. For brevity, feature names are shown only for the top positive/negative weight, and then for every other weight among the top 20. (D) Features show numerous differences between Cbf1 and Tye7. See also Figure S4 and Table S1.
We performed a cross-validation analysis to determine the best parameter values to be used by the regression algorithm (see Experimental Procedures). Using these parameter values, the linear regression models predicted the PBM log signal intensity values for both TFs with high accuracy using all 1-mer, 2-mer, and 3-mer features (Figure 4B). Regression models using just 1-mer features performed poorly (Figure S4), which suggests that individual base pairs in the flanking regions do not contribute independently to the DNA binding specificity. Adding 2-mer and 3-mer features improved the prediction accuracy, but including 4-mer features did not improve prediction accuracy further (see Extended Experimental Procedures), likely because such models have too many features compared to the number of training examples, and are thus prone to overfitting the training data.
Sequence features in the proximal and distal flanks contribute to DNA binding specificity
The regression analyses described above used a linear kernel SVR. The advantage of a linear kernel is that one can use linear SVR models to compute weights for all the features used in the regression. The resulting weights are readily interpretable, as they reflect to what degree each feature contributes to the predicted target values (i.e., PBM log signal intensities). Here, positive weights correspond to sequence features that have a positive contribution to the DNA binding signal, i.e., we can interpret such features as being preferred by a given TF, while features with negative weights have a negative effect on binding.
The feature weights for Cbf1 and Tye7 (Figure 4C, Table S1) indicate that sequence features in both the proximal and the distal flanks contribute to the predicted DNA binding specificities of these TFs. As expected, features closer to the E-box generally have an important contribution (i.e., large feature weights). For example, the nucleotide A at position 4, immediately next to the E-box, is strongly preferred by both Cbf1 and Tye7, consistent with prior reports on the binding preferences of these TFs (MacIsaac et al., 2006; Zhu et al., 2009). To determine how far away from the E-box the important features are located, we repeated the SVR analysis with flanking regions of different lengths (2 to 12 bps) to assess whether the overall prediction accuracy changes when shorter flanking regions are used. Briefly, for Cbf1 we obtained the best prediction accuracy (Pearson R2=0.745) when 11-bp flanks were used in the SVR analysis, while for Tye7 we obtained the best prediction accuracy (R2=0.898) when 5-bp flanks were used (Figure S4B). By comparison, models using just the 2-bp proximal flanks achieved accuracies of 0.694 and 0.836 for Cbf1 and Tye7, respectively. These correlations are expected since the 2-bp proximal flanks have important contributions to the DNA binding specificity. However, incorporating distal flanks allowed us to predict the PBM signal intensities even better: the prediction errors for the best Cbf1 and Tye7 models (using 11-bp and 5-bp flanks, respectively) are significantly lower than the prediction errors for models using 2-bp flanks (Wilcoxon P = 0.035 and 0.00091 for Cbf1 and Tye7, respectively). Thus, our results show that although the proximal flanks have a higher contribution to the predicted DNA binding signal compared to distal flanks, the latter are necessary for achieving the best prediction accuracy.
To further test the accuracy of our regression models, we introduced mutations at various positions in the proximal and distal flanks of the 30-bp genomic sites on our gcPBM (see Extended Experimental Procedures). We used wild type and mutated sequences to generate a new custom PBM (henceforth referred to as the “validation” PBM), and tested both Cbf1 and Tye7 on this new array. Our predictions from the SVR models agree very well with the measured PBM log signal intensities on the “validation” array (overall Pearson R2 was 0.84 for Cbf1 and 0.75 for Tye7; Figures S4C–F). Thus, both the Cbf1 and the Tye7 SVR models accurately predict the individual DNA binding specificities of these TFs.
Next, to investigate how the various sequence features contribute to differences in DNA binding specificity between Cbf1 and Tye7, we compared the feature weights computed from the regression models for these TFs (Figure 4D). Although the two sets of weights are positively correlated (R2=0.32), there are numerous differences between them, resulting from both proximal and distal flanks. For example, Tye7 disfavors the nucleotide C at position 4 (i.e., immediately downstream of the E-box), while Cbf1 actually prefers a C at this position (see feature “4-C” in the upper left quadrant of Figure 4D). Unlike this difference, which is apparent in their DNA binding site motifs (Figure 1A), most differences in feature weights are subtle, in that they cannot be inferred from the motifs, and the individual contributions of the corresponding features are small. However, taken together, these features can accurately predict the different DNA binding specificities of Cbf1 and Tye7, as illustrated by the accuracy of the SVR models on both our initial gcPBM and the “validation” PBM. This suggests that the features represented by the distal flanks might not correspond to direct recognition by Cbf1 and Tye7, but rather might contribute to TF-DNA binding specificity indirectly by influencing the three-dimensional DNA structure. To further investigate this hypothesis, we performed a detailed DNA shape analysis of the sequences bound by Cbf1 and Tye7 in gcPBMs.
DNA shape features are characteristic for bHLH binding sites
We used a high-throughput DNA shape prediction approach (Slattery et al., 2011) to analyze differential DNA shape preferences selected by Cbf1 and Tye7 as a function of the in vitro binding signal (i.e., PBM log signal intensity). This DNA shape prediction method derives structural features of DNA (e.g., groove width and helical parameters) by mining Monte Carlo trajectories using a sliding pentamer window (see Experimental Procedures). Groove width in B-DNA is measured over a region of four base pairs and thus is affected by the sequence composition of at least half a helical turn (Rohs et al., 2005). In contrast, helical parameters describe DNA shape at dinucleotide resolution and give rise to groove geometry (Joshi et al., 2007). We analyzed both groove geometry and helical parameters.
Minor groove width and propeller twist (Figure 5A) and roll and helix twist (Figure S5) reflect the unique shape of E-boxes (CAnnTG), with minor groove widening at both CpA (TpG) base pair steps due to weak stacking interactions and the tendency of these dinucleotides (at positions −2/−3 and +2/+3) to open the minor groove. Propeller twist, roll, and helix twist further indicate a distinct conformation of the E-box. Our analysis of these features shows differences between high and low affinity binding. For example, minor groove width tends to be wider for high compared to low binding affinity sites, and propeller twist can distinguish binding preferences of Tye7 versus Cbf1 (Figure 5A).
Figure 5.
DNA shape analysis. (A) Heat maps show the average minor groove width (left) and propeller twist (right) for sequences on the gcPBM. Sequences were sorted in decreasing order of gcPBM signal intensity for either Cbf1 (top) or Tye7 (bottom), and grouped into 50 bins. Average DNA shape parameters were computed within each bin. (B) Different proximal flanks surrounding the common CACGTG E-box are preferred by Tye7 and Cbf1. Sequences located in the upper left triangle are preferentially bound by Tye7 and 10-mers located in the lower right triangle are preferentially bound by Cbf1. Dashed lines indicate respective cutoffs of a difference ≥ 30 in rank between Tye7 preferred (red) and Cbf1 preferred (blue). Lighter colored dots exhibit larger differences. (C) DNA shape variation due to flanks surrounding CACGTG selected preferentially by Cbf1 (light blue) or Tye7 (light red), comparing. Asterisks (*) indicate positions with significant differences (P < 0.05, Mann-Whitney U-test) in the minor groove width (upper) or propeller twist (lower) between the sequences preferred by Cbf1 or Tye7. The symmetry of the box plots is due to the shape predictions having been performed for the combined flanks. (D) Incorporating DNA shape features improves binding intensity predictions in comparison to using DNA sequence (1-mers) alone. The improvement is similar to that obtained by adding 2-mer and 3-mer features. See also Figure S5.
DNA shape features in flanking regions are distinct for binding sites preferred by Cbf1 versus Tye7
Since our previous analysis of PBM data indicated that Tye7 and Cbf1 both bind preferentially to the E-box CACGTG (Zhu et al., 2009), we hypothesized that specificity for distinct binding sites arises from 5′ and 3′ flanking sequences. Therefore, we filtered the sequences derived from our gcPBM data based on their sharing of the E-box CACGTG, and then compared the ranked log signal intensities for Tye7 and Cbf1 for those probes. We next analyzed the groups of sequences bound preferentially either by Tye7 or Cbf1, defined as gcPBM probes with a difference ≥ 30 in rank between the two TFs (shown by dashed lines in Figure 5B). Next, for both sets of sequences, we predicted DNA structural features and analyzed them for variation in DNA shape due to different flanks. We performed this analysis for both strands of the double helix and averaged the results because of the palindromicity of the CACGTG E-box. Our results indicate that both of these TFs select sites with distinct minor groove geometry (Mann-Whitney U P = 0.03, 0.008, 8.7×10−7, and 5.08×10−7 at positions 6, 4, 3, and 2, respectively) and propeller twist (P = 0.02, 0.01, 0.04, 0.02, and 1.1×10−5 at positions 9, 8, 7, 4 and 2) (Figure 5C), due to different flanking regions of the E-box (positions −3 to +3) being selected by Tye7 versus Cbf1 (Figure 5B). We observed similar statistically significant distinctions in roll (between positions 6 to 3 and 2 to 1) and helix twist (between positions 12 to 11 and 4 to 2) (Figure S5G).
Incorporation of DNA shape features improves binding intensity predictions in comparison to using DNA sequence alone
If DNA shape distinguishes binding targets selected by Cbf1 and Tye7, the use of structural features should also improve binding affinity predictions. To test this hypothesis we incorporated structural features in our linear SVR approach. We found that adding DNA shape features (minor groove width, roll, propeller and helix twist) leads to an improvement in binding specificity predictions similar to that obtained by adding 2-mer and 3-mer features: R2=0.72 and 0.89 using 1-mers and DNA shape features (Figure 5D) compared to R2=0.74 and 0.88 using 1-mers, 2-mers and 3-mers, for Cbf1 and Tye7, respectively (Figures 4B and 5D). Incorporating DNA shape features in addition to 2-mers and 3-mers did not improve the prediction accuracy any further. This suggests that 2-mers and 3-mers implicitly contain structural information, while DNA shape implicitly contains interdependencies between nucleotides at different positions of the binding site. Using structural features instead of 2-mers and 3-mers has the advantage that the total number of features is much smaller, and thus regression algorithms other than SVR can be used successfully to learn accurate models of DNA binding specificity. To illustrate this point, we used L2-regularized linear regression and obtained highly accurate predictions: R2=0.7 and 0.87 for Cbf1 and Tye7, respectively, using 1-mers and DNA shape features (see Experimental Procedures).
Genomic sequences flanking the E-box motif contribute to explaining the differences in in vivo DNA binding between Cbf1 and Tye7
Both our regression analysis based on DNA sequence features and our DNA shape analysis show that Cbf1 and Tye7 interact differently with their putative genomic binding sites. To assess whether these differences contribute to differential DNA binding by these two TFs in vivo, we examined whether the DNA sequences preferred in vivo by a particular TF also have higher TF binding signal in vitro (Figure 6). Figure 6B shows a scatter plot of Cbf1 versus Tye7 in vitro binding signal for the 30-mer PBM probes selected from genomic regions bound in vivo by either of the two TFs (Harbison et al., 2004). We colored the data points based on in vivo specificity: blue for PBM probes selected from the 37 regions bound only by Cbf1 in vivo, red for PBM probes selected from the 67 regions bound only by Tye7 in vivo, and grey for PBM probes selected from the 11 genomic regions bound by both Cbf1 and Tye7 in vivo. Next, for each TF we compared the in vitro signal for PBM probes bound uniquely by only one TF in vivo (i.e., blue versus red data points), and found that DNA sequences preferred in vivo by a particular TF also have higher binding signal for that TF in vitro (Figure 6C) (Kolmogorov-Smirnov P = 0.00078 for Cbf1 and 0.003 for Tye7). We performed a similar analysis focusing on the PBM probes containing the E-box CACGTG and observed the same trend (Figure S6; Extended Experimental Procedures). Our results suggest that subtle differences in the intrinsic sequence preferences of Cbf1 and Tye7 observed in vitro on gcPBMs partially explain differential DNA binding in vivo observed in ChIP-chip data.
Figure 6.

Differences in the in vitro DNA binding preferences of Cbf1 and Tye7 are important for differential in vivo binding. (A) Overlap between sets of genomic regions bound by Cbf1 and Tye7 in ChIP-chip in rich medium (YPD). (B) Scatter plot of Tye7 versus Cbf1 PBM log signal intensity for 30-mer probes that occur in genomic regions bound in vivo only in Tye7_YPD (red), only in Cbf1_YPD (blue) or in both data sets (grey). (C) Cbf1 and Tye7 in vitro binding signal (i.e., natural logarithm of gcPBM probe intensity) for 30-mers probes selected from genomic regions bound only by Cbf1 (blue) or only by Tye7 (red) in vivo. The differences in PBM log signal intensity between the two sets of 30-mer probes are statistically significant by Kolmogorov-Smirnov (KS) tests. See also Figure S6.
DISCUSSION
This study shows, for the first time, that subtle differences in the intrinsic preferences of paralogous TFs for sequences flanking the core DNA binding site motif can contribute to differential DNA binding in vivo. Using the S. cerevisiae TFs Cbf1 and Tye7 as our model system we show that, when tested in vitro in their native genomic flanking sequences, putative DNA binding sites of Cbf1 and Tye7 are bound differentially by the two proteins. As expected, the differences between the intrinsic sequence preferences of the two TFs observed in vitro on our gcPBMs do not fully explain the differences in in vivo DNA binding observed in ChIP-chip data (Harbison et al., 2004). Other mechanisms might be used in vivo to provide additional specificity. For example, Cbf1 interacts with Met4 and Met28 to regulate genes involved in sulfur metabolism (Lee et al., 2010; Siggers et al., 2011). In addition, Cbf1 has chromatin remodeling properties (Kent et al., 2004) that may allow it to bind certain CACGTG sites that are inaccessible for Tye7 due to nucleosome occupancy. However, to fully understand how these different mechanisms are used, it is important to have a better characterization of the intrinsic sequence preferences of the two TFs.
The analyzed structural features characterized free DNA (i.e., DNA not bound by the proteins) and thus reflect the intrinsic properties of the E-box binding sites and their genomic sequence context. Analysis of DNA shape shows that a widening of the minor groove characterizes the E-box in its unbound state, as we observed for sites selected by Tye7 and Cbf1. The same observation was made for the crystal structures of E-boxes in complex with the yeast TF Pho4 (Shimizu et al., 1997) and mammalian bHLH TFs (Brownlie et al., 1997; Ma et al., 1994). This suggests that DNA shape features observed in complexes of bHLH factors and their DNA targets are inherent to DNA binding sites and thus may constitute previously under-appreciated, widely used signals in cis regulatory sequences recognized by TFs. This form of intrinsic DNA shape recognition was previously observed for Hox proteins (Joshi et al., 2007; Slattery et al., 2011) and other TFs (Rohs et al., 2009). In addition to reporting this observation for the first time for E-box binding sites, we show here that structural variations due to different flanking sequences of E-boxes are a source of differences in DNA binding specificity among bHLH TFs. Consequently, we demonstrate that the integration of DNA shape and sequence leads to improved binding intensity predictions, similar to the use of 2-mers and 3-mers, compared to sequence (1-mers) alone.
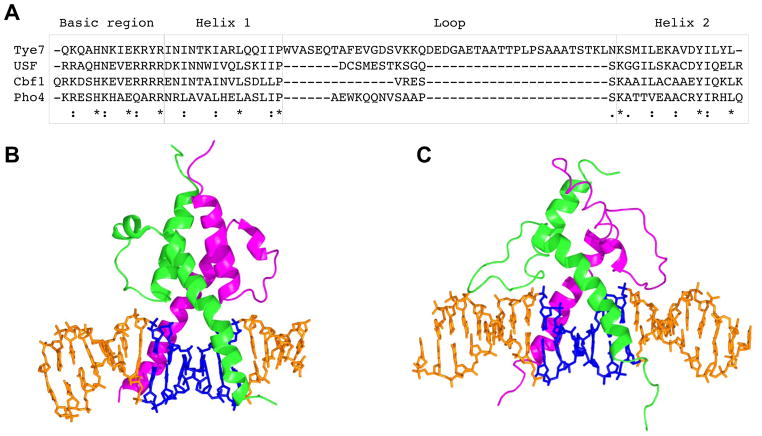
In this study we expressed both TFs as full-length proteins, so residues within or outside the DNA binding domain may play a role in the protein-DNA interactions. bHLH factors are known to select the E-box CAnnTG through DNA contacts by their His5 and Glu9 residues from each monomer of the bHLH dimers, which recognize the CpA (TpG) base pair steps (Shimizu et al., 1997). Based on co-crystal structures of a human bHLH factor and the yeast factor Pho4 bound to DNA (Shimizu et al., 1997), modeling and mutagenesis studies, we showed previously that the Arg13 side chain of bHLH factors selects C/G base pairs in the two central positions of the CACGTG E-box through the formation of a base-specific hydrogen bond with the guanine bases at positions −1 and +1 (De Masi et al., 2011). Since the yeast bHLH factors Tye7, Cbf1, and Pho4 all have His5, Glu9, and Arg13 residues, the CACGTG motif is the E-box that is most preferred by all of these TFs. However, the reason why Tye7, Cbf1, and Pho4 prefer different sequences flanking the common E-box motif CACGTG is likely due to the length and sequence variation of the loop that separates the H1 and H2 helices in the bHLH protein (Figure 7). Co-crystal structures are not available for either Cbf1 or Tye7 bound to DNA, but crystal structures of Pho4 (Shimizu et al., 1997) and the human homologue of Cbf1, the upstream stimulatory factor (USF), have been solved in complex with DNA (Ferre-D’Amare et al., 1994). The crystal structures of Pho4 and USF bound to DNA illustrate that the conformations of the respective loops between the H1 and H2 helices in both bHLH monomers can give rise to different DNA recognition in the regions flanking the E-box. The two loops of the Pho4 homodimer each form an additional α-helix, whereas the USF loops are fully extended (Figure 7). Although base-specific contacts by bHLH factors are restricted to the E-box, the extended loops of both USF monomers lead to phosphate and other nonspecific contacts further upstream and downstream from the E-box, which can also be detected in DNase I footprints (Hesselberth et al., 2009; Neph et al., 2012). We suggest that these additional contacts outside the E-box may result in the selection of different flanking sequences through DNA shape features. In addition, structural differences in the flanking regions affect the ability of DNA to deform upon protein binding in order to optimize bHLH-DNA contacts and protein-protein interactions within the bHLH dimer.
Figure 7.
Sequence and structure comparison of bHLH/DNA complexes. (A) Sequence alignment of S. cerevisiae Tye7, Cbf1, and Pho4, and human USF shows the sequence and length variation of the loops between α-helices H1 and H2. In complex with their target sites, (B) yeast Pho4 and (C) human USF form base-specific contacts with the E-box while the loops between the H1 and H2 helices of the bHLH motifs adopt different conformations. The bHLH-DNA complexes shown are based on crystal structures with PDB IDs (B) 1A0A and (C) 1AN4.
In summary, our combined experimental and computational analysis of DNA sequence and shape preferences of yeast bHLH factors demonstrates that Cbf1 and Tye7 share the same E-box as a result of highly specific base contacts in the major groove, while they prefer different DNA flanking sequences because of structural features that enhance bHLH loop-DNA phosphate contacts that optimize the induced fit within the complex. Thus, this study demonstrates that bHLH factors use a combination of two different mechanisms of protein-DNA recognition: “base readout” and “shape readout” (Harris et al., 2012; Rohs et al., 2010); base readout in the major groove conserves the E-box, while local DNA shape readout in the flanking regions appears to enable distinct DNA binding preferences among paralogous TFs. It will be interesting to investigate if other TF families utilize DNA shape readout in similar ways, as this could be an important mechanism through which closely related TFs recognize different DNA target sites and perform different regulatory roles in the cell.
EXPERIMENTAL PROCEDURES
Enrichment of DNA binding site motifs in ChIP-chip data
Using Cbf1 and Tye7 DNA binding motifs derived from both in vivo (ChIP-chip) (MacIsaac et al., 2006) and in vitro (PBM) (Zhu et al., 2009) data, we computed the AUC enrichment, as described previously (Gordân et al., 2009), for each motif in the ChIP-chip data sets Cbf1_YPD and Tye7_YPD, which correspond to Cbf1 and Tye7, respectively, tested in rich media (YPD) (Harbison et al., 2004). Briefly, from each ChIP-chip data set we selected the ‘bound’ and ‘unbound’ probes, defined as probes with P < 0.005 and P > 0.5, respectively. Next, for each probe we computed the probability of it being bound by a TF with a particular DNA motif. We used the scores for the ‘bound’ and ‘unbound’ probes to generate an ROC curve and took the area under the curve (AUC) as a measure of enrichment of the motif in the ChIP-chip data.
Protein expression and purification
GST-Cbf1 and GST-Tye7 (Zhu et al., 2009) were over-expressed in E. coli BL21 (DE3) cells (New England BioLabs), and purified by FPLC (AKTAprime plus) using GSTrap™ FF affinity columns (GE Healthcare). Anti-GST Western blots were performed to assess protein quality and concentration. See Extended Experimental Procedures for further details.
Genomic context protein binding microarray design
We designed a custom DNA oligonucleotide array in 4×44K format (Agilent Technologies, Inc.; AMADID #029393) containing putative Cbf1 and Tye7 DNA binding sites. Briefly, we represent three categories of 30-bp genomic sequences on our gcPBM: 1) “ChIP-chip bound” probes, 2) “ChIP-chip unbound” probes, and 3) negative control probes. “ChIP-chip bound” probes corresponded to genomic regions bound in vivo by Cbf1 or Tye7 (ChIP-chip P < 0.005 in rich medium (YPD) (Harbison et al., 2004)) contained at least two consecutive 8-mers with universal PBM E-score > 0.35 (Zhu et al., 2009). All putative binding sites occurred at the same position within the probes on the array. “ChIP-chip unbound” probes corresponded to genomic regions with ChIP-chip P > 0.5 and at least two consecutive 8-mers at a more stringent universal PBM E-score threshold of 0.4. Negative control probes corresponded to S. cerevisiae intergenic regions with a maximum 8-mer E-score < 0.3. We also designed probes that contain, within constant flanking regions, all 10-bp sequences that occur within the “ChIP-chip bound” probes and contain the E-box CACGTG, but are flanked by synthetic rather than native genomic sequence The reported PBM signal intensity for each probe is the median PBM signal intensity over 4 replicate spots. The “validation” array (Agilent Technologies, Inc.; AMADID #041711) contains 30-bp genomic sequences from our initial custom array, with 0 through 4 mutations designed at various positions in the genomic sequences. Details are provided in Extended Experimental Procedures.
Protein binding microarray experiments and data analysis
Custom-designed arrays were synthesized (Agilent Technologies, AMADID #029393 and #041711), converted to double-stranded DNA arrays by primer extension, and used in PBM experiments essentially as described previously (Berger and Bulyk, 2009; Berger et al., 2006). PBM data quantification was performed as previously described (Berger and Bulyk, 2009; Berger et al., 2006). See Extended Experimental Procedures for details.
Support Vector Regression analysis
Support Vector Regression (SVR) was run separately for Cbf1 and Tye7. For each TF, we first selected “ChIP-chip bound” and “ChIP-chip unbound” probes centered at the E-box CACGTG. To ensure that no additional binding sites occur in the regions flanking CACGTG, we selected probes (280 for Cbf1, and 312 for Tye7) for which the maximum PBM 8-mer E-score in the flanks was < 0.3. Next, for each selected sequence we computed the number of occurrences of each 1-mer, 2-mer and 3-mer in the combined flanks (Figure 4A), or the corresponding DNA shape features. We thus obtained sparse feature matrices for each of the two TFs. As target features for the SVR analyses, we used the natural logarithm of the Cbf1 and Tye7 PBM fluorescence signal intensities. We used the ε-SVR algorithm implemented in the libSVM toolkit (Chang and Lin, 2011) for all SVR analyses. We performed a grid search using 10-fold and leave-one-out cross-validation to determine the best values for parameters ε and C (see Extended Experimental Procedures). Using these parameters, we trained the final SVR models using all 280 sequences for Cbf1 and all 312 sequences for Tye7, and used them to predict the PBM log signal intensities for all probes on the “validation” array. We also performed an SVR analysis using the 312 sequences selected for Tye7, but shuffling the PBM log signal intensities; the best R2 on randomized sets of sequences was < 0.1 (Figure S4A).
High-throughput DNA shape prediction
DNA shape parameters were derived from a high-throughput (HT) prediction approach based on data-mining of 2,121 Monte Carlo (MC) predictions (Joshi et al., 2007; Rohs et al., 2005) for DNA fragments. Average groove width and helical parameters were calculated with a modified CURVES program (Lavery and Sklenar, 1989). The resulting structural features were used to describe the average conformation of each of the 512 unique pentamers. The average conformation at the central base pair (for groove width and propeller twist) or the two central base pair steps (for roll and helix twist) of each unique pentamer was used to characterize a pentamer. A query table for pentamers was assembled using these data and a sliding pentamer window was implemented to compute structural features for any DNA sequence. We validated our HT method for DNA shape predictions based on a comparison with all crystal structures of protein-DNA complexes available in Protein Data Bank with a DNA duplex of at least one helical turn (10 base pairs) and no chemical modifications as specified elsewhere (Bishop et al., 2011). Spearman’s rank correlations are 0.67 for minor groove width, 0.56 for propeller twist, 0.63 for roll, and 0.55 for helix twist. Comparison with solution-state NMR structures of unbound DNA derived based on residual dipolar coupling (Wu et al., 2003) yields excellent quantitative agreement with our predictions for most of the discussed parameters.
Statistical analysis of DNA shape parameters
For Cbf1 and Tye7 separately, the selected sequences were grouped into 50 bins according to their ranked natural log signal intensity from gcPBM data. To extract the effect of the flanking sequences, the probes were filtered by the criterion of sharing the E-box motif CACGTG. The signal intensity ranks for all those probes were compared, and flanks bound preferentially by Tye7 or Cbf1 were identified as a difference ≥ 30 in rank between the two TFs (Figure 5B). The statistical significance of differences in the predicted groove width and helical parameters of these two distinct groups at each position was determined by the Mann-Whitney U-test.
Regularized linear regression analysis using DNA sequence and shape features
We trained L2 regularized linear regression models using sequence (1-mer) features alone or in combination with shape features. 10-fold cross-validation was performed to assess their performance. In each round of cross-validation, the optimal regularization parameter λ was selected using an embedded 10-fold cross-validation on the training data set.
Supplementary Material
HIGHLIGHTS.
Cbf1 and Tye7 are paralogous TFs with virtually identical DNA binding site motifs
The two paralogous TFs bind different genomic target sites in vivo
The genomic context of putative DNA binding sites affects TF binding specificity
Structural analyses suggest genomic context influences TF binding through DNA shape
Acknowledgments
We thank Trevor Siggers for technical assistance and helpful discussions, and Alexander Hartemink for critical reading of the manuscript. This work was supported by NIH/NHGRI grant # R01 HG003985 (M.L.B.), funding from the Duke Institute for Genome Sciences and Policy (R.G.), the USC-Technion Visiting Fellows Program, and grant IRG-58-007-51 from the American Cancer Society (R.R.). R.G. was funded in part by an American Heart Association postdoctoral fellowship #10POST3650060. R.R. is an Alfred P. Sloan Research Fellow. The authors declare that they have no competing financial interests.
Footnotes
ACCESSION NUMBERS
The protein binding microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE44604.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arvey A, Agius P, Noble WS, Leslie C. Sequence and chromatin determinants of cell-type-specific transcription factor binding. Genome Res. 2012;22:1723–1734. doi: 10.1101/gr.127712.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. PNAS. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nature Protocols. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop EP, Rohs R, Parker SC, West SM, Liu P, Mann RS, Honig B, Tullius TD. A map of minor groove shape and electrostatic potential from hydroxyl radical cleavage patterns of DNA. ACS Chemical Biology. 2011;6:1314–1320. doi: 10.1021/cb200155t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie P, Ceska T, Lamers M, Romier C, Stier G, Teo H, Suck D. The crystal structure of an intact human Max-DNA complex: new insights into mechanisms of transcriptional control. Structure. 1997;5:509–520. doi: 10.1016/s0969-2126(97)00207-4. [DOI] [PubMed] [Google Scholar]
- Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser BW, Shokri L, Jaeger SA, Gisselbrecht SS, Singhania A, Berger MF, Zhou B, Bulyk ML, Michelson AM. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Development. 2012;139:1164–1174. doi: 10.1242/dev.077362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Davis RW. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2:27. [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Grove CA, Vedenko A, Alibes A, Gisselbrecht SS, Serrano L, Bulyk ML, Walhout AJ. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Research. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker H, Burges CJC, Kaufman L, Smola A, Vapnik V. Support vector regression machines. Adv Neur In. 1997;9:155–161. [Google Scholar]
- Ferre-D’Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. The EMBO Journal. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordân R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor-DNA interactions. Genome Research. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Murphy KF, McCord RP, Zhu C, Vedenko A, Bulyk ML. Curated collection of yeast transcription factor DNA binding specificity data reveals novel structural and gene regulatory insights. Genome Biol. 2011;12:R125. doi: 10.1186/gb-2011-12-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R, Mackoy T, Dantas Machado A, Xu D, Rohs R, Fenley M. Innovations in Biomolecular Modeling and Simulation. In: Schlick T, editor. Biomolecular Sciences Series. London, UK: Royal Society of Chemistry Publishing; 2012. pp. 53–80. [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. Journal of Biological Chemistry. 2004;279:27116–27123. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. Journal of Biomolecular Structure & Dynamics. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Lee TA, Jorgensen P, Bognar AL, Peyraud C, Thomas D, Tyers M. Dissection of combinatorial control by the Met4 transcriptional complex. Molecular Biology of the Cell. 2010;21:456–469. doi: 10.1091/mbc.E09-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard DA, Rajaram N, Kerppola TK. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. PNAS. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- Mann RS, Chan SK. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends in Genetics. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- Morin B, Nichols LA, Holland LJ. Flanking sequence composition differentially affects the binding and functional characteristics of glucocorticoid receptor homo- and heterodimers. Biochemistry. 2006;45:7299–7306. doi: 10.1021/bi060314k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka M, Shiraishi Y, Sugiura Y. Selected base sequence outside the target binding site of zinc finger protein Sp1. Nucleic Acids Res. 2001;29:4920–4929. doi: 10.1093/nar/29.24.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Park CS, Pepper AE, Eichinger G, Innis MA, Holland MJ. The GCR1 requirement for yeast glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene, which encodes a novel basic-helix-loop-helix protein. Molecular and Cellular Biology. 1995;15:2646–2653. doi: 10.1128/mcb.15.5.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajaram N, Kerppola TK. DNA bending by Fos-Jun and the orientation of heterodimer binding depend on the sequence of the AP-1 site. EMBO J. 1997;16:2917–2925. doi: 10.1093/emboj/16.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Annual Review of Biochemistry. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. The EMBO Journal. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Duyzend MH, Reddy J, Khan S, Bulyk ML. Non-DNA-binding cofactors enhance DNA-binding specificity of a transcriptional regulatory complex. Molecular Systems Biology. 2011;7:555. doi: 10.1038/msb.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Research. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. The EMBO Journal. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Delaglio F, Tjandra N, Zhurkin VB, Bax A. Overall structure and sugar dynamics of a DNA dodecamer from homo- and heteronuclear dipolar couplings and 31P chemical shift anisotropy. Journal of Biomolecular NMR. 2003;26:297–315. doi: 10.1023/a:1024047103398. [DOI] [PubMed] [Google Scholar]
- Zhou X, O’Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol Cell. 2011;42:826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Research. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.