Abstract
A key pathological feature of late-onset Alzheimer’s disease (LOAD) is the abnormal extracellular accumulation of the amyloid beta (Aβ) peptide. Thus altered Aβ degradation could be a major contributor to the development of LOAD. Variants in the gene encoding the Aβ-degrading enzyme, angiotensin-1 converting enzyme (ACE) therefore represent plausible candidates for association with LOAD pathology and risk. Following Alzgene meta-analyses of all published case-control studies, the ACE variants rs4291 and rs1800764 showed significant association with LOAD risk. Furthermore ACE haplotypes are associated with both plasma ACE levels and LOAD risk. We tested three ACE variants (rs4291, rs4343 and rs1800764) for association with LOAD in ten Caucasian case-control populations (n=8,212). No association was found using multiple logistic models (all p>0.09). We found no population heterogeneity (all p>0.38) or evidence for association with LOAD risk following meta-analysis of the ten populations for rs4343 (OR=1.00), rs4291 (OR=0.97) or rs1800764 (OR=0.99). Although we found no haplotypic association in our complete dataset (p=0.51), a significant global haplotypic p-value was observed in one population (p=0.007) due to an association of the H3 haplotype (OR=0.72, p=0.02) and a trend towards an association of H4 (OR=1.38, p=0.09) and H7 (OR=2.07, p=0.08) although these did not survive Bonferroni correction. Previously reported associations of ACE variants with LOAD will be diminished following this study. At best, ACE variants have modest effect sizes, which are likely part of a complex interaction between genetic, phenotypic and pharmacological effects that would be undetected in traditional case-control studies.
Keywords: Alzheimer Disease, Late Onset, Angiotensin-1 Converting Enzyme, Haplotype, Heterogeneity, Meta-Analysis
Introduction
Late-onset Alzheimer’s disease (LOAD; MIM 104300) is the most common form of dementia accounting for almost two-thirds of all dementia cases. Its key pathological features include the formation of intracellular neurofibrillary tangles comprised of microtubule-associated tau, abnormal extracellular accumulations of amyloid beta (Aβ) peptide in the form of characteristic senile plaques and in most LOAD cases, deposition of intracerebrovascular Aβ in the form of cerebral amyloid angiopathy [1, 2]. It is increasingly recognised that altered degradation and clearance of Aβ is likely to be of importance in the development and progression of LOAD [3] and that this may be contributed to by both environmental and genetic factors. Cumulative evidence from in vitro, in vivo and ex vivo studies now strongly support the role of ACE (EC 3.4.15.1), a zinc metalloprotease widely expressed in the brain, as an Aβ degrading enzyme (reviewed in [4]). Taken together with the observation that increased ACE levels and activity are observed in LOAD brains (reviewed in [5]) and are associated with increased plasma levels of Aβ [6] and reduced levels of Aβ in CSF [7, 8], these all point to the likely involvement of ACE in Aβ-related pathology in AD. This is further supported by evidence that variation in the gene encoding ACE (ACE; OMIM 106180), may play a role in LOAD pathology and modify LOAD risk. For example, the insertion/deletion (indel) of a 287bp Alu repeat in intron 16 (rs1799752 Alu I/D) of ACE, and perhaps the most widely studied for LOAD association, is predicted to explain 29–47% of the variation in plasma ACE levels [9–11]. In their meta-analysis of 39 case-control series, comprising 6,037 LOAD cases and 12,099 controls, Lehmann et al reported that homozygotes for the Alu deletion were at reduced risk of LOAD (p=0.0004), while heterozygotes were at increased risk [12], thus supporting a genetic association of ACE with LOAD. The fact that the indel does not account for all of the observed variation in ACE levels suggests that other functional ACE variants may be present and in turn associated with ACE levels and/or LOAD risk.
The APOE ε4 allele (107741) remains the most widely studied and accepted susceptibility gene for LOAD since its first report as a candidate gene almost 20 years ago [13, 14]. The remaining genetic component of AD risk may involve many genes, each with individually small-to-moderate effect sizes that interact to produce greater effects on disease susceptibility and/or disease modification. However, detection and confirmation of the involvement of genes with these effect sizes requires very large sample sizes. By example, over the last two decades 664 different genes and almost 3,000 variants have been investigated as susceptibility factors for LOAD risk [15] and until recently, the majority of these studies have been relatively underpowered, often resulting in inconclusive or inconsistent results for the majority of putative candidate genes. AlzGene (www.Alzgene.org) [15] was designed and established to resolve this problem to some extent by regularly performing meta-analyses of published data as it emerged to continually compile a list of “Top LOAD genes” that show the strongest associations in LOAD. A relatively constant member of this list has been ACE for which two (rs4291 and rs1800764) of the six variants studied show significant association with LOAD risk following the AlzGene meta-analyses based on total sample sizes of n=10,588 and n=4,756, respectively. Notably, rs1800764 has also been associated with elevated CSF Aβ42/Aβ40 ratio [7].
Despite the large number of reported independent genetic associations between ACE variants and LOAD in the last decade (22 out of 55 populations published to-date; for details see AlzGene), few studies utilised more comprehensive haplotype approaches [7, 8, 16–19]. Keavney and colleagues identified seven haplotypes in a Caucasian British population derived from data from ten polymorphisms spanning 26kb of ACE. From these haplotypes, they constructed a cladogram that contained three main branches (clades A, B and C), which accounted for 90% of the observed haplotypes. Clade A has since been associated with low plasma ACE levels and increased risk of LOAD [16], clades B and C with higher ACE levels [19–21] and clade C with increased risk for LOAD in families [18]. Kehoe and colleagues also analysed seven variants within ACE (rs4363, rs4362, rs4343, rs4331, rs4309, rs4291, rs1800764) that formed ten haplotypes with an LD structure that enabled the selection of three ‘tagging’ variants (rs4291, rs4343 and rs1800764) [8]. The most frequent haplotype (H1) contained the previously reported AD-associated (‘risk’) ACE indel I allele [22] while the H2 haplotype contained the (‘protective’) D allele [8]. Some indication that the indel was not the only functional ACE variant involved in LOAD pathogenesis came from H5 (also containing the I allele) which was also associated with a reduced risk of LOAD ref [8].
In line with previous haplotype and cladistic approaches described we have used the three tagging variants rs4291, rs4343 and rs1800764 to investigate the association of ACE with LOAD in our large multi-centre cohort comprising ten case-control series, nine of which 3,930 LOAD cases and 4,282 controls. This represents the largest study to-date to investigate the effects of ACE haplotypes in LOAD.
Materials and Methods
European Patient Samples
Informed consent was obtained from all subjects included in this study, which was approved by the local Ethics Committee. This European Caucasian cohort combined three case control sample collections; 1) the Alzheimer’s Research Trust (ART) Collaborative Sample Collection (1,197 LOAD patients and 886 controls) supplied from six ART network centres across the UK, 2) the Medical Research Council (MRC) Collaborative Sample Collection collected from both community and hospital settings in the UK (816 LOAD patients and 959 controls) and 3) a Swedish sample collection (156 LOAD patients and 59 controls). It must be noted that nine of the ten case-control series used in this cohort do not overlap with those previously published in case-control association studies of ACE variants. The remaining series (Oxford) has previously been reported with respect to the ACE indel (rs1799752) but not for the three tagging SNPs investigated here. A summary of patient details from each centre is shown in Table 1.
Table 1.
Details of samples used in this study.
| Ethnicity | Sample Collection |
Series | N |
Mean Age |
% Female |
%E4+ |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOAD | CTRL | Total | LOAD | CTRL | LOAD | CTRL | LOAD | CTRL | ||||
| Caucasian European | ART | Belfast | 238 | 236 | 474 | 78.3 | 76.1 | 0.66 | 0.65 | 0.58 | 0.28 | |
| Bonn | 175 | 198 | 373 | 75.9 | 70.9 | 0.76 | 0.49 | 0.55 | 0.27 | |||
| Nottingham/Manchester | 381 | 102 | 483 | 71.0 | 72.6 | 0.54 | 0.34 | 0.62 | 0.23 | |||
| Oxford | 173 | 205 | 378 | 72.7 | 77.3 | 0.56 | 0.57 | 0.64 | 0.25 | |||
| Southampton | 230 | 145 | 375 | 80.6 | 75.8 | 0.58 | 0.49 | 0.47* | 0.26 | |||
| MRC | UK | 816 | 959 | 1,775 | 81.5 | 76.6 | 0.72 | 0.64 | 0.63 | 0.24 | ||
| Sweden | Sweden | 156 | 59 | 215 | 77.4 | 73.9 | 0.70 | 0.41 | 0.68 | 0.31 | ||
| Caucasian North American |
Mayo Clinic | Autopsy-confirmed | 592 | 374 | 966 | 81.2 | 75.7 | 0.60 | 0.43 | 0.60 | 0.23 | |
| Jacksonville | 608 | 602 | 1210 | 78.1 | 78.0 | 0.62 | 0.60 | 0.62 | 0.26 | |||
| Rochester | 561 | 1,402 | 1,963 | 79.5 | 78.4 | 0.62 | 0.54 | 0.56 | 0.25 | |||
| Caucasian | All | Total | 3,930 | 4,282 | 8,212 | 78.6 | 76.9 | 0.64 | 0.56 | 0.61 | 0.25 | |
APOE E4 status was unknown for 50% of LOAD samples from Southampton
The number of LOAD patients (LOAD) and controls (CON), mean age-at-diagnosis, percentage that are female and percentage that possess at least one copy of the APOE ε4 allele are given for each individual and pooled series. Mean age is given as age-at-diagnosis/entry with the standard deviation (SD) from the mean in parentheses.
US Patient samples
A total of 4,139 Caucasian samples were obtained with written consent from the Mayo Clinic, USA. These samples included 592 autopsy-confirmed AD patients and 374 autopsy-confirmed controls (AUT), 1,169 clinically diagnosed LOAD patients and 976 controls from Mayo Clinic Jacksonville (JS) and Mayo Clinic, Rochester (RS). None of the samples used in this cohort overlap with those previously published in case-control association studies of ACE variants. Further information regarding these samples can be found in Table 1.
LOAD Diagnosis
The majority of samples were diagnosed possible or probable AD (n=3,215) or control (n=3,968) using NINCDS-ADRDA criteria [23]. The remaining samples were histopathologically confirmed as definite AD (n=1,091) or control (n=476) using NINCDS-ADRDA (AUT) or CERAD criteria (ART) [24]. All patients with evidence of an autosomal dominant AD trait, where a first-degree relative had been diagnosed with AD or where there was evidence for other causes of dementia were excluded.
Genotyping
Genotyping data from the ART samples was obtained using fluorescently-labelled (VIC or FAM) allele-specific TaqMan probes that were designed by ABI; all assays performed by Geneservice (Cambridge, UK). In addition to assay controls incorporated by Geneservice, 15% of the samples assayed were sequenced for genotype at source, 10% of samples were assayed in duplicate for quality assurance. Data were only accepted when there was 100% concordance between duplicates. All genotype plots were subjected to quality control upon receipt and assays were only accepted when call rates were above 95%. A detailed description of the ascertainment and assessment of the MRC sample collection has been reported previously [25]. The data from the Swedish samples was generated using the Dynamic Allele-Specific Hybridization (DASH) method as described elsewhere [8]. The Mayo Clinic samples were genotyped using TaqMan® SNP Genotyping Assays in an ABI PRISM® 7900HT Sequence Detection System with 384-Well Block Module from Applied Biosystems. All variants passed the p>0.01 cut-off for deviation from Hardy-Weinberg equilibrium as suggested by Wigginton et al. when investigating >100 samples [26].
Single variant analysis
Odds ratios and 95% CI were calculated by binary logistic regression (allelic dose model) using the --logistic command in PLINK software [27]. The covariates age-at-onset (where unknown, age-at-death minus the 8 year average disease duration was used), carriage of the APOE ε4 allele and sex were added into the model using the --covar command. The total dataset was also tested for association by binary logistic regression under dominant and recessive models adjusted for covariates using StatsDirect v2.5.8. For meta-analyses, summary ORs, 95% CI and Breslow-Day tests were calculated under the DerSimonian and Laird (1986) random-effects model using StatsDirect v2.5.8 software.
Haplotype association
Haplotype frequencies were estimated using the expectation-maximization approach implemented in the haplo.em function of Haplo.stats v1.2.2 [28] using R programming software. Global haplotype association and individual haplotype score tests adjusted for APOE ε4 dose, sex and age-at-diagnosis were performed using the haplo.score function of Haplo.stats v1.2.2.
Results
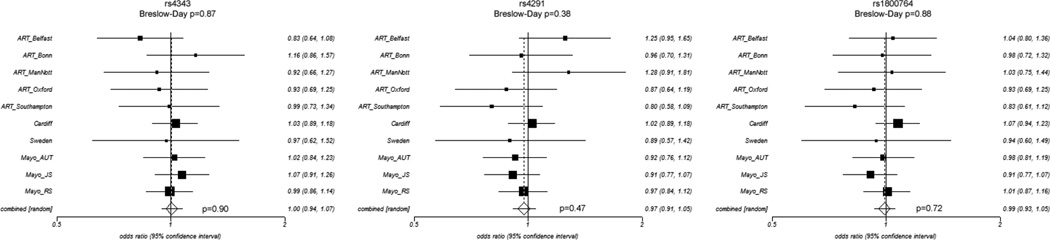
We tested three ACE variants for association with LOAD in our seven European and three North American case-control series (series details are shown in Table 1). Genotype and allele counts for each series are shown in Table 2. We first tested for association with LOAD in each case-control series by logistic regression using an additive/allelic dosage model correcting for sex, age-at-diagnosis and possession of at least one copy of the APOE ɛ4 allele as covariates (Table 2). None of the variants were associated with LOAD risk in any series (all p>0.09). We also tested for association the three variants in all ten series pooled (n=8,212) but again found no association (all p>0.18). Since some genetic variants may exert dominant or recessive effects we also performed logistic regression for the total dataset using these models but found no association with LOAD risk (all p>0.36). We also tested for association of all three variants with LOAD in individuals not possessing the APOE ε4 allele in all series (1,495 LOAD, 3,175 controls) but found no association (all p>0.13; data not shown). In order to determine whether the lack of observation could be attributed to population heterogeneity, we performed meta-analyses of each variant testing for association in the combined data from all ten series using a DerSimonion-Laird random effects model to estimate a pooled odds ratio and testing for heterogeneity between series using the Breslow-Day test (Fig. 1). We found no evidence for population heterogeneity (all p>0.38) or for association of rs4343 (OR=1.00, p=0.90), rs4291 (OR=0.97, p=0.47) or rs1800764 (OR=0.99, p=0.72) with LOAD risk.
Table 2.
Association of ACE variants rs4291, rs4343 and rs1800764 with LOAD
| Series | MAF | LOAD | CON | Logistic Regression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LOAD:CON | Maj | Het | Min | Maj | Het | Min | OR (95%CI) | p | |||
| rs4343 (G:A) | Belfast (474) | 0.51:0.56 | 60 | 113 | 65 | 50 | 109 | 77 | 0.82 (0.63–1.07) | 0.14 | |
| Bonn (373) | 0.52:0.48 | 42 | 84 | 49 | 56 | 93 | 49 | 1.17 (0.85–1.62) | 0.33 | ||
| Notts./Manch. (483) | 0.49:0.51 | 92 | 197 | 87 | 28 | 42 | 31 | 0.90 (0.64–1.25) | 0.52 | ||
| Oxford (378) | 0.47:0.48 | 50 | 84 | 38 | 57 | 98 | 50 | 0.88 (0.64–1.21) | 0.43 | ||
| Southampton (375) | 0.45:0.45 | 68 | 116 | 46 | 45 | 67 | 32 | 1.01 (0.72–1.42) | 0.93 | ||
| MRC (1,775) | 0.50:0.49 | 205 | 358 | 200 | 233 | 444 | 216 | 1.05 (0.90–1.22) | 0.51 | ||
| Sweden (226) | 0.48:0.49 | 40 | 81 | 35 | 19 | 22 | 18 | 1.22 (0.75–1.97) | 0.42 | ||
| AUT (966) | 0.47:0.46 | 166 | 283 | 129 | 112 | 167 | 85 | 1.02 (0.84–1.25) | 0.88 | ||
| JS (1,210) | 0.48:0.49 | 159 | 302 | 132 | 172 | 295 | 125 | 1.05 (0.95–1.16) | 0.30 | ||
| RS (1,963) | 0.48:0.46 | 144 | 259 | 145 | 336 | 693 | 344 | 1.04 (0.87–1.24) | 0.67 | ||
| All Additive (8,212) | 0.49:0.49 | 1026 | 1877 | 926 | 1108 | 2030 | 1027 | 0.99 (0.93–1.05) | 0.75 | ||
| All Dominant | 1.02 (0.91–1.14) | 0.75 | |||||||||
| All Recessive | 1.02 (0.90–1.14) | 0.80 | |||||||||
| Rs4291 (A:T) | Belfast (474) | 0.37:0.32 | 96 | 110 | 32 | 114 | 91 | 28 | 1.28 (0.96–1.70) | 0.09 | |
| Bonn (373) | 0.35:0.36 | 71 | 81 | 19 | 83 | 87 | 27 | 0.84 (0.60–1.20) | 0.35 | ||
| Notts./Manch. (483) | 0.37:0.31 | 159 | 158 | 60 | 48 | 44 | 10 | 1.31 (0.92–1.87) | 0.13 | ||
| Oxford (378) | 0.35:0.39 | 71 | 75 | 22 | 75 | 97 | 29 | 0.94 (0.67–1.31) | 0.88 | ||
| Southampton (375) | 0.37:0.42 | 92 | 106 | 32 | 50 | 65 | 28 | 0.74 (0.52–1.05) | 0.09 | ||
| MRC (1,775) | 0.37:0.36 | 323 | 341 | 117 | 364 | 416 | 118 | 1.00 (0.85–1.16) | 0.91 | ||
| Sweden (226) | 0.37:0.40 | 59 | 78 | 19 | 26 | 19 | 14 | 0.77 (0.47–1.23) | 0.27 | ||
| AUT (966) | 0.37:0.38 | 242 | 256 | 85 | 143 | 171 | 57 | 0.93 (0.76–1.14) | 0.50 | ||
| JS (1,210) | 0.37:0.39 | 236 | 292 | 78 | 217 | 294 | 88 | 0.96 (0.80–1.16) | 0.58 | ||
| RS (1,963) | 0.35:0.35 | 241 | 246 | 70 | 586 | 628 | 178 | 0.94 (0.80–1.10) | 0.42 | ||
| All Additive (8,212) | 0.36:0.37 | 1590 | 1743 | 534 | 1706 | 1912 | 577 | 0.99 (0.92–1.05) | 0.67 | ||
| All Dominant | 0.95 (0.86–1.06) | 0.36 | |||||||||
| All Recessive | 0.97 (0.84–1.12) | 0.68 | |||||||||
| rs1800764 (T:C) | Belfast (474) | 0.41:0.40 | 82 | 117 | 39 | 92 | 98 | 45 | 1.03 (0.76–1.35) | 0.84 | |
| Bonn (373) | 0.43:0.44 | 53 | 92 | 29 | 66 | 91 | 41 | 0.86 (0.61–1.20) | 0.37 | ||
| Notts./Manch. (483) | 0.43:0.42 | 124 | 183 | 70 | 32 | 52 | 16 | 1.08 (0.76–1.53) | 0.66 | ||
| Oxford (378) | 0.44:0.46 | 55 | 84 | 34 | 59 | 104 | 42 | 0.96 (0.69–1.33) | 0.96 | ||
| Southampton (375) | 0.47:0.52 | 60 | 124 | 46 | 39 | 62 | 44 | 0.85 (0.61–1.20) | 0.37 | ||
| MRC (1,775) | 0.44:0.43 | 262 | 378 | 170 | 316 | 451 | 176 | 1.07 (0.93–1.25) | 0.34 | ||
| Sweden (226) | 0.43:0.45 | 46 | 84 | 25 | 22 | 19 | 16 | 0.77 (0.48–1.26) | 0.30 | ||
| AUT (966) | 0.47:0.48 | 169 | 276 | 137 | 104 | 180 | 87 | 1.02 (0.80–1.14) | 0.78 | ||
| JS (1,210) | 0.45:0.47 | 179 | 308 | 113 | 167 | 296 | 130 | 0.95 (0.80–1.14) | 0.57 | ||
| RS (1,963) | 0.43:0.43 | 183 | 261 | 109 | 462 | 655 | 272 | 1.00 (0.86–1.15) | 0.96 | ||
| All Additive (8,212) | 0.44:0.44 | 1213 | 1907 | 772 | 1359 | 2008 | 869 | 1.00 (0.94–1.07) | 0.95 | ||
| All Dominant | 1.02 (0.92–1.14) | 0.69 | |||||||||
| All Recessive | 0.97 (0.86–1.10) | 0.64 | |||||||||
Designated major:minor alleles for each variant are shown in parentheses after the variant name in the first column. The number of samples in each series is shown in parentheses after the series name. The minor allele frequency (MAF) and genotype counts in LOAD and controls (CON) for major allele homozygotes (Maj), heterozygotes (Het) and minor allele homozygotes (Min) are provided for each series. Odds ratios (ORs), 95% confidence intervals (CI) and p-values (p) were calculated for each series using a binary logistic regression additive model. The total pooled data (All) was also tested for association using dominant and recessive models. All logistic regression models included age-at-onset, sex and APOE ε4 allele as covariates.
Figure 1. Forest plots for meta-analysis for each variant.
ORs (boxes) and 95%CI (whiskers) are plotted for each series and shown on the right of each plot. Combined OR is the overall OR calculated by the meta-analysis using a random effects model. P-values from Breslow-Day tests of heterogeneity and meta-analysis are provided.
In an attempt to replicate previous findings that the two most common haplotypes (H1 and H2) are significantly associated with opposing risk for LOAD [8], we constructed haplotypes using the three tagging variants. As shown in Table 3, the haplotype frequencies were comparable across all series and are consistent with previous studies [8]. In order to limit the number of tests used, global association of the six haplotypes in each series were tested and individual haplotypes only tested for association for populations with a global p-value <0.05. A significant global haplotypic p-value was observed in the MRC sample only (p=0.007), largely to the protective association of H3 (OR=0.72, p=0.02) and the trend towards a risky association of H4 (OR=1.38, p=0.09) and H7 (OR=2.07, p=0.08), associations that were not previously observed by Kehoe and colleagues [8]. The global haplotypic p-value for our complete dataset (n=7,557) was not significant (p=0.51) and the individual haplotypic ORs observed in our complete dataset for the two most common haplotypes gave weaker, ORs compared to those observed by Kehoe et al in their complete dataset (H1; OR=1.0 vs OR=1.2, H2; OR=0.96 vs 0.80 [8]). Therefore, these data do not replicate the previously reported association of the Alu indel [12], which tags H1.
Table 3.
Association of ACE haplotypes with LOAD
| Series | Haplotype* | Frequency |
OR | 95%CI | p-value | |
|---|---|---|---|---|---|---|
| LOAD | CTRL | |||||
|
ART global p-value = 0.27 |
H1 - AAT | 973 (0.46) | 810 (0.47) | 0.96 | 0.83 – 1.10 | |
| H2 - GTC | 722 (0.34) | 596 (0.34) | 1.04 | 0.90 – 1.21 | ||
| H3 - GAT | 236 (0.11) | 157 (0.09) | 1.25 | 0.99 – 1.59 | ||
| H4 - GAC | 236 (0.11) | 270 (0.16) | 0.78 | 0.57 – 1.07 | ||
| H7 - AAC | 44 (0.02) | 35 (0.02) | 0.89 | 0.52 – 1.52 | ||
| H5 - ATC | 30 (0.01) | 29 (0.02) | 0.76 | 0.41 – 1.41 | ||
|
MRC global p-value = 0.007 |
H1 - AAT | 671 (0.47) | 760 (0.47) | 1.00 | 0.85 – 1.18 | 0.99 |
| H2 - GTC | 500 (0.35) | 551 (0.34) | 1.05 | 0.88 – 1.25 | 0.59 | |
| H3 - GAT | 129 (0.09) | 174 (0.11) | 0.72 | 0.54 – 0.96 | 0.02 | |
| H4 - GAC | 87 (0.06) | 83 (0.05) | 1.38 | 0.95 – 1.99 | 0.09 | |
| H7 - AAC | 20 (0.01) | 24 (0.02) | 2.07 | 0.92 – 4.63 | 0.08 | |
| H5 - ATC | 23 (0.02) | 19 (0.01) | 0.88 | 0.40 – 1.93 | 0.75 | |
|
Sweden global p-value = 0.86 |
H1 - AAT | 144 (0.47) | 54 (0.47) | 1.37 | 0.57 – 3.28 | |
| H2 - GTC | 115 (0.37) | 45 (0.39) | 0.70 | 0.18 – 2.76 | ||
| H3 - GAT | 32 (0.10) | 9 (0.08) | 1.18 | 0.72 – 1.95 | ||
| H4 - GAC | 13 (0.04) | 4 (0.03) | 1.02 | 0.15 – 7.01 | ||
| H7 - AAC | 6 (0.02) | 2 (0.02) | 0.81 | 0.50 – 1.31 | ||
| H5 - ATC | 0 (0.00) | 0 (0.00) | NA | NA | ||
|
Mayo Clinic global p-value = 0.31 |
H1 - AAT | 1478 (0.45) | 2043 (0.45) | 1.01 | 0.91 – 1.12 | |
| H2 - GTC | 1140 (0.34) | 1593 (0.35) | 0.92 | 0.83 – 1.02 | ||
| H3 - GAT | 350 (0.11) | 439 (0.10) | 1.05 | 0.88 – 1.24 | ||
| H4 - GAC | 212 (0.06) | 272 (0.06) | 1.03 | 0.83 – 1.28 | ||
| H7 - AAC | 79 (0.02) | 83 (0.02) | 1.45 | 1.00 – 2.09 | ||
| H5 - ATC | 46 (0.01) | 61 (0.01) | 1.38 | 0.85 – 2.21 | ||
|
All global p-value = 0.51 |
H1 - AAT | 3267 (0.46) | 3667 (0.46) | 1.00 | 0.93 – 1.07 | |
| H2 - GTC | 2477 (0.34) | 2785 (0.35) | 0.97 | 0.90 – 1.04 | ||
| H3 - GAT | 746 (0.11) | 779 (0.10) | 1.03 | 0.92 – 1.17 | ||
| H4 - GAC | 420 (0.06) | 472 (0.06) | 1.03 | 0.88 – 1.20 | ||
| H7 - AAC | 152 (0.02) | 139 (0.02) | 1.28 | 0.97 – 1.68 | ||
| H5 - ATC | 96 (0.01) | 114 (0.01) | 1.05 | 0.75 – 1.47 | ||
Order of variants in haplotype is as follows rs4343, rs4291, rs1800764
Haplotypes are numbered according to their frequency in the Kehoe et al study [8] (only haplotypes with a frequency >1% in this study are shown). Haplotype frequencies are shown for the total dataset and in each of the individual series. A haplotype score test was used to calculate a “global p-value: for the association of the haplotypes in the total dataset and in each of the individual series. ORs, 95% confidence intervals and p-values are shown for the individual haplotypes in the MRC series only due to the significant global p-value.
Discussion
We have conducted a large case-control study of three haplotype tagging variants in the LOAD candidate gene, ACE, that has previously been associated with LOAD risk [8, 29–34]. Meta-analyses of ten case-control series totalling 3,930 LOAD and 4,282 controls showed no population heterogeneity (all p>0.38) or evidence for association of rs4343 (OR=1.00 p=0.90), rs4291 (OR=0.97 p=0.47) or rs1800764 (OR=0.99 p=0.72) with LOAD risk.
We also tested for association of six ACE haplotypes with LOAD but found no evidence for association in the total dataset (global p=0.51). However, we did observe a significant global haplotypic p-value of 0.007 in one of the series (the MRC population - 1,430 LOAD patients and 1,611 controls) from the UK, which was largely due to a novel protective association of H3, the haplotype containing the major allele (G-A-T) at all three variants (9% LOAD, 11% Controls, OR=0.72, p=0.02). However, this association was not observed in any of the other case-control populations (all p>0.07) or in the pooled dataset (p=0.58). Indeed the directionality of ORs observed in all other populations was opposite to that seen for H3 (OR=1.03–1.25) but none produced significant findings. The modest p-value (p=0.03) for association of H3 with LOAD, which would not survive Bonferroni correction for the six haplotypes studied (p<0.008) along with the lack of association of H3 in the other nine populations studied here or in previously published studies suggests that the significance of this association should be treated with caution.
The lack of association in any of our ten series for the two ACE variants that have been significantly associated with LOAD following AlzGene meta-analyses is perhaps not surprising. None of the eight Caucasian populations used previously to study rs4291 and included in AlzGene showed significant association for AD and only one [35] of the seven Caucasian populations used to study rs1800764 showed significant association (OR=0.86 95% CI 0.74–0.99). Despite this, AlzGene reported significant association for both rs4291 (OR=0.87, 95%CI 0.80–0.95) and rs1800764 (OR=0.84, 95%CI 0.77.0.92) prior to this study. This is largely due to the fact that there was a consistently similar directionality of the ORs in the majority of the Caucasian populations previously used to study rs4291 and rs1800764 and the resulting increase in sample size achieved by analyzing the studies together provides sufficient power to detect association. When the present data is eventually incorporated into Alzgene these overall associations will likely diminish further towards the null. However, the fact that we also observed same direction ORs in seven out of ten series for rs4291 and six out of ten series for rs1800764, raises the possibility that a true association of modest effect size (OR~0.90) is present, but which requires even larger studies to gain sufficient power for detection.
It is possible that the initial association may have, by chance, been the result of an over-estimation of the effect size of these variants that has since diminished in subsequent follow-up case-control studies. For example, in the case of rs4291, the initial study reported ORs ranging from 0.76–0.84 in four Caucasian populations each consisting of ~400 subjects [8]. In comparison, the four subsequent Caucasian studies (all of equal or larger size than the initial study populations) reported ORs ranging from 0.76–1.00 [31, 32, 36, 37] and here we report ORs ranging from 0.80–1.28 in ten populations of equal or larger size than the initial study thus further diminishing the effect size. The same appears true for rs1800764 where the initial study reported ORs ranging from 0.74–0.84 [8], compared to ORs ranging from 0.80–1.09 in subsequent follow-up studies [31, 38, 39] and in the ORs 0.83–1.07 reported here. This further supports the need for multiple, large follow-up studies and meta-analyses of all data to reduce the likelihood of an over-estimation of the effect size.
Failure to detect association could also be explained by these variants merely tagging one or more truly functional variants. In such instances the corresponding degree of linkage disequilibrium between these variants could differ between series leading to weaker and/or opposing effects. However, given the Caucasian background of all these series and the lack of significant population heterogeneity or association with LOAD risk for any variant in any individual population, this possibility seems unlikely.
If ACE variants modify LOAD risk this effect may be dependent on interactions with other environmental/phenotypic background. For example, ACE mediates hypertensive effects by its function on angiotensin I to convert it to the vasoactive angiotensin II [4]. Thus the co-occurrence of hypertension in sub-groups of AD patients and controls and this being treated to varying extents by drugs that target the pathway in which ACE is functional is likely to be a confounding variable in studies for ACE. Indeed studies of AD brain tissue have shown that while ACE genotypes did not influence levels or activity of brain ACE [40], rs4343 and rs1800764 have been associated with soluble Aβ levels [41] and exposure of neuronal SH-SY5Y cell lines to oligomeric Aβ1–42 for 24 hours resulted in significant increases in ACE protein level and activity [41]. These collectively suggest that along with previous evidence of elevated ACE activity in AD brain [40, 42] that there could be phenotypic-specific post-translational changes to ACE that contribute to AD pathogenesis. Further information supporting this are the findings of Ellul and colleagues [43]. They noted in a longitudinally assessed clinical cohort, that drugs affecting the Renin Angiotensin System, in which ACE is very important, can slow the rates of deterioration of AD and could serve as a confounder in clinical outcome measurement in clinical trials. The possibility of phenotype-specific pharmacogenetic considerations are also reinforced by findings from a recent GWAS [44] of two young-onset hypertension populations totalling 1,023 subjects that reported eight ACE variants were significantly associated with ACE activity and rs4343 showing the strongest association (p=3.0×10−25). In the same study an association between blood type and ACE activity in an independent young-onset hypertension family study (n=428) was reported and showed a potential differential blood pressure response to anti-hypertensive treatment (ACE inhibitors) in subjects dependent on ACE genotype. This latter observation is particularly important in view of findings from a number of observational studies where anti-hypertensive treatments such as inhibitors of ACE activity (i.e. ACE-inhibitors) or of angiotensin II function (i.e. angiotensin II receptor antagonists – ARAs) appeared to be protective against the development of and/or progression of LOAD and Mild Cognitive Impairment (MCI) [45–51].
These data do not replicate previously reported haplotype associations with LOAD risk in a large case-control series of 3,930 LOAD patients and 4,282 controls. However, it is clear that when considered alongside data from other disciplines, these findings contribute just one important part of what appears to be a highly complex interaction between ACE genetics, phenotype and pharmacological effects in AD and which traditional case-control studies are not equipped to unpick. Larger studies which would include richer phenotypic data that would allow for more accurate adjustment for confounders and where possible incorporation of additional measurements specific to candidate gene function (e.g. qPCR, protein based measurements, etc), would likely increase the chances of unpicking the real genetic involvement of many current candidate genes.
Acknowledgements
Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD) were used in this study. We thank contributors, including the Alzheimer’s Disease Centers who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible. We also thank Jonathan A. Prince (Center for Genomics and Bioinformatics, Karolinska Institute, Stockholm, Sweden) for supplying the genotype data for the case-control series from Sweden. This work was supported by grants from the US National Institutes of Health, NIA R01 AG18023 (Steven G. Younkin); Mayo Alzheimer’s Disease Research Center, P50 AG16574 (Steven G. Younkin); The Mayo Clinic sample collection was funded by the Mayo Clinic Mayo Alzheimer’s Disease Patient Registry, U01 AG06576 and US National Institute on Aging, AG25711, AG17216, AG03949. Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24AG21886) awarded by the National Institute on Aging (NIA), were used in this study. This project was also generously supported by the Robert and Clarice Smith Postdoctoral Fellowship (M.M.C.); Robert and Clarice Smith and Abigail Van Buren Alzheimer’s Disease Research Program (Steven G. Younkin) and by the Palumbo Professorship in Alzheimer’s Disease Research (Steven G. Younkin). K.M. is funded by the Alzheimer’s Research Trust and the Big Lottery Fund. P.G.K. is funded by the Sigmund Gestetner Foundation.
References
- 1.Walsh DM, Selkoe DJ. Deciphering the molecular basis of memory failure in Alzheimer's disease. 0896-6273. 2004;44:181–193. doi: 10.1016/j.neuron.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Love S, Miners S, Palmer J, Chalmers K, Kehoe P. Insights into the pathogenesis and pathogenicity of cerebral amyloid angiopathy. Front Biosci. 2009;14:4778–4792. doi: 10.2741/3567. [DOI] [PubMed] [Google Scholar]
- 3.Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer's disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kehoe PG, Miners S, Love S. Angiotensins in Alzheimer's disease - friend or foe? Trends Neurosci. 2009;32:619–628. doi: 10.1016/j.tins.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 5.Kehoe PG. Angiotensins and Alzheimer's disease: a bench to bedside overview. Alzheimers Res Ther. 2009;1:3. doi: 10.1186/alzrt3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vardy ER, Rice PJ, Bowie PC, Holmes JD, Catto AJ, Hooper NM. Plasma angiotensin-converting enzyme in Alzheimer's disease. J Alzheimers Dis. 2009;16:609–618. doi: 10.3233/JAD-2009-1002. [DOI] [PubMed] [Google Scholar]
- 7.Kauwe JS, Wang J, Mayo K, Morris JC, Fagan AM, Holtzman DM, Goate AM. Alzheimer's disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics. 2009;10:13–17. doi: 10.1007/s10048-008-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kehoe PG, Katzov H, Feuk L, Bennet AM, Johansson B, Wiman B, de Faire U, Cairns NJ, Wilcock GK, Brookes AJ, Blennow K, Prince JA. Haplotypes extending across ACE are associated with Alzheimer's disease. Hum Mol Genet. 2003;12:859–867. doi: 10.1093/hmg/ddg094. [DOI] [PubMed] [Google Scholar]
- 9.Cambien F, Alhenc-Gelas F, Herbeth B, Andre JL, Rakotovao R, Gonzales MF, Allegrini J, Bloch C. Familial resemblance of plasma angiotensin-converting enzyme level: the Nancy Study. Am J Hum Genet. 1988;43:774–780. [PMC free article] [PubMed] [Google Scholar]
- 10.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- 12.Lehman DM, Fu DJ, Freeman AB, Hunt KJ, Leach RJ, Johnson-Pais T, Hamlington J, Dyer TD, Arya R, Abboud H, Goring HH, Duggirala R, Blangero J, Konrad RJ, Stern MP. A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-beta-D glucosaminidase is associated with type 2 diabetes in Mexican Americans. 0012-1797. 2005;54:1214–1221. doi: 10.2337/diabetes.54.4.1214. [DOI] [PubMed] [Google Scholar]
- 13.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 14.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. 0028-3878. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 15.Bertram L, McQueen M, Mullin K, Blacker D, Tanzi R. 2006 Available at: http://www.alzgene.org. Accessed [xxx]
- 16.Katzov H, Bennet AM, Kehoe P, Wiman B, Gatz M, Blennow K, Lenhard B, Pedersen NL, de Faire U, Prince JA. A cladistic model of ACE sequence variation with implications for myocardial infarction, Alzheimer disease and obesity. Hum Mol Genet. 2004;13:2647–2657. doi: 10.1093/hmg/ddh286. [DOI] [PubMed] [Google Scholar]
- 17.Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, Farrer LA. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am J Hum Genet. 2006;78:871–877. doi: 10.1086/503687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schjeide BM, McQueen MB, Mullin K, DiVito J, Hogan MF, Parkinson M, Hooli B, Lange C, Blacker D, Tanzi RE, Bertram L. Assessment of Alzheimer's disease case-control associations using family-based methods. Neurogenetics. 2009;10:19–25. doi: 10.1007/s10048-008-0151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keavney B, McKenzie CA, Connell JM, Julier C, Ratcliffe PJ, Sobel E, Lathrop M, Farrall M. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum Mol Genet. 1998;7:1745–1751. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- 20.Soubrier F, Martin S, Alonso A, Visvikis S, Tiret L, Matsuda F, Lathrop GM, Farrall M. High-resolution genetic mapping of the ACE-linked QTL influencing circulating ACE activity. Eur J Hum Genet. 2002;10:553–561. doi: 10.1038/sj.ejhg.5200847. [DOI] [PubMed] [Google Scholar]
- 21.Rieder MJ, Taylor SL, Clark AG, Nickerson DA. Sequence variation in the human angiotensin converting enzyme. Nat Genet. 1999;22:59–62. doi: 10.1038/8760. [DOI] [PubMed] [Google Scholar]
- 22.Kehoe PG, Russ C, McIlory S, Williams H, Holmans P, Holmes C, Liolitsa D, Vahidassr D, Powell J, McGleenon B, Liddell M, Plomin R, Dynan K, Williams N, Neal J, Cairns NJ, Wilcock G, Passmore P, Lovestone S, Williams J, Owen MJ. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat Genet. 1999;21:71–72. doi: 10.1038/5009. [DOI] [PubMed] [Google Scholar]
- 23.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. 0028-3878. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 24.Mirra SS, Hart MN, Terry RD. Making the diagnosis of Alzheimer's disease. A primer for practicing pathologists. Arch Pathol Lab Med. 1993;117:132–144. [PubMed] [Google Scholar]
- 25.Morgan AR, Hamilton G, Turic D, Jehu L, Harold D, Abraham R, Hollingworth P, Moskvina V, Brayne C, Rubinsztein DC, Lynch A, Lawlor B, Gill M, O'Donovan M, Powell J, Lovestone S, Williams J, Owen MJ. Association analysis of 528 intra-genic SNPs in a region of chromosome 10 linked to late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:727–731. doi: 10.1002/ajmg.b.30670. [DOI] [PubMed] [Google Scholar]
- 26.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez R, Alvarez V, Lahoz CH, Martinez C, Pena J, Sanchez JM, Guisasola LM, Salas-Puig J, Moris G, Vidal JA, Ribacoba R, Menes BB, Uria D, Coto E. Angiotensin converting enzyme and endothelial nitric oxide synthase DNA polymorphisms and late onset Alzheimer's disease. J Neurol Neurosurg Psychiatry. 1999;67:733–736. doi: 10.1136/jnnp.67.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards TL, Pericak-Vance M, Gilbert JR, Haines JL, Martin ER, Ritchie MD. An association analysis of Alzheimer disease candidate genes detects an ancestral risk haplotype clade in ACE and putative multilocus association between ACE, A2M, and LRRTM3. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:721–735. doi: 10.1002/ajmg.b.30899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghebranious N, Mukesh B, Giampietro PF, Glurich I, Mickel SF, Waring SC, McCarty CA. A Pilot Study of Gene/Gene and Gene/Environment Interactions in Alzheimer Disease. Clin Med Res. 2010 doi: 10.3121/cmr.2010.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helbecque N, Codron V, Cottel D, Amouyel P. An age effect on the association of common variants of ACE with Alzheimer's disease. Neurosci Lett. 2009;461:181–184. doi: 10.1016/j.neulet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Kolsch H, Jessen F, Freymann N, Kreis M, Hentschel F, Maier W, Heun R. ACE I/D polymorphism is a risk factor of Alzheimer's disease but not of vascular dementia. Neurosci Lett. 2005;377:37–39. doi: 10.1016/j.neulet.2004.11.062. [DOI] [PubMed] [Google Scholar]
- 34.Mattila KM, Rinne JO, Roytta M, Laippala P, Pietila T, Kalimo H, Koivula T, Frey H, Lehtimaki T. Dipeptidyl carboxypeptidase 1 (DCP1) and butyrylcholinesterase (BCHE) gene interactions with the apolipoprotein E epsilon4 allele as risk factors in Alzheimer's disease and in Parkinson's disease with coexisting Alzheimer pathology. J Med Genet. 2000;37:766–770. doi: 10.1136/jmg.37.10.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Craig DW, Coon KD, Dunckley T, Bandy D, Lee W, Chen K, Beach TG, Mastroeni D, Grover A, Ravid R, Sando SB, Aasly JO, Heun R, Jessen F, Kolsch H, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Papassotiropoulos A, Stephan DA, Huentelman MJ. Evidence for an association between KIBRA and late-onset Alzheimer's disease. Neurobiol Aging. 2010;31:901–909. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruandet A, Richard F, Tzourio C, Berr C, Dartigues JF, Alperovitch A, Amouyel P, Helbecque N. Haplotypes across ACE and the risk of Alzheimer's disease: the three-city study. J Alzheimers Dis. 2008;13:333–339. doi: 10.3233/jad-2008-13310. [DOI] [PubMed] [Google Scholar]
- 37.Cousin E, Mace S, Rocher C, Dib C, Muzard G, Hannequin D, Pradier L, Deleuze JF, Genin E, Brice A, Campion D. No replication of genetic association between candidate polymorphisms and Alzheimer's disease. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O'Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren-Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ. Association of CR1, CLU and PICALM with Alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shulman JM, Chibnik LB, Aubin C, Schneider JA, Bennett DA, De Jager PL. Intermediate phenotypes identify divergent pathways to Alzheimer's disease. PLoS One. 2010;5:e11244. doi: 10.1371/journal.pone.0011244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miners JS, Ashby E, Van Helmond Z, Chalmers KA, Palmer LE, Love S, Kehoe PG. Angiotensin-converting enzyme (ACE) levels and activity in Alzheimer's disease, and relationship of perivascular ACE-1 to cerebral amyloid angiopathy. Neuropathol Appl Neurobiol. 2008;34:181–193. doi: 10.1111/j.1365-2990.2007.00885.x. [DOI] [PubMed] [Google Scholar]
- 41.Miners JS, van Helmond Z, Raiker M, Love S, Kehoe PG. ACE variants and association with brain Aβ levels in Alzheimer's disease. Am J Transl Res. 2011;3:71–78. [PMC free article] [PubMed] [Google Scholar]
- 42.Miners S, Ashby E, Baig S, Harrison R, Tayler H, Speedy E, Prince JA, Love S, Kehoe PG. Angiotensin-converting enzyme levels and activity in Alzheimer's disease: differences in brain and CSF ACE and association with ACE1 genotypes. Am J Transl Res. 2009;1:163–177. [PMC free article] [PubMed] [Google Scholar]
- 43.Ellul J, Archer N, Foy CM, Poppe M, Boothby H, Nicholas H, Brown RG, Lovestone S. The effects of commonly prescribed drugs in patients with Alzheimer's disease on the rate of deterioration. J Neurol Neurosurg Psychiatry. 2007;78:233–239. doi: 10.1136/jnnp.2006.104034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung CM, Wang RY, Chen JW, Fann CS, Leu HB, Ho HY, Ting CT, Lin TH, Sheu SH, Tsai WC, Chen JH, Jong YS, Lin SJ, Chen YT, Pan WH. A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J. 2010 doi: 10.1038/tpj.2009.70. [DOI] [PubMed] [Google Scholar]
- 45.Khachaturian AS, Zandi PP, Lyketsos CG, Hayden KM, Skoog I, Norton MC, Tschanz JT, Mayer LS, Welsh-Bohmer KA, Breitner JC. Antihypertensive medication use and incident Alzheimer disease: the Cache County Study. Arch Neurol. 2006;63:686–692. doi: 10.1001/archneur.63.5.noc60013. [DOI] [PubMed] [Google Scholar]
- 46.Li NC, Lee A, Whitmer RA, Kivipelto M, Lawler E, Kazis LE, Wolozin B. Use of angiotensin receptor blockers and risk of dementia in a predominantly male population: prospective cohort analysis. BMJ. 2010;340:b5465. doi: 10.1136/bmj.b5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohrui T, Matsui T, Yamaya M, Arai H, Ebihara S, Maruyama M, Sasaki H. Angiotensin-converting enzyme inhibitors and incidence of Alzheimer's disease in Japan. J Am Geriatr Soc. 2004;52:649–650. doi: 10.1111/j.1532-5415.2004.52178_7.x. [DOI] [PubMed] [Google Scholar]
- 48.Ohrui T, Tomita N, Sato-Nakagawa T, Matsui T, Maruyama M, Niwa K, Arai H, Sasaki H. Effects of brain-penetrating ACE inhibitors on Alzheimer disease progression. 0028-3878. 2004;63:1324–1325. doi: 10.1212/01.wnl.0000140705.23869.e9. [DOI] [PubMed] [Google Scholar]
- 49.Rozzini L, Chilovi BV, Bertoletti E, Conti M, Del Rio I, Trabucchi M, Padovani A. Angiotensin converting enzyme (ACE) inhibitors modulate the rate of progression of amnestic mild cognitive impairment. Int J Geriatr Psychiatry. 2006;21:550–555. doi: 10.1002/gps.1523. [DOI] [PubMed] [Google Scholar]
- 50.Rozzini L, Vicini Chilovi B, Trabucchi M, Padovani A. Antihypertensive medications influence the rate of conversion from mild cognitive impairment to Alzheimer disease. Arch Neurol. 2008;65:993–994. doi: 10.1001/archneur.65.7.993. author reply 994–995. [DOI] [PubMed] [Google Scholar]
- 51.Sink KM, Leng X, Williamson J, Kritchevsky SB, Yaffe K, Kuller L, Yasar S, Atkinson H, Robbins M, Psaty B, Goff DC., Jr. Angiotensin-converting enzyme inhibitors and cognitive decline in older adults with hypertension: results from the Cardiovascular Health Study. Arch Intern Med. 2009;169:1195–1202. doi: 10.1001/archinternmed.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]