Abstract
Cellular microRNAs play a key role in the post-transcriptional regulation of almost every cellular gene regulatory pathway and it therefore is not surprising that viruses have found ways to subvert this process. Several viruses encode microRNAs that directly downregulate the expression of innate immune factors, including proteins involved in promoting apoptosis and recruiting immune effector cells. Viruses have also evolved the ability to downregulate or upregulate specific cellular miRNAs in order to enhance their replication. This perspective provides an overview of our current knowledge of the complex interplay of viruses with the microRNA machinery of cells.
First identified as regulators of larval development in nematodes1,2, it is now known that microRNAs (miRNAs) play key roles in the regulation of almost every important cellular process in all multicellular eukaryotes3. Human cells encode over 1000 miRNA species, and these have been implicated in cellular differentiation, innate immunity, apoptosis and oncogenic transformation, as well as many other cell fate decisions3. Almost all cellular miRNAs are first transcribed as capped, polyadenylated primary miRNA (pri-miRNA) transcripts that can encompass one or a cluster of ~22-nt miRNAs4. These miRNAs occupy the upper part of an ~33-bp imperfect stem that is crowned by a large (≥10 nt) unstructured loop and flanked by single stranded RNA. This ~80-nt RNA structure is recognized by the nuclear microprocessor, consisting of the RNase III enzyme Drosha and the dsRNA binding protein DGCR8, which cleaves the stem ~22 bp from the stem:loop junction to generate the ~60-nt long pre-miRNA hairpin intermediate5,6. Pre-miRNAs are bound by the nuclear export factor Exportin 5 (Exp5), which transports them into the cytoplasm7. Here, the pre-miRNA is bound by a second RNase III enzyme, called Dicer, which cleaves the pre-miRNA at the stem-loop junction8. The resultant miRNA duplex intermediate then interacts with one of the four mammalian Argonaut (Ago) proteins, which incorporates one RNA strand to form the RNA induced silencing complex (RISC), while the second RNA strand is degraded9–11. Discrimination between the strands is thought to be regulated by the stability of the ends of the duplex, with the strand whose 5’ end is less tightly base paired being favored for incorporation into RISC12,13. However, this discrimination is rarely complete, so that both the major, miRNA strand and minor, star strand of the duplex intermediate are often detectable in RISC.
Once loaded into RISC, the miRNA serves as a guide RNA to target RISC to mRNAs bearing sequence complementarity14. If this complementarity is essentially complete, RISC binding can induce endonucleolytic cleavage and marked mRNA destabilization. However, if the target mRNA is only partially complementary, RISC binding induces translational inhibition, sometimes followed by deadenylation and mRNA destabilization15,16. For mRNA targets that are only partially complementary, the key is the complementarity between the so-called miRNA seed region—positions 2 to 8 from the 5’ end—and the mRNA target17,18. As a result, miRNAs that share a common seed region have very similar mRNA targets, even if the rest of the miRNA is different in sequence.
Because seven nucleotides of sequence complementarity are sufficient, at least in principle, to allow inhibition of an mRNA by a given miRNA, it is easy to calculate that each miRNA has the potential to target large numbers of cellular mRNAs, even allowing for the fact that many complementary target sites on mRNAs may be occluded by RNA secondary structure or by bound proteins. Initial efforts to identify the “targetome” of a given miRNA by relying on bioinformatics gave long lists of potential mRNA targets and did not prove to be particularly reliable, though these methods continue to improve. A more sophisticated approach uses assays that measure global gene expression in the presence or absence of a given miRNA, for example using mRNA microarrays, to identify mRNAs whose expression is specifically inhibited. If these mRNAs also contain a computationally predicted target site for that miRNA, then this provides a priori evidence in favor of the hypothesis that this is indeed a target19. More recently, methods have been developed to directly recover and deep sequence RISC binding sites on mRNAs by using mRNA:protein crosslinking followed by immunoprecipitation with a RISC component, such as Ago220–23. These “CLIP” approaches have proven to be a very powerful tool for the global recovery of RISC target sites, with the major remaining difficulty being the accurate identification of which miRNA is actually responsible for mediating RISC recruitment.
Discovery and expression pattern of viral microRNAs
Given the small size of miRNAs, their lack of antigenicity and their ability to post-transcriptionally inhibit the expression of specific mRNA species, they would seem to represent ideal tools for use by viruses to inhibit the expression of proteins that might act as inhibitors of viral replication, including especially mediators of antiviral innate immunity. The first viral miRNAs were discovered in 2003 in human B cells latently infected with the γ-herpesvirus Epstein-Barr virus (EBV)24, and subsequent papers identifying miRNAs in several other human and animal herpesviruses rapidly followed25 (Table 1). In addition, several human and animal polyomaviruses have been found to each encode a single pre-miRNA, while human adenoviruses have been proposed to encode two pre-miRNAs25. On the other hand, no miRNAs have been identified in human papillomaviruses or poxviruses, which are also DNA viruses26,27, and no human RNA virus, including human immunodeficiency virus (HIV-1), hepatitis C virus (HCV) and influenza B virus, has so far been shown to express any viral miRNAs28–30. This may relate to the fact that excision of a pre-miRNA stem-loop from an RNA virus genome would result in the cleavage of that genome, which might inhibit virus replication. Indeed, insertion of pri-miRNA stem-loops into the genome of HIV-1-based vectors does reduce vector titre, especially if multiple stem-loops are inserted (B.R. Cullen, unpublished observations).
Table 1.
MicroRNAs encoded by human viruses
Because the large majority of known viral miRNAs are encoded by herpesviruses, I will focus this review on this viral genus and in particular on EBV, Kaposi’s sarcoma-associated herpesvirus (KSHV) and human cytomegalovirus (HCMV), with only occasional reference to other human or animal virus species. In the case of EBV, we now know that this virus encodes 25 pre-miRNAs located in two distinct clusters, i.e., the 3 pre-miRNA BHRF1 cluster and the 22 pre-miRNA BART cluster24,31,32 (Fig. 1). Expression of these clusters is differential, depending on the latency stage of the virus. In primary B cells, EBV infection results in establishment of latency III, marked by high level expression of the three miR-BHRF1 miRNAs and moderate expression of the miR-BART miRNAs. In contrast, cells recovered from EBV-induced nasopharyngeal carcinomas (NPCs), as well as in B-cell primary effusion lymphomas (PELs), where EBV is in the latency II stage, are characterized by high levels of EBV miR-BART miRNA expression and a total lack of miR-BHRF1 miRNA expression31. Mutational analysis of the EBV genome has demonstrated that the miR-BHRF1 miRNAs, which are expressed at high levels in the transformed lymphoblastoid cell lines (LCLs) that result from primary B-cell infection, are important but not essential for LCL outgrowth and that LCLs lacking these viral miRNAs grow more slowly33,34. In contrast, the miR-BART miRNAs do not affect B-cell transformation by EBV in culture. As there is currently no in vitro model to study transformation of epithelial cells by EBV, it remains unclear whether the miR-BART miRNAs play a role in the initiation or maintenance of epithelial cell-derived tumors, such as NPCs.
Figure 1. Schematic representation of the genome structure of the human herpesviruses EBV, KSHV and HCMV.
This figure provides an overview of the repeat elements and viral miRNAs present in these viruses.
The oncogenic human γ-herpesvirus KSHV encodes a single cluster of 12 pre-miRNAs that are expressed at high levels in latently KSHV-infected B cells28,35 (Fig. 1). Ten of the 12 viral pre-miRNAs are located in an intron, while the other two, miR-K10 and miR-K12, are unusually located in the viral K12 open reading frame (ORF) and in the K12 mRNA 3’ untranslated region (3’UTR), respectively. Processing of these two miRNAs appears to be inefficient, thus allowing expression of the viral K12 protein36. Whether any of these miRNAs are important for B-cell transformation by KSHV remains unclear, as there is currently no in vitro model that accurately measures KSHV-mediated transformation. Nevertheless, several mRNA targets for the KSHV miRNAs have been identified and these data strongly suggest that this is indeed likely to be the case. Of interest, while all 12 KSHV pre-miRNAs are expressed during latency, this miRNA cluster also contains a lytic viral promoter, located immediately 5’ to the viral K12 ORF, that strongly induces expression of not only the K12 protein but also the viral miR-K10 and miR-K12 miRNAs during lytic re-activation36. However, it remains unclear what roles these two miRNAs play during productive KSHV replication.
Finally, HCMV is quite different from both EBV and KSHV in that the 12 known HCMV pre-miRNAs are scattered over the entire viral genome and, hence, transcribed by several different promoter elements28,37 (Fig. 1). Curiously, no report of HCMV miRNA expression during viral latency has so far appeared. However, all 12 HCMV pre-miRNAs are expressed at substantial levels during HCMV lytic replication. As there is no animal model available to analyze HCMV replication and pathogenicity in vivo, it is unclear whether these miRNAs indeed play a critical role in the viral life cycle. However, analysis of mutants of the related mouse CMV (MCMV) have shown that viral miRNAs promote MCMV replication and persistence in salivary glands, a key organ in terms of viral transmission38.
Virally-encoded microRNAs as mediators of immune evasion
As noted above, obvious potential targets for virally encoded miRNAs include factors that form part of the innate antiviral immune response, including factors that promote apoptosis or cell cycle arrest, or factors that help to recruit immune effector cells to virus infected cells. A number of these have been proposed, but instead of providing a comprehensive list of these proposed targets, I will here focus on mRNA targets for miRNAs encoded by EBV, KSHV or HCMV that are supported by several different lines of evidence and/or by data from more than one laboratory and that also illustrate the kinds of viral miRNA targets that have the potential to enhance viral replication or pathogenicity in vivo (Table 2).
Table 2.
Selected cellular innate immunity factors inhibited by EBV, KSHV or HCMV-encoded microRNAs
| Cellular factor | Targeted by | Predicted phenotype |
| PUMA | EBV miR-BART5 and miR-BART19 | Reduced apoptosis in response to DNA damage |
| BIM | EBV miR-BART4 and miR-BART15 | Reduced apoptosis |
| MICB | HCMV miR-UL112 KSHV miR-K7 EBV miR-BART1, BART3, BART5 and BART9 |
Reduced killing by NK cells |
| TWEAKR | KSHV miR | Reduced apoptosis |
| IRAK1 | KSHV miR | Reduced TLR signaling, weaker response to proinflammatory cytokines |
| MYD88 | KSHV miR | Reduced TLR signaling, weaker response to pro-inflammatory cytokines |
| TGBRII | KSHV miR | Prevention of TGFβ-induced apoptosis |
| Caspase 3 | KSHV miR-K1, miR-K3 and miR-K4 | Inhibition of apoptosis induced by pro-inflammatory stimuli |
| p21 | KSHV miR | Reduced cell cycle arrest after DNA damage |
| ERAP1 | HCMV miR | Inhibition of CTL killing due to poor antigen presentation |
| RANTES | HCMV miR | Reduced recruitment of immune effector cells |
See text for more detailed discussion of the potential effects exerted by these viral miRNAs, as well as relevant references.
One of the first cellular mRNA targets reported for an EBV miRNA was the p53 up-regulated mediator of apoptosis (PUMA), which was proposed to be targeted by EBV miR-BART539. This in turn was proposed to protect EBV-infected NPC cells from apoptosis induced by DNA damaging reagents. EBV infection is, in fact, known to induce a cellular DNA damage response and attenuation of this response has been shown to substantially increase the efficiency of transformation of B cells by EBV40. A second target for the EBV miRNA miR-BART19 in the 3’ UTR of PUMA mRNAs has recently been identified by CLIP21, further supporting the importance of this mRNA target.
Another pro-apoptotic cellular gene product that has been shown to be targeted by EBV miRNAs is BCL2L11/BIM, which appears to be targeted by both miR-BART4 and miR-BART1520,41. This activity was proposed to correlate with the observed inhibition of apoptosis by miR-BART miRNAs in the human gastric carcinoma cell line AGS.
A third, well supported target for the EBV miR-BART miRNA is the major histocompatibility complex class I-related chain B (MICB), a stress-induced ligand of the natural killer (NK) cell activating receptor NKG2D that plays a key role in promoting NK-mediated killing of virus infected cells42. MICB was actually first identified as a target for the HCMV-encoded miRNA miR-UL11243, but more recent data suggest the MICB is also targeted by KSHV miR-K7 and by EBV BART miRNAs42. As expected, miRNA-mediated downregulation of MICB has been shown to protect cells from NK cell killing in vitro42. CLIP analysis has more recently documented binding sites for miR-BART1, miR-BART3, miR-BART5 and miR-BART9 in the MICB 3’UTR20,21. The fact that MICB is not only targeted by miRNAs encoded by several herpesvirus species but also by several distinct EBV miRNAs strongly suggests that attenuation of MICB function is highly advantageous to viruses in vivo. Indeed, in the case of HCMV, MICB expression is also inhibited by a virally encoded protein, UL16, which acts synergistically with miR-UL11243.
An interesting final point is that EBV miRNAs not only target cellular mRNAs but also viral mRNAs. For example, the EBV miR-BART2 miRNA lies directly antisense to the mRNA encoding the viral DNA polymerase BALF524. This antisense location has been observed previously for several viral miRNAs, including the single pre-miRNAs encoded by all members of the polyomavirus family, which are encoded antisense to the viral large T antigen (TAg)25,44, and the miR-H2 miRNA encoded by Herpes Simplex Virus Type 1, which lies antisense to the ICP0 protein45. In the case of the polyomavirus SV40, it was proposed that expression of this miRNA late in the viral replication cycle inhibited T antigen expression and thereby reduced killing of SV40 infected cells by cytotoxic T lymphocytes (CTLs) in vivo 44. In contrast, inhibition of ICP0 expression by miR-H2 and inhibition of BALF5 expression by miR-BART2 has been proposed to stabilize viral latency45,46, though neither of these proposals has so far been experimentally validated.
The miRNAs encoded by KSHV have been perhaps the most intensely studied viral miRNAs, and a number of mRNA targets for KSHV miRNAs have been defined (Table 2). In addition to MICB, as noted above, these include:
The Tumor Necrosis Factor-Like Weak Inducer of Apoptosis receptor protein TWEAKR, which is targeted by miR-K1047. This targeting, which reduces the sensitivity of expressing cells to pro-apoptotic stimuli, has been further validated by CLIP23.
IL-1 receptor associated kinase 1 (IRAK1) and MYD88, which are components of the human Toll-like receptor (TLR)/IL-1 receptor signaling cascade, are downregulated by KSHV miR-K9 and miR-K5, respectively48. The expression of these two miRNAs in endothelial cells was shown to inhibit the production of the pro-inflammatory cytokines IL-6 and IL-8 after IL-1α treatment, and this activity may inhibit the immune clearance of KSHV-infected cells in vivo48.
The KSHV miRNA miR-K10 has also been proposed to target the transforming growth factor-β type II receptor (TGBRII), which was shown to inhibit TGF-β induced apoptosis in culture49. This may again promote the survival of KSHV-infected endothelial cells in culture.
Another protein involved in apoptosis that is targeted by KSHV miRNAs is the key effector protein Caspase 3, whose expression is attenuated by miR-K1, miR-K3 and miR-K450. When the activity of these viral miRNAs was inhibited, both Caspase 3 and apoptosis levels increased significantly.
A final cellular mRNA target for KSHV miRNAs that merits discussion is the cyclin-dependent protein kinase inhibitor p2151. As noted above, infection of cells by herpesviruses induces a DNA damage response that activates not only pro-apoptotic proteins but can also induce cell-cycle arrest via the p53-mediated induction of p21 expression40. The KSHV miRNA miR-K1 binds two targets in the 3’ UTR of the p21 mRNA and this can prevent cell-cycle arrest by factors that activate p5351. Therefore, miR-K1 may play an important role in KSHV-mediated oncogenesis in vivo.
As noted above, HCMV strongly inhibits the expression of MICB using both protein and miRNA-mediated mechanisms43. Other cellular factors targeted by HCMV miRNAs include ERAP1, an aminopeptidase that is important for appropriate presentation of antigenic viral peptides to CTLs52. As a result, the relevant HCMV miRNA, termed miR-US4-1, was able to reduce the susceptibility of HCMV-infected cells to CTL-mediated killing. Finally, the HCMV miRNA miR-UL148D has been shown to target mRNAs encoding the important chemokine RANTES, which attracts immune cells to sites of inflammation and tissue damage53. Blocking RANTES expression obviously could prevent the recruitment of immune effector cells to infected cells in vivo.
Viral subversion of cellular microRNA function
Cellular miRNAs are highly conserved during evolution—for example, the let-7 miRNA family is conserved from nematodes to humans54—and many mRNA targets for cellular miRNAs are also conserved3. In this way, cells are able to use miRNAs to regulate logically coherent sets of mRNA species despite the fact that miRNA seed targets are only ~7 nt in length and hence predicted to occur by chance every 47=16,384 nt. In contrast, viral miRNAs presumably promote viral replication at the expense of the host and viral miRNA targets are therefore, if anything, subject to negative selection. As a result, one might have predicted that viral miRNA would have evolved to be perfectly complementary to perhaps only a single deleterious cellular mRNA that they would then strongly inhibit by endonucleolytic cleavage. In fact, however, viral miRNAs do not generally show perfect complementarity to any mRNAs, the main exception being the antisense viral mRNAs mentioned previously. One can therefore hypothesize that viral miRNAs have likely evolved seed sequences that target non-conserved 3’UTR sequences present on a small number of functionally relevant cellular mRNAs, as well as a larger number of other cellular mRNAs whose downregulation at least does not significantly inhibit viral replication.
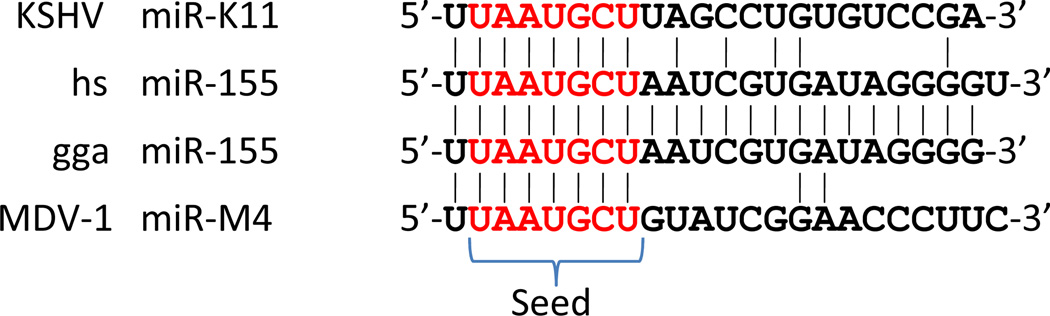
While de novo evolution of a useful viral miRNA, especially in the face of possible host cell adversarial evolution, is therefore challenging, viruses do have the option of simply expressing a miRNA that mimics a cellular miRNA, if that cellular miRNA has an activity that is potentially helpful to the virus. Analysis of the seed sequences of known viral and cellular miRNAs has in fact revealed that the former have a tendency to share the seed sequences of cellular miRNAs that is substantially higher than predicted by chance alone25. While shared seed sequences obviously predict a shared miRNA targetome, this has only been demonstrated for a small number of viral miRNAs55. The most intensively investigated example of viral mimics of a cellular miRNA arises in the case of cellular miR-155, a miRNA that is transiently induced upon activation of both lymphoid and myeloid cells and that has been shown to be important during antigenic stimulation of T- and B-lymphocytes56,57. Moreover, there is extensive evidence linking the constitutive expression of miR-155 to oncogenic transformation of, especially, lymphoid cells58–60. The first viral miR-155 mimic to be described was KSHV miR-K11, which was shown by two groups to both share the full seed sequence of miR-155 (Fig. 2) and to also target a very similar set of cellular mRNAs23,61,62. Indeed, miR-K11 can even substitute for miR-155 in promoting hematopoiesis in vivo63! More recently, a second miR-155 analog, miR-M4, was identified in the avian α-herpesvirus Marek’s Disease Virus Type 1 (MDV1)64. MDV1 causes T-cell lymphomas in chickens and miR-M4 is highly expressed in these tumors. Strikingly, an MDV1 mutant lacking miR-M4 was found to replicate normally in culture but failed to induce any tumors in vivo, while a “revertant” MDV1 mutant in which cellular miR-155 was substituted in place of miR-M4 regained oncogenic potential. This striking result therefore suggests that the miR-155 analog encoded by KSHV is also likely to play a critical role in the transformation of infected human B cells in vivo.
Figure 2. Sequence alignment of miR-155 with viral mRNA mimics.
Alignment of human (hs) and chicken (gga) miR-155 with the sequences of the viral miR-155 mimics KSHV miR-K11 and MDV-1 miR-M4.
While a virus can certainly evolve a viral miRNA that mimics the function of a cellular miRNA, it is also possible for a virus to simply induce the expression of a cellular miRNA in infected cells. While viral infection has been shown to affect cellular mRNA expression in a number of experimental systems, and this has been proposed to potentially facilitate viral replication65, this is perhaps most clearly demonstrated in the case of EBV. EBV strongly induces several cellular miRNAs after infection of primary B cells in culture, and this infection, as noted above, leads to the outgrowth of transformed LCLs66,67. One cellular miRNA that is particularly strongly induced is miR-155, which increases by ~200-fold within days of EBV infection66–68. This is probably due to activation of cellular NF-κB by the EBV LMP1 oncoprotein, which in turn results in increased pri-miR-155 transcription 69. EBV does not encode a miR-155 analog, so this finding suggested the possibility that EBV had evolved an alternative mechanism to inhibit the cellular mRNAs targeted by miR-155 and promote B-cell growth and transformation. Indeed, inhibition of miR-155 function in both EBV- infected LCLs as well as EBV-positive diffuse large B-cell lymphoma cells was found to induce cell cycle arrest and strongly promote apoptosis70. These data therefore reveal that miR-155 plays a critical role in the transformation of lymphoid cells by several human and non-human oncogenic herpesviruses and suggest that manipulation of the pattern of miRNA expression by viruses may play a critical role in viral replication and pathogenesis in several different disease contexts.
Cellular microRNAs as regulators of viral replication
As noted above in the case of EBV, viruses can certainly use cellular miRNAs to their advantage and indeed, in the case of Hepatitis C virus (HCV), it is known that the liver-specific cellular miRNA miR-122 is essential for productive virus replication71. Interestingly, the ability of miR-122 to enhance HCV replication results from a non-conventional activity of miR-122, which binds two sites near the HCV genomic RNA 5’ end and appears to prevent recognition of this 5’ end, which bears a terminal triphosphate, by cellular innate immune factors72. One can also envision that cellular miRNAs could act to significantly inhibit viral replication. Cellular miRNAs are expressed in a highly tissue specific manner and the pattern of miRNA expression can therefore vary dramatically across not only different tissue types but also, for example, depending on whether a cell is growth arrested or actively dividing3. Each cell expresses dozens of miRNAs, each of which has the potential to bind to and inhibit viral mRNA species. The question of how viruses are able to replicate in the face of this potential innate cellular restriction mechanism is therefore of considerable interest.
One possible explanation is that viruses could simply block cellular miRNA function, either selectively or globally. Two examples are now known of viruses that use RNA “decoys” to bind and destabilize a specific cellular miRNA73,74. Interestingly the same miRNA, miR-27, is inhibited by both the simian Herpesvirus saimiri and MCMV. In the latter case, restoration of miR-27 expression was found to inhibit MCMV replication74, although this seemed to be due to targeting of cellular, not viral, mRNA species.
Two virus families have also been found to globally block miRNA expression or function. The best documented case is the global degradation of miRNAs induced by members of the poxvirus family, which encode a protein that induces 3’ polyadenylation of miRNAs, which then induces degradation by the host cell26. In the case of adenovirus, it has been demonstrated that the viral non-coding VA1 RNA competitively inhibits both Exp5-mediated pre-miRNA export and Dicer-mediated pre-miRNA processing, thus strongly inhibiting miRNA biogenesis75,76.
Despite these interesting examples of virus-mediated inhibition of cellular miRNA function, it is clear that the majority of virus-infected cells retain the ability to use miRNAs to regulate mRNA function. Indeed, several groups have used this fact to control virus tropism by engineering perfect target sites for particular tissue-specific cellular miRNAs into the viral genome, thus preventing the virus from being able to replicate effectively in that tissue77–79. This approach shows considerable promise for the development of attenuated virus vaccines and in the design of oncolytic viruses that can grow in, and kill, cancer cells but spare adjoining normal cells. We are then left with the question of how viruses normally evade inhibition by cellular miRNAs, given that a cursory bioinformatic analysis reveals that viruses often contain large numbers of potential seed-target sites for cellular miRNAs, including miRNAs expressed in their normal target tissues80. One hypothesis that can explain why this is not, apparently, a general problem is that viral RNA transcripts are highly structured and that potential miRNA target sites are therefore occluded. Indeed, several RNA viruses, including HIV-1, appear to be highly structured, while others, such as poliovirus, appear to be fairly unstructured81,82. As poliovirus RNAs are uncapped, and the lack of a cap structure has been reported to prevent translational inhibition by miRNAs83,84, it is interesting to speculate that there might be a correlation between a high level of viral RNA secondary structure and predicted susceptibility to inhibition by RISC. Regardless, it will certainly be interesting to determine whether cellular miRNAs are indeed able to bind viral mRNAs effectively and to what degree this binding affects viral replication either positively, as seen for HCV71, or negatively, as one would predict from the repressive activity that is normally characteristic of miRNAs80,85. Clearly, our understanding of how viruses interact with the cellular miRNA machinery remains at an early stage, and research in this area may still present us with a number of surprises.
ACKNOWLEDGMENTS
Research in my laboratory is supported by NIH grants R21-AI088327, R01-AI067968 and R01-DA030086.
Footnotes
COMPETING INTERESTS STATEMENT
The author declares that he has no competing financial interests.
REFERENCES
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 7.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of premicroRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 9.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 12.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 20.Skalsky RL, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley KJ, et al. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haecker I, et al. Ago HITS-CLIP expands understanding of Kaposi's sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012;8:e1002884. doi: 10.1371/journal.ppat.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottwein E, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. This is the first report of viral miRNAs
- 25.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Backes S, et al. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe. 2012;12:200–210. doi: 10.1016/j.chom.2012.05.019. This paper describes a unique viral mechanism that globally degrades cellular miRNAs.
- 27.Cai X, Li G, Laimins LA, Cullen BR. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J Virol. 2006;80:10890–10893. doi: 10.1128/JVI.01175-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 29.Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio. 2010;1:e00204–e00210. doi: 10.1128/mBio.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai X, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu JY, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feederle R, et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7:e1001294. doi: 10.1371/journal.ppat.1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seto E, et al. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. These two papers33,34 provide the first demonstration that viral miRNAs play an important role in virusmediated tranformation of human cells.
- 35.Cai X, et al. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci U S A. 2011;108:11229–11234. doi: 10.1073/pnas.1105799108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. 2012;86:226–235. doi: 10.1128/JVI.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dölken L, et al. Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands. PLoS Pathog. 2010;6:e1001150. doi: 10.1371/journal.ppat.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choy EY, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikitin PA, Luftig MA. At a crossroads: human DNA tumor viruses and the host DNA damage response. Future virology. 2011;6:813–830. doi: 10.2217/fvl.11.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. This paper reveals that three different herpesviruses, KSHV, EBV and HCMV, all use distinct miRNAs to target the same immune factor.
- 43.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 45.Umbach JL, et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barth S, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. 2010;84:12139–12151. doi: 10.1128/JVI.00884-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abend JR, et al. Kaposi's Sarcoma-Associated Herpesvirus MicroRNAs Target IRAK1 and MYD88, Two Components of the Toll-Like Receptor/Interleukin-1R Signaling Cascade, To Reduce Inflammatory-Cytokine Expression. J Virol. 2012;86:11663–11674. doi: 10.1128/JVI.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei X, et al. A Kaposi's Sarcoma-Associated Herpesvirus MicroRNA and Its Variants Target the Transforming Growth Factor beta Pathway To Promote Cell Survival. J Virol. 2012;86:11698–11711. doi: 10.1128/JVI.06855-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suffert G, et al. Kaposi's sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis. PLoS Pathog. 2011;7:e1002405. doi: 10.1371/journal.ppat.1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gottwein E, Cullen BR. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol. 2010;84:5229–5237. doi: 10.1128/JVI.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S, et al. Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol. 2011;12:984–991. doi: 10.1038/ni.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, et al. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012;8:e1002577. doi: 10.1371/journal.ppat.1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 55.Kincaid RP, Burke JM, Sullivan CS. An RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 57.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci U S A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 61.Gottwein E, et al. A viral microRNA functions as an ortholog of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Skalsky RL, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. These two papers61, 62 reported the first example of a viral miRNA mimic of a cellular miRNA.
- 63.Boss IW, et al. A KSHV encoded ortholog of miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2R{gamma}null mice. J Virol. 2011 doi: 10.1128/JVI.05558-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhao Y, et al. Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas. PLoS Pathog. 2011;7:e1001305. doi: 10.1371/journal.ppat.1001305. This is the first report documenting a critical role for a viral miRNA in cell transformation in vivo.
- 65.Ho BC, et al. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe. 2011;9:58–69. doi: 10.1016/j.chom.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Yin Q, et al. MicroRNA-155 is an Epstein-Barr Virus induced gene that modulates Epstein Barr virus regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Forte E, et al. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growth promoting in EBV-infected B cells. J Virol. 2012;86:6889–6898. doi: 10.1128/JVI.07056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu F, et al. Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence. J Virol. 2008;82:10436–10443. doi: 10.1128/JVI.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gatto G, et al. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. This is the first report showing that a cellular miRNA is critical for virus replication.
- 72.Machlin ES, Sarnow P, Sagan SM. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci U S A. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cazalla D, Yario T, Steitz JA. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Buck AH, et al. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andersson MG, et al. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–1283. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 78.Ylösmäki E, et al. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J Virol. 2008;82:11009–11015. doi: 10.1128/JVI.01608-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 81.Davis M, Sagan SM, Pezacki JP, Evans DJ, Simmonds P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian RNA viruses. J Virol. 2008;82:11824–11836. doi: 10.1128/JVI.01078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the capbinding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 84.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lecellier CH, et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 86.Jurak I, et al. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J Virol. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang S, Patel A, Krause PR. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J Virol. 2009;83:1433–1442. doi: 10.1128/JVI.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol. 2009;83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuddenham L, Jung JS, Chane-Woon-Ming B, Dolken L, Pfeffer S. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol. 2012;86:1638–1649. doi: 10.1128/JVI.05911-11. [DOI] [PMC free article] [PubMed] [Google Scholar]