Abstract
NPC2 is a small lysosomal glycoprotein that binds cholesterol with submicromolar affinity. Deficiency in NPC2 is the cause of Niemann Pick type C2 disease, a fatal neurovisceral disorder characterized by accumulation of cholesterol in lysosomes. Here we report the crystal structure of bovine NPC2 bound to cholesterol-3-O-sulfate, an analog that binds with greater apparent affinity than cholesterol. Structures of both apo- and sterol-bound NPC2 were observed within the same crystal lattice, with an asymmetric unit containing one molecule of apoNPC2 and two molecules of sterol-bound NPC2. As predicted from a previously determined structure of apoNPC2, the sterol binds in a deep hydrophobic pocket sandwiched between the two β sheets of NPC2, with only the sulfate substituent of the ligand exposed to solvent. In the two available structures of apoNPC2, the incipient ligand-binding pocket, which ranges from a loosely packed hydrophobic core to a small tunnel, is too small to accommodate cholesterol. In the presence of sterol, the pocket expands, facilitated by a slight separation of the β strands and substantial reorientation of some side chains, resulting in a perfect molding of the pocket around the hydrocarbon portion of cholesterol. A notable feature is the repositioning of two aromatic residues at the tunnel entrance that are essential for NPC2 function. The NPC2 structures provide evidence of a malleable binding site, consistent with the previously documented broad range of sterol ligand specificity.
Intracellular cholesterol trafficking is central to the distribution of dietary cholesterol for utilization in membrane synthesis, synthesis of sterol hormones and derivatives, creation of lipid storage droplets, and regulation of de novo cholesterol biosynthesis. Dietary cholesterol, in the form of cholesterol esters in low-density lipoprotein (LDL) particles, enters cells via LDL receptor-mediated endocytosis. LDL particles are transported to lysosomes where acid lipase converts cholesterol esters to free cholesterol. This cholesterol then leaves lysosomes and is subsequently targeted to other intracellular and extracellular locations to meet specific cellular needs.
Insight into the proteins that mediate egress of cholesterol from lysosomes has come from identification of genes involved in Niemann-Pick type C disease, a fatal recessive hereditary disorder characterized by accumulation of cholesterol in lysosomes. Two genes are associated with the disease: NPC1 and NPC2. Similar phenotypes have been observed in mice deficient in either one or both genes, leading to the hypothesis that NPC1 and NPC2 function together in a single pathway to transport cholesterol out of lysosomes. NPC1 is a large transmembrane protein that contains an intramembranous sterol-sensing domain (SSD) and belongs to the resistance-nodulation-division (RND) permease superfamily. NPC2 is a 130-amino acid lysosomal glycoprotein and belongs to the MD-2-related lipid recognition (ML) domain family of lipid binding proteins (1).
NPC2, which is found in lysosomes (2) and secretory fluids such as milk (3) and epididymal fluid (4), binds sterols with submicromolar affinity (5–7). The crystal structure of apoNPC2 (7) indicated a β-sandwich fold with topology identical to that of the three other structurally characterized members of the ML domain family, GM2 activator protein (8) and the dust mite allergens Der f 2 and Der p 2 (9, 10). However, unlike the structures of these proteins in the absence of ligands, apoNPC2 lacks a large preformed internal hydrophobic cavity for lipid binding. In its place is a loosely packed hydrophobic core that was proposed to be an incipient sterol-binding pocket that expands to accommodate cholesterol. Further support for the proposed binding site was provided by mutagenesis studies that showed three conserved residues lining the pocket to be essential for cholesterol binding (6) and by the identification of a mutated residue at the base of the pocket in a patient with late-onset NPC2 disease (11).
We have determined the crystal structure of bovine NPC2 bound to cholesterol-3-O-sulfate. The ligand-binding pocket is positioned as previously postulated and is formed by subtle repositioning of β strands and side chains within the hydrophobic interior, creating a tunnel that appears to be molded to fit around the sterol. The malleable nature of the pocket suggested by this tight fit is consistent with the observed tolerance of structural and chemical variations in both the sterol ligand and in residues that line the binding cavity. NPC2 exhibits a unique composite of specific features observed in different lipid binding proteins from diverse families.
EXPERIMENTAL PROCEDURES
Sterol Binding Analysis
Delipidated Endo-H-deglycosylated bovine NPC2 was prepared as previously described (7, 12). For each analysis, 68 μM NPC2 in 12.5 mM ammonium acetate, pH 4.5 was incubated with 1.5 mol eq of the indicated sterol (addition of 4.8% vol of 2.5 mM sterol dissolved in dimethyl sulfoxide) for 30 min at 20 °C. For competition assays, NPC2 was incubated with cholesterol for 30 min, cholesterol sulfate was added and incubation was continued an additional 30 min. Sterol binding was assessed by shifts in retention time of NPC2 during cation exchange chromatography on a Mono S column (GE Healthcare) as previously described (12) using 3.4 nmol (50 μg) NPC2 per injection.
Protein Complex Preparation and Crystallization
Deglycosylated NPC2 in 10 mM ammonium acetate, pH 5.0 was concentrated to 5 mg/mL and 1.2 mol eq of cholesterol sulfate (1.2 mg/mL in dimethyl sulfoxide) was added. After 10 min at 20 °C, the mixture was filtered (0.2 μm) and applied to a Mono S 5/50 GL column (GE Healthcare) and the complex was eluted using a linear 60-column volume gradient of 0.01 M to 0.5 M ammonium acetate, pH 5.0. The complex was concentrated to 10–15 mg/mL and buffer exchanged into 10 mM ammonium acetate, pH 5.0. Crystals were grown using hanging drop vapor diffusion with equal volumes of protein complex and reservoir (2.0 M ammonium sulfate, 0.1 M sodium acetate, pH 5.0, 1 mM cetyltrimethylammonium bromide). Rod-shaped crystals with a width of ~0.1 mm appeared in 1 week.
Data Collection and Structure Determination
A crystal was cryoprotected in 1.7 M ammonium sulfate, 85 mM sodium acetate, pH 5.0, 15% glycerol and frozen in a 100 K nitrogen stream. A complete dataset at 1.81 Å resolution was collected from a single crystal at beam line X29C at the National Synchrotron Light Source at Brookhaven National Laboratory. Data were processed using the HKL Suite (13). The structure was solved by molecular replacement using PHASER (14) with the structure of apo NPC2 (PDB code 1NEP) as a search model. Three protein molecules (A, B, and C) were found in the asymmetric unit. Subsequent rigid-body refinement using CNS (15) gave an R factor of 43.6% using 20–2.5 Å data. Strong electron density was observed in 2Fo−Fc and Fo−Fc maps at the predicted sterol binding sites in molecules B and C, but not in molecule A. Two cholesterol sulfate molecules were added to the model and it was subjected to multiple rounds of manual rebuilding using XtalView (16) and refinement using CNS with 5% of the data reserved for calculation of Rfree. The final model includes residues 1–130 of NPC2 molecules A, B, and C with N-acetylglucosamine modification at Asn39, 2 cholesterol sulfate, 6 acetate, 3 sulfate, 2 glycerol, and 371 water molecules. The final model has good stereochemistry, with 89.9% of the residues in the most favored regions of the Ramachandran plot and none in the disallowed regions. Data collection and refinement statistics are summarized in Table 1. Figure 3A was generated using MolScript (17); all other structural images were generated using PyMOL (http//pymolsourceforge.net).
TABLE 1.
Crystallographic data and refinement
| Parameter | Value |
|---|---|
| Data Collection | |
| Space group | C2 |
| Unit cell a, b, c (Å) | 122.38, 62.16, 72.45 |
| Unit cell α, β, γ (°) | 90.00, 98.62, 90.00 |
| Wavelength (Å) | 1.1 |
| Resolution limits (Å) | 50 - 1.81 (1.87 - 1.81)* |
| Rsym† | 0.107 (0.350) |
| Average I/σ(I) | 17.9 (4.3) |
| Completeness (%) | 98.2 (93.4) |
| Redundancy | 3.8 |
|
| |
| Refinement | |
| Resolution range (Å) | 20 - 1.81 |
| No. reflections | 48,241 |
| Rcryst‡/Rfree§ | 0.195/0.214 |
| No. protein atoms | 3081 |
| No. ligand/non-H2O solvent atoms | 157 |
| No. water atoms | 371 |
| Rmsd bond lengths (Å) | 0.006 |
| Rmsd bond angles (°) | 1.35 |
| Ave. B value, protein (Å2) | 22.9 |
| Ave. B value, heteroatoms (Å2) | 33.8 |
Values in parentheses are for reflections in the highest resolution shell.
Rsym = ΣhklΣi|I(hkl)i − <I(hkl)>|/ΣhklΣi<I(hkl)i> over i observations.
Rcryst = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|.
Value of Rcryst for 5% of randomly selected reflections excluded from refinement.
FIGURE 3. Backbone displacements in apo- and sterol-bound NPC2.
(A) NPC2 is shown in ribbon representation (blue) with cholesterol sulfate (carbon in gold, oxygen in red, sulfur in yellow) bound in the protein interior between strands βD and βE. The three disulfide bonds in NPC2 (yellow) and the N-acetyl glucosamine remnant of glycosylation at N39 (carbon in black, oxygen in red, nitrogen in blue) are shown in stick and ball-and-stick representation, respectively. (B) Cα traces of superimposed models of sterol-bound NPC2 (MolB, blue) and two apoNPC2 structures (MolA, green and PDB 1NEP, gold) are shown with regions of the largest displacements (rmsd >0.4 Å) highlighted in red in the model of sterol-bound NPC2.
RESULTS AND DISCUSSION
Cholesterol Sulfate Binding to NPC2
Numerous attempts to crystallize bovine NPC2 bound to cholesterol were unsuccessful. NPC2 formed a stable 1:1 complex with cholesterol that showed no detectable loss of ligand over several weeks (data not shown). Crystals were readily obtained under a variety of conditions, but all crystals had cubic morphology similar to apoNPC2 crystals (7), and upon structure determination, all were found to lack cholesterol. Thus it appeared that this crystal lattice selectively favored apoNPC2.
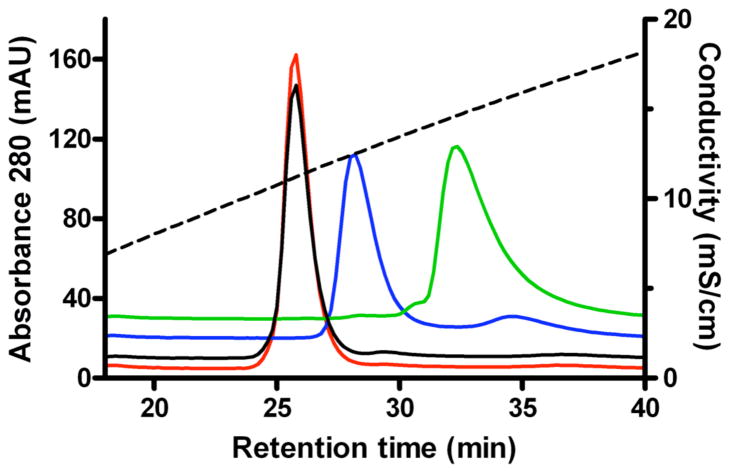
Human and bovine NPC2 bind a variety of cholesterol derivatives, including cholesterol-3-O-sulfate (12, Fig. 1). Cholesterol sulfate binds tightly, with an apparent affinity greater than that of cholesterol. NPC2 selectively binds cholesterol sulfate from an equimolar mixture of the two sterols (data not shown) and cholesterol sulfate can completely displace previously bound cholesterol from NPC2 (Fig. 1).
FIGURE 1. Chromatographic analysis of ligand binding to NPC2.
Bovine NPC2 was incubated in the absence (green) or presence of cholesterol (blue), cholesterol-3-O-sulfate (red), or cholesterol followed by cholesterol-3-O-sulfate (black) and subjected to cation exchange chromatography as described in Materials and Methods. Decreased retention time is correlated with ligand binding (12).
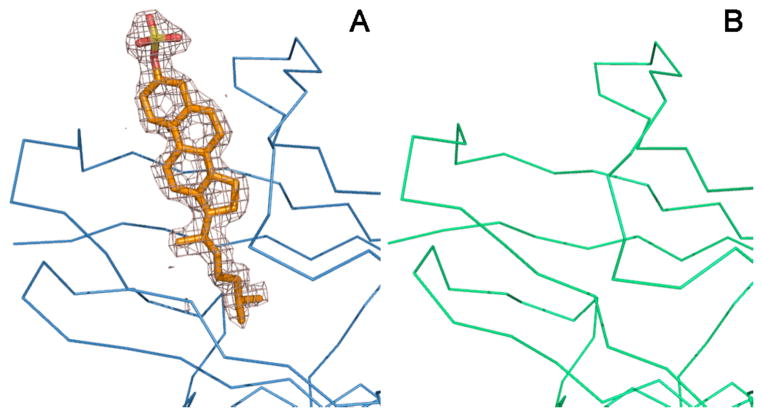
Crystallization of a cholesterol sulfate-NPC2 complex produced primarily cubic crystals that, as before, were ascertained to lack sterol. Under similar conditions, crystals of a different morphology were occasionally obtained. A single dataset to 1.81 Å resolution was collected from a rare rod-like crystal that was subsequently found to represent a new crystal form. Using the structure of apoNPC2 as a search model, the structure was solved by molecular replacement methods. Crystallographic data and refinement statistics are summarized in Table 1. The asymmetric unit contained three molecules of NPC2 (MolA, B, and C). Difference electron density maps revealed strong density compatible with cholesterol sulfate located within the proposed ligand binding sites of two NPC2 protomers (MolB and C) but not in the third (MolA) (Fig. 2). The lack of ligand in MolA can be rationalized by inspection of crystal packing. The protruding sulfate of a bound cholesterol sulfate ligand in Mol A cannot be accommodated within the crystal lattice due to steric collisions with symmetry-related NPC2 protomers.
FIGURE 2. Electron density at the ligand-binding site.
A difference electron density map (Fo − Fc) calculated at 1.8 Å resolution with phases from the final model with ligand omitted and contoured at 2.5σ reveals electron density consistent with cholesterol-3-O-sulfate within the interior of MolB (A) but not in MolA (B). The Cα traces for NPC2 MolB (blue) and MolA (green) and a stick representation of cholesterol sulfate in MolB (carbon in gold, oxygen in red, sulfur in yellow) from the final model are shown superimposed with the density maps.
Apo- and Sterol-Bound States of NPC2
Both apo and sterol-bound structures exhibit the same 7-stranded β-sandwich fold, stabilized by three disulfide bonds connecting residues Cys8-Cys121, Cys23-Cys28 and Cys74-Cys80 (Fig. 3A). An N-acetylglucosmine moiety, the residual of EndoHf deglycosylation, modifies Asn39, the single site of glycosylation in bovine NPC2. Cholesterol sulfate binds as previously postulated in a deep hydrophobic tunnel that penetrates into the protein core from an opening rimmed by βD and the βE-βF loop with only the sulfate substituent at the 3-hydroxyl of the sterol exposed to solvent.
The conformation of apoNPC2 (MolA) is distinct from that of sterol-bound NPC2 and also different than the previously determined structure of apoNPC2 (7). The conformations of the two NPC2 proteins with bound sterol are very similar, with an rmsd of 0.37 Å for all 130 aligned Cα atoms. Henceforth, MolB will be used as a representative of cholesterol sulfate-bound NPC2. Pair-wise alignment of the two apoNPC2 structures and cholesterol sulfate-bound NPC2 in all combinations yielded rmsds of 0.8–1.1 Å for all Cα atoms. Superpositioning of the three structures reveals that substantial backbone displacements (rmsd > 0.4 Å) are localized to a small region near the sterol-binding pocket, corresponding to portions of βB (residues 31–37), βD (residues 61–70), βE (residues 93–102) and adjacent loops (Fig. 3B). Within this region, the two apoNPC2 structures are more “closed” than sterol-bound NPC2 with a closer positioning of the β strands from opposing β sheets. The apoNPC2 structure observed in MolA is intermediate between the fully “closed” previously described apoNPC2 structure (7) and the fully “open” structure exhibited by sterol-bound NPC2. Surface residues near the binding pocket of MolA make several intermolecular contacts with a symmetry related MolB, raising the possibility that the crystal lattice stabilizes the intermediate conformation of MolA.
The Sterol-Binding Pocket
Comparison of surfaces and volumes of the binding pockets in the different apoNPC2 and sterol-bound NPC2 structures indicates that the pocket is fully formed only in the presence of bound ligand (Fig. 4). The incipient binding site in closed apoNPC2 contains only a cluster of small cavities with a total volume of 180 Å3, while the intermediate conformation of apoNPC2 contains an almost continuous cavity with a total volume of 310 Å3, calculated using the CASTp web server (18). The cavity in sterol-bound NPC2 is 720 Å3, just large enough to accommodate the hydrophobic portion of cholesterol, which occupies a total volume of 740 Å3. The cavity is nearly identical in both sterol-bound NPC2 molecules, with similar conformations of cholesterol sulfate (rmsd of 0.13 Å for all sterol atoms) and tunnels that are molded tightly around the hydrophobic portion of the ligand. The creation of a molded-to-fit hydrophobic tunnel by expansion of a minimal incipient cavity has been previously observed in a glycosphingolipid transfer protein (GLTP), which binds the lipid chains of lactosylceramide in a tunnel sandwiched between two α-helical layers (19). However, this mode of interaction is relatively rare among lipid binding proteins which typically contain large preformed cavities that have volumes substantially greater than the ligands and can often accommodate a variety of lipids and different binding orientations (8–10, 20–25).
FIGURE 4. Sterol binding cavities of apo- and sterol-bound NPC2.

The ligand-binding sites in (A) apoNPC2 (PDB 1NEP), (B) apoNPC2 (MolA) and (C) cholesterol-3-O-sulfate-bound NPC2 (MolB) are shown in identical orientations in surface representation with a cross-sectioned slab removed to allow viewing of the internal ligand-binding tunnel.
The tunnel in NPC2 is almost exclusively hydrophobic, formed primarily by the side chains of residues V20, L30, Y36, V38, V55, G57, V59, V64, F66, L94, P95, V96, Y100, P101, I103, V105, V107, W109, W122, I124, I126, and V128. Approximately half of these residues are positioned differently in the closed and open tunnels, either as a result of backbone displacements alone or in combination with side chain reorientations. The other residues occupy similar positions in both structures. Interestingly, with only two exceptions (V20 and F66), these two classes of static and displaced residues are segregated onto different faces of the β sandwich (Fig. 5A). Thus, the tunnel appears to be constructed from one fixed and one mobile wall.
FIGURE 5. Ligand-binding site residues.

A Cα trace of NPC2 (blue) bound to cholesterol-3-O-sulfate (depicted as transparent spheres with carbon in white, oxygen in red, sulfur in yellow) is shown with side chains of residues that form the sterol-binding tunnel depicted in stick representation. (A) Residues that have similar positions in apo- and sterol-bound NPC2 are shown in red. Residues that occupy different positions in the closed and open states are shown in green. (B) Residues that form the binding site (blue) are colored to indicate features that reflect their malleability. Residues that have been identified in mutagenesis studies (6) to be critical to function (F66, V96, Y100) are colored red and those tolerant to non-conservative substitutions (V64, W122) are colored yellow. A residue identified in a patient with late onset disease (V20) (11) is colored orange. Residues that are not strictly conserved among mammalian NPC2 proteins (V38, V59, V64, I103, V105, I127, V129) are colored green.
Cholesterol sulfate is positioned in the tunnel with the iso-octyl chain deeply buried at the base of the tunnel and curved slightly toward the face of the tetracyclic rings from which the two methyl groups protrude. The sulfate moiety sits at the mouth of the tunnel, fully accessible to solvent. It is presumed that cholesterol binds in an analogous manner with the 3β-hydroxyl group exposed. The oxygen of cholesterol sulfate that corresponds to the hydroxyl of cholesterol forms no hydrogen bonds with the protein. Notably, there are no interactions with the side chain of K97, which was previously postulated to interact with the hydroxyl group of bound sterol (6) by analogy with hydrogen bonds between the hydroxyl groups and basic side chains in other sterol-binding proteins (22, 26). In NPC2, the lysine side chain is oriented away from the tunnel entrance with the side chain of Y100 blocking its access to the ligand (Fig. 6). This is consistent with the observation that substitution of alanine for K97 has little effect on sterol binding (6).
FIGURE 6. Entrance to the sterol-binding tunnel.

Apo- (A) and sterol-bound NPC2 (B) proteins are shown as transparent surfaces (carbon in white, oxygen in red, nitrogen in blue) with side chains of residues that form the entrance to the ligand-binding tunnel depicted in stick representation on a Cα frame. Cholesterol sulfate is shown in stick representation (carbon in gold, oxygen in red, sulfur in yellow).
Likewise, the sulfate makes no contacts with the protein. Thus, unlike the case of the retinoic acid-related orphan receptor α (RORα) ligand binding domain, for which increased affinity for cholesterol sulfate relative to cholesterol was attributed to three additional hydrogen bonds between the protein and the sulfate (27), the crystal structure of NPC2 provides no structural rationale for the apparent higher affinity for cholesterol sulfate. It therefore seems likely that the higher apparent affinity of cholesterol sulfate relative to cholesterol reflects its greater solubility, providing a higher concentration of non-micellar ligand available for binding to the protein (12, 28).
Gating of the Sterol-Binding Pocket
In the absence of ligand, the mouth of the hydrophobic tunnel is essentially closed, with only a small pore leading into the loosely packed hydrophobic interior (Fig. 6). Upon sterol binding, the aperture opens to a diameter of ~8 Å. Six hydrophobic amino acids, V59, V64, F66, Y100, P101, and I103 form an apolar rim at the tunnel entrance in both the closed and open states. The opening of the mouth is achieved primarily by the repositioning of residues F66, Y100, P101, and I103, a consequence of the wider spacing of βD and the βE-βF loop and the flipping of side chains of F66 and I103 from inward to outward orientations.
It seems unlikely that sterol binding occurs via iso-octyl tail-first insertion into the incipient cavity, with the ligand drilling its way into the hydrophobic interior of the protein. Furthermore, given the low solubility of cholesterol (critical micelle concentration ~25 nM, 28), it can be assumed that ligands do not bind directly from solution but rather are transferred between NPC2 and membranes or other proteins. The orientation of sterols in bilayers, with the 3β-hydroxyl at the membrane surface, is opposite that which would be most amenable to tail-first transfer into the binding tunnel of an NPC2 molecule docked at the membrane surface. However, alternative lipid aggregates such as hexagonal phases in which polar head groups are clustered at the center with hydrophobic regions extending outward would present lipids for binding in a tail-first orientation. Such an orientation, if accessible to cholesterol, might facilitate transfer. Indeed, formation of lipid hexagonal phases is promoted by low pH, as exists in lysosomes where NPC2 resides (29).
It is most likely that ligand binding occurs via lengthwise insertion of the sterol into a cleft formed by separation of βD and βE, the edge strands of the two β sheets, followed by closure of the cleft that becomes molded around the ligand to create a perfectly fitting tunnel. An open cleft is presumably among the dynamic conformational repertoire sampled by NPC2 in solution. However, it is possible that cleft opening might be facilitated by association of this surface of NPC2 with a hydrophobic environment such as could be provided by interaction with another protein, membrane or lipid aggregate. Indeed, it has been shown that NPC2 interacts with membranes and that negatively charged phospholipid vesicles greatly enhance the rate of cholesterol transfer from NPC2 in vitro, leading to the conclusion that cholesterol transfer involves NPC2-membrane collisions (30).
One of the most noticeable differences between NPC2 and many other lipid transfer proteins is the absence of a “lid” that in different conformational states might either close the ligand-binding cavity or promote interactions with membranes. Such lids, which vary greatly in size and structure, are very common among lipid transfer proteins. In the structurally related GM2 activator protein, the lid is formed by a large apolar loop that in its extended form is proposed to dip into the hydrophobic region of membranes to facilitate lipid extraction (31, 32). A similar “bulldozer” mechanism for lipid extraction has been proposed for the helical lid of yeast phosphatidylinositol-transfer protein Sec14 (33). In other proteins, lid regions are proposed to promote interactions with membrane surfaces. The oxysterol-binding protein-related protein Osh4 contains an N-terminal lid rich in lysine residues that closes the cavity in the ligand-bound state and is hypothesized to spring open upon interaction with phospholipid phosphate groups at the membrane surface to facilitate cargo transfer. The cavity in fatty acid-binding proteins (FABPs) is capped by a helix-turn-helix motif that has been shown to be critical for membrane-protein interactions (34).
The absence of a preformed binding cavity in NPC2 perhaps correlates with the absence of a lid-like structure formed by large loops or terminal extensions. However, the lack of a lid also precludes the role of such an appendage in facilitating interactions with membrane surfaces or interiors. The fairly even distribution of charged and hydrophobic residues on the surface of NPC2 provides no obvious clue to alternative mechanisms for interaction of the protein with membranes.
Malleability of the Sterol-Binding Pocket
The molded-to-fit nature of the sterol-binding pocket implies malleability. This is supported by the documented tolerance of sterol-binding function to variations in both the protein and the ligand. Within the protein, evolutionary divergence and both engineered and naturally occurring mutations have established that some substitutions in binding-site residues can be accommodated without loss of function. Only 15 of the 22 residues that form the binding tunnel are invariant among 16 sequenced mammalian NPC2 proteins. Variation of hydrophobic residues (that appears to be uncompensated) occurs at 7 positions (V38, V59, V64, I103, V105, I126, V128) (Fig. 5B). Additional data are provided by mutagenesis studies that have probed the importance of several highly conserved binding-site residues. While substitution of alanine for F66 or Y100, the two aromatic residues at the tunnel entrance, or substitution of phenylalanine for V96, midway within the tunnel, severely compromised cholesterol binding and NPC2 function in cultured cells, substitution of phenylalanine for V64 or alanine for W122 had little effect (6). Additionally, a mutation encoding replacement of V20 with methionine has been identified in a patient with late-onset disease (11), indicating a partial loss of function when this longer chain residue is substituted at the base of the tunnel. Given the tight fit of the tunnel to ligand, remodeling of the tunnel must be invoked to explain retention of ligand binding in the presence of many of these substitutions. Indeed, direct evidence of malleability is provided by the occurrence of different rotameric states of V38 in the two cholesterol sulfate NPC2 complexes, resulting in minor alterations in the contours of the binding tunnels and corresponding small differences in the curvature of the iso-octyl chains of the bound ligands.
Recent investigations of ligand binding to NPC2 have revealed a fairly broad specificity for sterols and derivatives (12). Direct evidence of binding was obtained for a variety of animal and plant sterols, indicating tolerance for the degree and placement of double bonds and/or alkyl substituents on the tetracyclic rings and/or hydrocarbon tails (e.g. 5α-cholestan-3-β-ol, dehydroergosterol, desmosterol, lathosterol, β-sitosterol, and stigmasterol). Binding of oxysterols (e.g. 24-hydroxycholesterol) was also observed, indicating some tolerance for polar substituents within the hydrophobic tunnel. However, substantially more polar ligands (e.g. bile acids) failed to bind. Substitutions at the 3-hydroxyl had various effects (e.g. cholesteryl sulfate, cholesteryl acetate, and 5α-cholestan-3-one bound, while thiocholesterol, cholesteryl bromide, and longer chain cholesteryl esters did not). The latter cannot be rationalized by the crystal structure, which shows no specific contacts between NPC2 and the 3-hydroxyl. It should be noted that binding of sterols to proteins reflects the relative preference of the sterol for association with the protein compared to association with itself to form micelles (28). Thus it is possible that lack of binding of some ligands might reflect the solubility of the ligands rather than discrimination by the binding site of the protein. NPC2 appears to be relatively specific for sterols, as no binding of fatty acids or glycosphingolipids was observed (12).
The accommodation of substitutions in protein residues and ligands requires substantial remodeling of the static tunnel observed in the crystal structure. This is not difficult to envision if the protein and hydrophobic portion of the ligand are considered in continuum to comprise the hydrophobic core of the protein. Protein interiors display substantial tolerance to substitution of hydrophobic residues, utilizing minor positional adjustments of residues within the core to optimize packing interactions (35). There are no hydrogen bonds formed between the sterol and NPC2 that might constrain specific geometries or chemical composition, thus enabling a substantial degree of malleability in packing arrangements. Clearly, there are limits to the variations that can be accommodated, and the mutagenesis and binding studies performed to date have begun to define the allowable range. We conclude that the physiologically relevant ligands bound by NPC2 are likely to be dictated by the specific subcellular repertoire of sterols available for binding rather than by stringent selectivity of the binding cavity.
Acknowledgments
We thank L. Gu for assistance and advice with structure refinement; the staff at beamline X29C at the National Synchrotron Light Source for support with data collection; D. Mayor at Delaware Valley College for raw milk; and D. Sleat for comments on the manuscript. This work was supported in part by grants from the National Institutes of Health (DK54317 to P.L.) and the Ara Parseghian Medical Research Foundation (A.M.S. and P.L). A.M.S. is an investigator of the Howard Hughes Medical Institute.
The abbreviations used are
- LDL
low density lipoprotein
- ML
MD-2-related lipid recognition
- PDB
Protein Data Bank
- rmsd
root mean square deviation
Footnotes
This work was supported in part by grants from the National Institutes of Health (DK54317 to P.L.) and the Ara Parseghian Medical Research Foundation (A.M.S. and P.L). A.M.S. is an investigator of the Howard Hughes Medical Institute.
The atomic coordinates and structure factors have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Databank = PDB # 2HKA.
References
- 1.Inohara N, Nuñez G. Trends Biochem Sci. 2002;27:219–221. doi: 10.1016/s0968-0004(02)02084-4. [DOI] [PubMed] [Google Scholar]
- 2.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 3.Larsen LB, Ravn P, Boisen A, Berglund L, Petersen TE. Eur J Biochem. 1997;243:437–441. doi: 10.1111/j.1432-1033.1997.0437a.x. [DOI] [PubMed] [Google Scholar]
- 4.Kirchhoff C, Osterhoff C, Young L. Biol Reprod. 1996;54:847–856. doi: 10.1095/biolreprod54.4.847. [DOI] [PubMed] [Google Scholar]
- 5.Okamura N, Kiuchi S, Tamba M, Kashima T, Hiramoto S, Baba T, Dacheux F, Dacheux JL, Sugita Y, Jin YZ. Biochim Biophys Acta. 1999;1438:377–387. doi: 10.1016/s1388-1981(99)00070-0. [DOI] [PubMed] [Google Scholar]
- 6.Ko DC, Binkley J, Sidow A, Scott MP. Proc Natl Acad Sci USA. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedland N, Liou HL, Lobel P, Stock AM. Proc Natl Acad Sci USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright CS, Li SC, Rastinejad F. J Mol Biol. 2000;304:411–422. doi: 10.1006/jmbi.2000.4225. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa S, Hatanaka H, Yuuki T, Iwamoto N, Kojima S, Nishiyama C, Ogura K, Okumura Y, Inagaki F. J Biol Chem. 1998;273:356–360. doi: 10.1074/jbc.273.1.356. [DOI] [PubMed] [Google Scholar]
- 10.Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller GA, Rule GS, Benjamin DC. J Mol Biol. 2002;318:189–197. doi: 10.1016/S0022-2836(02)00027-X. [DOI] [PubMed] [Google Scholar]
- 11.Klünemann HH, Elleder M, Kaminski WE, Snow K, Peyser JM, O’Brien JF, Munoz D, Schmitz G, Klein HE, Pendlebury WW. Ann Neurol. 2002;52:743–749. doi: 10.1002/ana.10366. [DOI] [PubMed] [Google Scholar]
- 12.Liou HL, Dixit SS, Xu S, Tint GS, Stock AM, Lobel P. J Biol Chem. 2006;281:36710–36723. doi: 10.1074/jbc.M608743200. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 14.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 15.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.McRee DE. J Mol Graphics. 1992;10:44–46. [Google Scholar]
- 17.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 18.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malinina L, Malakhova ML, Teplov A, Brown RE, Patel DJ. Nature. 2004;430:1048–1053. doi: 10.1038/nature02856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacchettini JC, Scapin G, Gopaul D, Gordon JI. J Biol Chem. 1992;267:23534–23545. [PubMed] [Google Scholar]
- 21.Cowan SW, Newcomer ME, Jones TA. J Mol Biol. 1993;230:1225–1246. doi: 10.1006/jmbi.1993.1238. [DOI] [PubMed] [Google Scholar]
- 22.Tsujishita Y, Hurley JH. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 23.Choinowski T, Hauser H, Piontek K. Biochemistry. 2000;39:1897–1902. doi: 10.1021/bi992742e. [DOI] [PubMed] [Google Scholar]
- 24.Young AC, Scapin G, Kromminga A, Patel SB, Veerkamp JH, Sacchettini JC. Structure. 1994;2:523–534. doi: 10.1016/s0969-2126(00)00052-6. [DOI] [PubMed] [Google Scholar]
- 25.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. Cell. 1997;89:685–692. doi: 10.1016/s0092-8674(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 27.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 28.Haberland ME, Reynolds JA. Proc Natl Acad Sci USA. 1973;70:2313–2316. doi: 10.1073/pnas.70.8.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Bambeke F, Kerkhofs A, Schanck A, Remacle C, Sonveaux E, Tulkens PM, Mingeot-Leclercq MP. Lipids. 2000;35:213–223. doi: 10.1007/BF02664772. [DOI] [PubMed] [Google Scholar]
- 30.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 31.Wright CS, Zhao Q, Rastinejad F. J Mol Biol. 2003;331:951–964. doi: 10.1016/s0022-2836(03)00794-0. [DOI] [PubMed] [Google Scholar]
- 32.Wendeler M, Hoernschemeyer J, Hoffmann D, Kolter T, Schwarzmann G, Sandhoff K. Eur J Biochem. 2004;271:614–627. doi: 10.1111/j.1432-1033.2003.03964.x. [DOI] [PubMed] [Google Scholar]
- 33.Sha B, Phillips SE, Bankaitis VA, Luo M. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 34.Wu F, Corsico B, Flach CR, Cistola DP, Storch J, Mendelsohn R. Biochemistry. 2001;40:1976–1983. doi: 10.1021/bi002252i. [DOI] [PubMed] [Google Scholar]
- 35.Matthews BW. Adv Protein Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]