Abstract
A20 is an anti-inflammatory protein linked to multiple human autoimmune diseases and lymphomas. A20 possesses a deubiquitinating motif and a zinc finger, ZF4, that binds ubiquitin and supports its E3 ubiquitin ligase activity. To understand how these activities mediate A20’s physiological functions, we generated two lines of gene-targeted mice, abrogating either A20’s deubiquitinating activity (Tnfaip3OTU mice) or A20’s ZF4 (Tnfaip3ZF4 mice). Both Tnfaip3OTU and Tnfaip3ZF4 mice exhibited increased responses to TNF and sensitivity to colitis. A20’s C103 deubiquitinating motif restricted both K48- and K63-linked ubiquitination of receptor interacting protein 1 (RIP1). A20’s ZF4 was required for recruiting A20 to ubiquitinated RIP1. A20OTU proteins and A20ZF4 proteins complemented each other to regulate RIP1 ubiquitination and NFκB signaling normally in compound mutant Tnfaip3OTU/ZF4 cells. This complementation involved homodimerization of A20 proteins, and we have defined an extensive dimerization interface in A20. These studies reveal how A20 proteins collaborate to restrict TNF signaling.
Introduction
Ubiquitination has emerged as a potent and complex mechanism for regulating cell signaling (Pickart and Fushman, 2004). Attachment of either single ubiquitin molecules or polymeric ubiquitin chains to signaling proteins induces their association with degradative proteasomes, lysosomal compartments, or other proteins that propagate signals toward nuclear transcription factors (Chen and Sun, 2009). These diverse outcomes are largely specified by polyubiquitin chains whose units are polymerized via epsilon amino groups of distinct lysine residues (or the N-terminal amino group) on ubiquitin (e.g., K11, K48, K63). Distinct types of polyubiquitin chains can be recognized by ubiquitin binding proteins via combinations of a variety of ubiquitin interacting motifs (Sims et al, 2009, Sims and Cohen, 2009). Unanchored ubiquitin chains, chains that are not covalently attached to signaling proteins, are also important in the propagation of signaling (Xia et al, 2009). Ubiquitination events are regulated by enzymes that orchestrate the attachment or removal of ubiquitin chains from proteins. Ubiquitin chains are built with combinations of E1, E2 and E3 enzymes, while these chains are degraded or removed by deubiquitinating enzymes (DUBs). While these broad outlines of ubiquitination have been partly dissected in cell-free studies, the mechanisms by which ubiquitin modifying enzymes recognize and modify ubiquitinated signaling complexes and how their functions are integrated in cells are poorly understood.
A20 is a potent regulator of several innate immune signals, including tumor necrosis factor (TNF), Toll-like receptor (TLR), nucleotide oligomerizatin domain (NOD) containing proteins and CD40 triggered NF-κB signals (Opipari et al, 1990, Lee et al, 2000, Boone et al, 2004, Hitotsumatsu et al, 2008, Tavares et al, 2010). A20 protein is encoded by the Tnfaip3 gene. A20 deficient (Tnfaip3−/−) mice develop spontaneous inflammation and perinatal lethality, which is largely abrogated by elimination of MyD88 adaptor-dependent signals (Lee et al, 2000, Turer et al, 2008). Single nucleotide polymorphisms (SNPs) of the human TNFAIP3 gene are strongly linked to susceptibility to rheumatoid arthritis, systemic lupus erythematosus, and psoriasis as well as multiple other inflammatory and autoimmune diseases (Plenge et al, 2007, Thompson et al, 2007, Musone et al, 2008, Graham et al, 2008, Nair et al, 2009, Ma and Malynn, 2012). In addition, biallelic mutations of this gene are pathogenetic in a variety of human lymphomas (Compagno et al, 2010, Kato et al, 2010, Malynn and Ma, 2010). Hence, the biological and clinical functions of this protein are of great interest.
In vitro studies suggest that A20 restricts NF-κB signals via deubiquitinating (DUB) activity, ubiquitin binding activity, and/or E3 ligase activity (Wertz et al, 2004; Bosanac et al, 2010). The N-terminus of A20 contains an ovarian tumor (OTU) domain that mediates its DUB activity. A20’s C103 based DUB activity preferentially cleaves K11, K48 and/or K63-linked ubiquitin chains, but not linear ubiquitin chains (Boone et al, 2004, Wertz et al, 2004, Bosanac et al, 2010, Lin et al, 2008, Komander and Barford, 2008). A20 appears to remove K63 chains from receptor interacting protein 1 (RIP1) and tumor necrosis factor receptor-associated factor 6 (TRAF6, providing potential mechanisms for how A20 may restrict signaling pathways utilizing these proteins (Boone et al, 2004, Wertz et al, 2004; Lin et al, 2008, Komander and Barford, 2008). A20 may also utilize its C103 DUB motif to inhibit E2-E3 enzyme interactions, thereby limiting synthesis of ubiquitin chains (Shembade et al, 2010). However, studies with N-terminal A20 constructs containing the C103 motif suggests that this half of the protein does not restrict tumor necrosis factor (TNF) induced NF-κB signaling (Heyninck and Beyaert, 1999). In addition, none of these studies utilized cells bearing physiologically expressed A20 protein. Thus, the physiological roles of A20’s DUB activity in restricting NF-κB signals are unclear.
The C-terminal half of the A20 protein contains seven zinc fingers. The fourth finger, ZF4, has been shown to bind ubiquitin chains and support E3 ligase activity (Wertz et al, 2004, Bosanac et al, 2010). Ubiquitin binding by this motif resembles ubiquitin binding by a similar zinc finger in the E3 ubiquitin ligase Rabex 5, a guanine nucleotide exchange factor (Lee et al, 2006, Penengo et al, 2006, Mattera et al, 2006). A20’s ZF4 based E3 ligase activity may support K48 ubiquitination of RIP1 or ubiquitination of E2 enzymes such as ubiquitin conjugating enzyme-5 (Ubc5) or Ubc13 (Wertz et al, 2004, Shembade et al, 2010). The localization of both ubiquitin binding and E3 ligase activity to ZF4 suggests that these functions are intimately related, however this relationship is incompletely understood. Moreover, as with A20’s C103 based deubiquitination, the physiological functions of the ZF4 motif and its relationship to A20’s C103 have not been investigated in vivo.
A20 expression is dynamically induced by NF-κB dependent signals, and A20 expression is precisely regulated to maintain cellular homeostasis (Krikos et al, 1992, Lee et al, 2000). Progressively higher heterologous A20 expression inhibits TNF induced NF-κB signaling in a dose dependent fashion, and hypomorphic expression of endogenous A20 renders murine cells hypersensitive to various ligands (Werner et al, 2008, Tavares et al, 2010, Hammer et al, 2011). Hypomorphic expression or function of A20 may also confer susceptibility to human disease (Musone et al, 2008, Adrianto et al., 2011). Hence, to define the physiological functions of A20’s ubiquitin modifying functions, we have generated gene-targeted mice bearing either a point mutation that abrogates A20’s DUB activity or point mutations that abrogate A20’s ZF4 based E3 ligase or ubiquitin binding activity. These gene targeted mice should express A20 at physiological and properly regulated expression levels. We have used these mice to determine the physiological functions of these motifs in regulating innate immune signals.
Results
Generation of Tnfaip3OTU and Tnfaip3ZF4 mice
To determine the physiological functions of A20’s DUB activity, we recombineered a gene targeting construct encoding a cysteine to alanine mutation at amino acid residue 103, the catalytic cysteine in A20’s OTU domain required for its deubiquitinating function, as well as an intronic LoxP flanked neomycin selection cassette (Suppl. Fig. 1A) (Boone et al, 2004; Wertz et al, 2004, Lin et al, 2008, Komander and Barford, 2008). This construct was introduced into PRXB6T (C57BL/6J inbred) embryonic stem (ES) cells. After identification of properly targeted ES cells that underwent homologous recombination, LoxP flanked neomycin sequences were deleted in vitro by transfection with a Cre expression construct. Selected ES cells bearing the C103A point mutation and lacking the neomycin cassette were then used to generate germline mice, hereafter referred to as Tnfaip3OTU mice.
To abrogate A20’s ZF4-based E3 ligase and ubiquitin binding activity, we generated a second construct encoding tandem cysteine to alanine point mutations in this motif: Cys 609 and Cys 613. These mutations abrogate A20’s E3 ligase activity (Wertz et al, 2004). This construct was also introduced into PRXB6T ES cells, targeted ES cells were selected, Cre-mediated removal of the LoxP-neomycin cassette was performed in vitro, and the final selected ES cells bearing the ZF4 tandem point mutations were used to generate Tnfaip3ZF4 mice (Suppl. Fig. 1B).
In contrast to Tnfaip3−/− mice that develop perinatal cachexia and lethality, homozygous Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice were both grossly normal for at least 4 months of life (data not shown). Thus, neither A20’s C103 nor its ZF4 motifs are required for preventing spontaneous cachexia and premature death. Flow cytometric analyses of lymphoid tissues from 2 month old mice revealed that both Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice contained normal numbers of lymphocytes (data not shown). As these mice age to 6 months, they gradually developed splenomegaly and accumulated modestly increased numbers of myeloid cells and lymphocytes, suggesting that both A20’s C103 and ZF4 motifs regulate immune homeostasis (Fig. 1A).
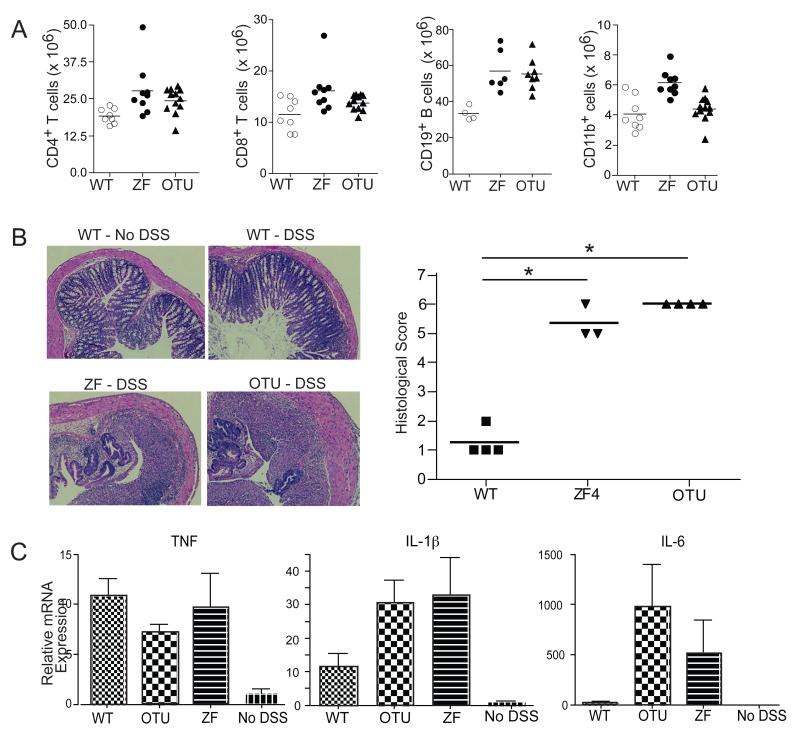
Figure 1. Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice exhibit mild immune dysplasia with age and fail to restrict inflammation in DSS colitis.
(A) Flow cytometric quantitation of splenic T (CD4+ and CD8+), B (CD19+), and myeloid (CD11b+) cells from older (6 month old) mice of indicated genotypes. Data are representative of 3-4 mice per genotype. (B, C) DSS responses of young adult (2 month old) Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice. (B) Hematoxylin eosin stain of colon sections and computed histological scores of wildtype, Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice treated with 3% oral DSS for 5 days. (C) Quantitative PCR (QPCR) analyses of expression of indicated cytokine mRNAs, normalized to actin mRNA, from intestinal tissues of the same mice described in (B). * indicates p < 0.05 by ANOVA. Bars represent means and standard deviations. Significant (p <0.05) differences noted in between WT and mutant cells in IL-6 and IL-1β but not TNF production. Data are representative of 3 independent experiments using at least 3 mice per genotype. See also Figure S1.
A20 is an inducible molecule that may be particularly important for restricting inflammatory signals. Accordingly, we challenged Tnfaip3OTU/OTU and A20ZF4/ZF4 mice with oral dextran sulfate sodium (DSS). DSS treatment caused greater intestinal inflammation in both Tnfaip3OTU/OTU and A20ZF4/ZF4 mice compared to wild type control mice, as measured by a combinatorial histological score (Fig. 1B). Both mutant mice strains also induced greater expression of interleukin-1 (IL-1) and IL-6, but not TNF, in intestinal tissues (Fig. 1C). Thus, both A20’s C103 and ZF4 motifs restrict inflammatory responses in vivo.
A20’s C103 and ZF4 motifs restrict TNF responses
As A20 regulates TNF signals, we examined the role of A20’s C103 and ZF4 motifs in regulating TNF responses. Injection of a sublethal dose of TNF into Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice induced greater amounts of serum IL-6 and monocyte chemotactic protein-1 (MCP-1) than in control mice, suggesting that both C103 and ZF4 are required for restricting TNF responses in vivo (Fig. 2A). To more directly determine how these motifs restrict TNF signals, we stimulated embryonic fibroblasts (MEFs) from Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 mice with TNF in vitro. Both Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells produced elevated amounts of the NF-κB dependent IL-6 and A20 mRNAs within one hour of TNF stimulation, consistent with increased NF-κB signaling in these cells (Fig. 2B). The amounts of A20 protein were induced by TNF to a greater degree in Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells than control cells, suggesting that increased Tnfaip3 mRNA in these cells led to increased A20 protein (Fig. 2C). Moreover, these results suggest that both A20OTU and A20ZF4 mutant proteins are similarly stable as wild type A20 protein. The relative amounts of NF-κB dependent mRNAs produced by these cells correlated with the degree of NF-κB signaling reflected by phospho-IκBα and IκBα protein levels as well as IKK kinase assays (Fig. 2C, 2D). Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells produced less of the NF-κB dependent mRNAs IL-6 and cellular inhibitor of apoptosis protein 2 (cIAP2), and exhibited less NF-κB signaling than Tnfaip3−/− cells, suggesting that neither A20’s C103 motif nor its ZF4 motif are singly responsible for all of A20’s functions during TNF signaling (Suppl. Fig. 2A, 2B). Immunoblotting studies of pJNK, p38, and pERK kinase signaling revealed normal signaling activity in Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells (Fig. 2E). Thus, A20’s C103 and ZF4 motifs regulate TNF responses by regulating the kinetics of NF-κB signaling.
Figure 2. A20’s OTU and ZF4 motifs restrict TNF induced NF-κB signals.
(A) ELISA analyses of serum production of IL-6 and MCP-1 in mice of indicated genotypes after intraperitoneal injection of TNF. * indicates p < 0.05. Data are representative of 3 independent experiments of 3 mice per genotype. (B) QPCR analyses of A20 and IL-6 mRNA expression by MEFs of indicated genotypes at indicated times after TNF treatment. Data are normalized to actin mRNA. Data are represented as means +/− standard deviations. Significant differences (p < 0.05) present between WT/WT and both ZF4/ZF4 and OTU/OTU mutant cells at 1 and 2 hours. (C) Immunoblot analyses of A20, pIκBα, and IκBα expression by MEFs of indicated genotypes at indicated times after TNF treatment. Ratios of pIκbα/IκBα expression are shown below as reflection of NF-κB signaling activity for selected time points. Actin expression is shown as loading control. (D) IKK kinase assay using lysates from TNF induced MEFs of indicated genotypes at indicated time points. Quantitation of pIκBα amounts normalized to IKKβ expression in IPs is shown below. (E) Immunoblot analyses of JNK, p38, and pERK signaling in TNF stimulated MEFs of indicated genotypes. See also Figure S2.
RIP1 ubiquitination supports TNF induced NF-κB signaling, and A20 restricts RIP1 ubiquitination (Ea et al, 2006, Wu et al, 2006, Wertz et al, 2004). Accordingly, we measured TNF receptor (TNFR) induced RIP1 ubiquitination in Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells by immunoprecipitating TNFR complexes and immunoblotting for RIP1. Greater amounts of ubiquitinated RIP1 were associated with TNFR1 in both Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells compared to wild type (WT) cells 10 and 15 minutes after TNF treatment (Fig. 3A, Suppl. Fig 2C). As distinct types of ubiquitin chains are associated with diverse outcomes of modified proteins, and as A20 has been shown to restrict K63-linked polyubiquitin chains and build K48 chains, our results raised the question of what types of ubiquitin chains are present on RIP1 in these cells. We characterized the ubiquitin chains on TNFR associated RIP1 molecules by performing serial TNFR and RIP1 immunoprecipitations (IPs) on TNF stimulated cells followed by immunoblotting with ubiquitin linkage-specific antibodies. These studies revealed that both Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells contained increased amounts of both K48 and K63-linked ubiquitin chains on RIP1 molecules (Fig. 3B). These results suggest that A20’s C103 deubiquitinating motif restricts both K48 and K63-linked ubiquitination of RIP1.
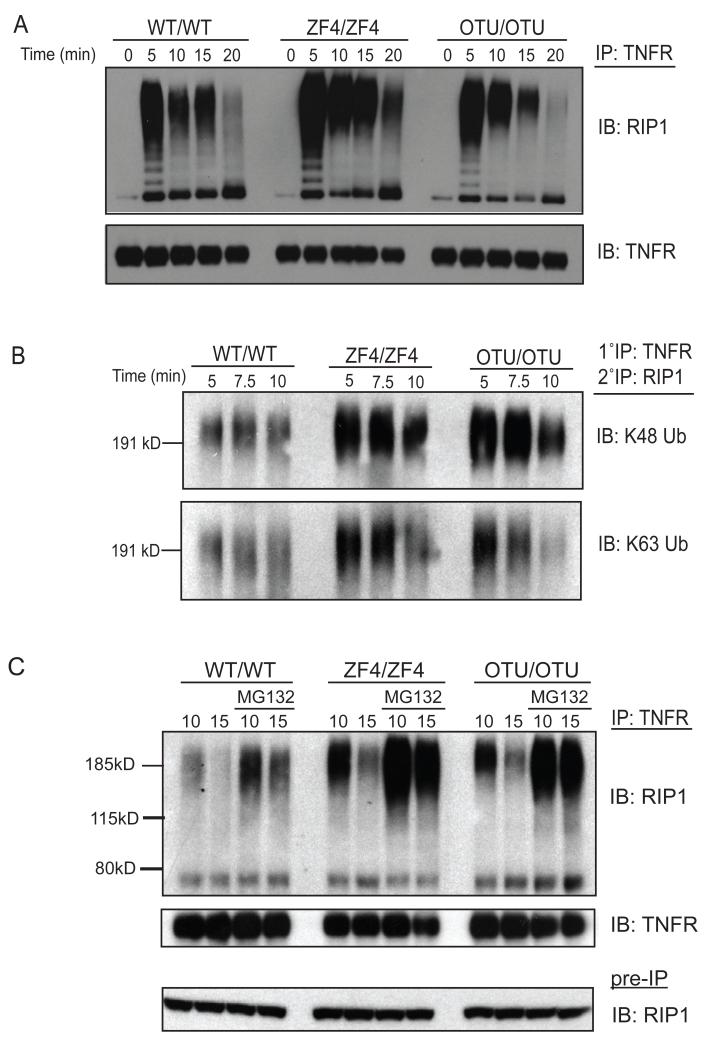
Figure 3. A20’s OTU and ZF4 motifs restrict TNF induced RIP1 ubiquitination.
(A) Immunoblot analyses of RIP1 ubiquitination in TNFR IPs from indicated cells at indicated time points after TNF treatment. TNFR protein expression in IPs shown below as control. Note higher molecular weight forms of RIP1 reflecting ubiquitinated RIP1. (B) Immunoblot analysis of K48- and K63-linked ubiquitin chains on RIP1 molecules after sequential IP with first anti-TNFR and then anti-RIP1 (2°) of lysates from TNF treated cells. (C) Immunoblot analyses of TNFR IPs as in (B), with the exception that indicated samples were treated with MG-132. Pre-IP quantities of RIP1 protein shown below as control. All experiments were performed at least three times.
As A20’s ZF4 motif has been proposed to ligate K48-ubiquitin chains to RIP1, the presence of increased ubiquitinated RIP1 in A20ZF4/ZF4 cells could be explained by decreased ubiquitin mediated turnover of RIP1 proteins in these cells (Wertz et al, 2004). We thus assayed RIP1 ubiquitination of TNF stimulated cells in the presence of the proteasome inhibitor MG-132. These experiments revealed that both Tnfaip3OTU/OTU and Tnfaip3ZF4/ZF4 cells continued to exhibit increased RIP1 ubiquitination (Fig. 3C). Thus, A20’s ZF4 motif is unexpectedly required for restricting TNF induced RIP1 ubiquitination.
A20’s ZF4 motif recruits A20 to ubiquitinated RIP1 in TNFR signaling complexes
Increased ubiquitination of RIP1 in A20ZF4/ZF4 cells is not readily explained by a reduction in A20’s ZF4 based ligation of ubiquitin chains on RIP1. We thus investigated alternative mechanisms by which mutation of A20’s ZF4 might cause increased RIP1 ubiquitination. A20’s ZF4 resembles a zinc finger in Rabex-5 that binds ubiquitin, and ZF4 directly binds ubiquitin chains (Lee et al, 2006, Penengo et al, 2006, Mattera et al, 2006, Bosanac et al, 2010). Accordingly, we asked whether our ZF4 mutation abrogates A20’s ability to bind ubiquitin. Binding studies with recombinant C-terminal A20 proteins and ubiquitin chains demonstrated that A20 binds K63-linked ubiquitin chains and that the dual cysteine to alanine substitutions we generated in A20’s ZF4 motif (A20ZF4 proteins) eliminated A20’s ability to bind these chains (Fig. 4A). To determine whether this ubiquitin binding activity is important for A20’s ability to bind physiologically ubiquitinated RIP1 proteins, we incubated lysates from TNF stimulated A20−/− cells with either GST-A20 or mutant GST-A20ZF4 proteins, and asked whether RIP1 molecules bound to these A20 proteins. These experiments revealed that wild type GST-A20 preferentially bound to ubiquitinated rather than unmodified RIP1 proteins (compare WT GST-A20 with input lysate, Fig. 4B). By contrast, A20ZF4 mutant proteins interacted only with unmodified RIP1 proteins (Fig. 4B). Preferential co-precipitation of ubiquitinated RIP1 proteins with wild type A20 protein in these assays was unlikely to reflect A20 ZF4 dependent E3 ligase activity upon RIP1 as these experiments were performed in the presence of N-ethyl maleimide (NEM) at 4°C, precluding E3 ligase activity. These studies indicate that A20 utilizes its ZF4 motif to bind ubiquitinated RIP1.
Figure 4. A20 requires ZF4 to bind K63-linked ubiquitin chains and ubiquitinated RIP1.
(A) Binding of recombinant GST-A20 proteins to recombinant K63-linked ubiquitin chains. Recombinant C-terminal (AA 370-776) GST-WT and A20ZF4 mutant proteins were incubated with increasing concentrations of recombinant K63-linked ubiquitin chains. Glutathione bead bound proteins were then analyzed by immunoblotting for ubiquitin (top panel). Immunoblot for GST proteins are shown as controls (bottom panel). (B) GST pull down of ubiquitinated RIP1 from cell lysates. Cell lysates from TNF stimulated A20−/− MEFs were incubated with the indicated C-terminal GST-A20 proteins, after which glutathione bead bound proteins were analyzed by immunoblotting for RIP1. Input cell lysates are shown in right two lanes. Immunoblot for GST proteins shown below as controls (bottom panel). (C) Recruitment of endogenous A20 proteins to TNFR signaling complexes. MEFs of the indicated genotypes were stimulated with TNF for the indicated times, immunoprecipitated with anti-TNFR antibody, and analyzed by immunoblotting for endogenous A20 proteins. Immunoblot for TNFR protein in IPs shown below as control. All experiments were performed at least two times.
To further investigate the recruitment of A20 proteins to TNFR signaling proteins, we measured the recruitment of endogenous A20 proteins to TNFR complexes in Tnfaip3ZF4/ZF4, Tnfaip3OTU/OTU and WT cells. Despite being expressed at higher amounts than A20OTU and WT A20 proteins, A20ZF4 proteins were recruited poorly to TNFR complexes after TNF stimulation (Fig. 4C). By contrast, A20OTU proteins are recruited nearly normally to TNFR complexes (Fig. 4C). Taken together, these findings indicate that A20 uses its ZF4 motif to bind ubiquitinated RIP1 in TNFR complexes. As poor recruitment of A20ZF4 proteins to ubiquitinated TNFR signaling complexes would prevent A20’s OTU based DUB function from removing ubiquitin chains from RIP1, this mechanism can explain why Tnfaip3ZF4/ZF4 cells exhibit increased RIP1 ubiquitination.
A20OTU proteins complement A20ZF4 proteins in dimers during TNF responses of Tnfaip3OTU/ZF4 compound mutant cells
A20’s C103 based deubiquitination activity may be biochemically coupled to its ZF4 based E3 ubiquitin ligase activity. For example, A20 might exchange K63-linked chains for K48 linked chains on RIP1. To better understand how A20’s C103 and ZF4 motifs may coordinate A20’s ubiquitin dependent functions, we interbred Tnfaip3OTU/OTU with Tnfaip3ZF4/ZF4 mice and analyzed TNF responses of cells from the resulting compound Tnfaip3OTU/ZF4 mice. Compound mutant Tnfaip3OTU/ZF4 cells should express physiologically regulated A20 proteins divided equally between A20OTU and A20ZF4 proteins. Compound mutant Tnfaip3OTU/ZF4 mice exhibited less myeloid expansion than Tnfaip3ZF4 mice, suggesting that A20OTU complementation of A20ZF4 proteins can also rescue TNF dependent homeostasis in vivo (Fig. 5A). In contrast to either Tnfaip3OTU/OTU or Tnfaip3ZF4/ZF4 mouse embryonic fibroblasts (MEFs), stimulation of Tnfaip3OTU/ZF4 cells with TNF resulted in normal amounts of the NF-κB dependent mRNAs IL-6 and A20 over a variety of TNF doses (Fig. 5B). Thus, A20OTU proteins and A20ZF4 proteins complement each other in trans in Tnfaip3OTU/ZF4 cells.
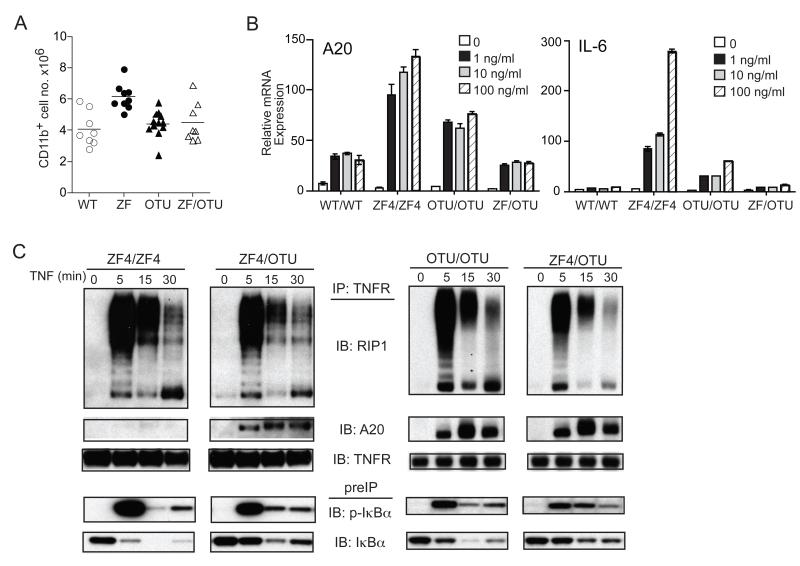
Figure 5. A20OTU mutant proteins rescue recruitment defects and NF-κB signaling defects of A20ZF4 proteins in compound Tnfaip3OTU/ZF4 MEFs.
(A) Flow cytometric analyses of splenic myeloid cells from compound Tnfaip3OTU/ZF4 and control mice. (B) Compound Tnfaip3OTU/ZF4 MEFs exhibit normal TNF responses. QPCR analyses of IL-6 and A20 mRNA expression in TNF treated MEFs of the indicated genotypes. TNF doses are indicated. Error bars represent standard deviations. (C) Compound Tnfaip3OTU/ZF4 cells rescue RIP1 ubiquitination, A20 recruitment, and NF-κB signaling defects seen in Tnfaip3ZF4/ZF4 cells. Immunoblot analyses of TNFR associated RIP1 ubiquitination and A20 recruitment in TNF stimulated cells of the indicated genotypes after TNF stimulation for the indicated time points. TNFR immunoblot of TNFR IP shown as IP loading control. Immunoblots of pIκBα and Iκbα expression in pre-IPs shown as indicators of NF-κB signaling. Lines represent means in A; bars represent mean +/− standard deviations in B. All experiments were performed at least three times.
The observation that A20OTU and A20ZF4 proteins complement each other to normally regulate NF-κB signaling in Tnfaip3OTU/ZF4 cells suggests that A20OTU proteins rescue the aberrant RIP1 ubiquitination and defective homing of A20ZF4 proteins observed in Tnfaip3ZF4/ZF4 cells. To test this prediction, we assayed RIP1 ubiquitination in TNFR immunoprecipitates from Tnfaip3OTU/ZF4 as well as Tnfaip3ZF4/ZF4 and Tnfaip3OTU/OTU cells. These experiments revealed that RIP1 ubiquitination in compound mutant Tnfaip3OTU/ZF4 cells was reduced when compared to Tnfaip3ZF4/ZF4 cells (Fig. 5C). Thus, A20OTU and A20ZF4 proteins collaborate to properly regulate RIP1 ubiquitination in Tnfaip3OTU/ZF4 cells.
As A20ZF4 proteins are recruited poorly to TNFR signaling complexes, the presence of normal NF-κB signaling in Tnfaip3OTU/ZF4 cells also raises the interesting possibility that A20OTU proteins may dimerize with A20ZF4 proteins and recruit the latter to TNFR signaling complexes in Tnfaip3OTU/ZF4 cells. Accordingly, we tested the recruitment of A20 proteins to TNFR immunoprecipitates in TNF stimulated Tnfaip3OTU/ZF4, Tnfaip3ZF4/ZF4, and Tnfaip3OTU/OTU cells. These experiments revealed that A20 proteins were recruited normally to TNFR complexes in Tnfaip3OTU/ZF4 cells, in marked contrast to Tnfaip3ZF4/ZF4 cells (Fig. 5C, left panel). One possible interpretation of this result is that A20OTU proteins, which exhibit normal recruitment to TNFR, are selectively recruited to TNFR complexes in Tnfaip3OTU/ZF4 cells. However, the kinetics of RIP1 ubiquitination and NF-κB signaling in compound heterozygote Tnfaip3OTU/ZF4 cells were normal--in contrast to Tnfaip3OTU/OTU cells--suggesting that A20 proteins at the TNFR complex in Tnfaip3OTU/ZF4 cells are not predominantly A20OTU proteins (Fig. 5C). The more likely explanation for normal A20 protein recruitment and NF-κB signaling in Tnfaip3OTU/ZF4 cells is that A20OTU proteins form hetero-oligomers with A20ZF4 proteins, allowing the intact ZF4 domains of A20OTU proteins to recruit A20OTU/ZF4 oligomers to TNFR complexes. The successful recruitment of A20OTU/ZF4 oligomers to TNFR signaling complexes may then allow A20 to properly regulate RIP1 ubiquitination.
The apparent ability of A20OTU mutant proteins to recruit A20ZF4 proteins to TNFR signaling complexes suggests that A20 proteins dimerize under physiological conditions. To directly test this idea, we transfected wild type or mutant A20 proteins bearing distinct epitope tags into cells, immunoprecipitated with one tag and immunoblotted with the alternative tag. These studies revealed that full-length A20 proteins, including A20OTU and A20ZF4 proteins, co-precipitated comparably as oligomers (Fig. 6A). This result suggests that A20 proteins oligomerize in cells in a manner that requires neither A20’s C103 nor its ZF4 motifs. Thus, ZF4 mediated ubiquitin binding is not required for A20 oligomerization.
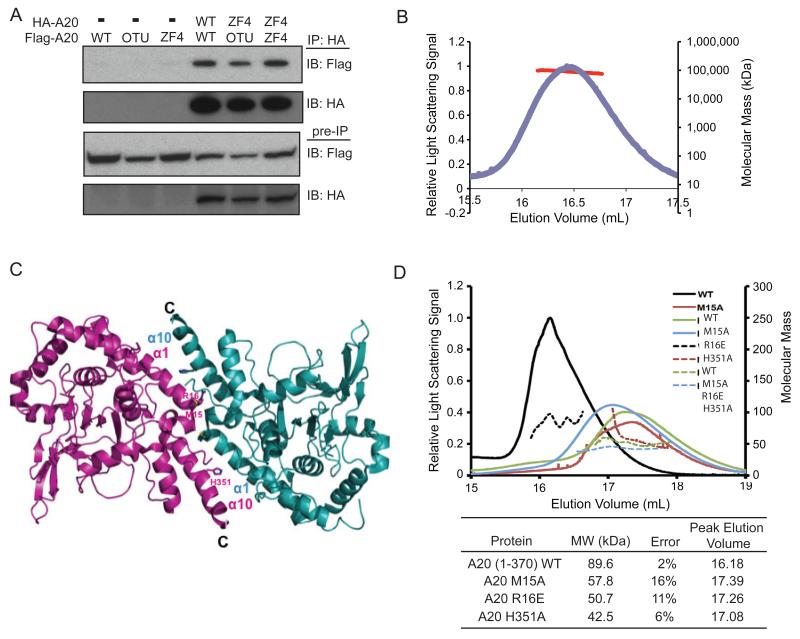
Figure 6. A20 proteins form dimers.
(A) Co-precipitation of HA-A20 proteins with FLAG-A20 proteins from cells. Immunoprecipitation of HA-A20 followed by immunoblotting of FLAG-A20 proteins from co-transfected cells. Pre-IP expression of transfected proteins are shown below as controls. (B) Multi-angle light scattering (MALS) analysis of N-terminal (residues 1-370) A20 proteins. The calculated mass of the His-A20 monomer is 45.5 kDa, and the measured mass is 89.6 kDa. (C) Ribbon diagram of conserved dimers of N-terminal A20 proteins. Distinct A20 monomers are shown in Magenta and Cyan. α1 and α10 helices forming the intermolecular interface are labeled. (D) MALS analyses of N-terminal A20 proteins bearing predicted dimerization mutations (M15A, R16E, and M351A). Upper panel: relative light scattering intensities (solid lines) and molecular mass distribution (dashed lines) of wild type (WT), A20 (residues 1-370), and indicated dimerization mutants are plotted as functions of elution volume (mL) on a Superdex 200 10/30 column. Lower panel: tabulated measured molecular mass, experimental errors and peak elution volumes of WT and mutant A20 proteins. Experiments were performed at least three times.
The oligomerization of A20 proteins in cells may involve a number of A20 binding partners. A20 proteins might also directly form complexes in vitro. To test the latter hypothesis, we determined the molecular mass of recombinant A20 proteins by gel chromatography and multi-angle light scattering (MALS). An N-terminal A20 protein bearing the OTU domain (residues 1-370) formed dimers in solution with a measured mass of ~89 kDa, while the calculated monomer mass of this protein is 45.5 kDa (Fig. 6B). Moreover, re-examination of the OTU portion of the A20 protein in two crystal structures, P32 (Lin et al, 2008) and P21 (Komander and Barford, 2008), revealed that there were six and four molecules, respectively, per crystallographic asymmetric unit. These ten molecules formed five conserved dimers, with interfaces formed mostly by the α1 and α10 helices of the OTU structure (Table 1, Fig. 6C). The dimer interface is extensive, burying ~800Å2 surface areas per monomer. Residues that bury the largest surface areas include M15, R16 and H351, and mutation of these residues, M15 to alanine (M15A), R16 to glutamate (R16E), or H351 to alanine (H351A), compromised A20 dimerization, with H351A having the most drastic effect (Fig. 6D). Thus, A20 proteins directly form homodimers. Taken together, these findings indicate that A20’s ZF4 motif recruits A20 dimers to ubiquitinated RIP1 signaling complexes during TNF signaling. These dimers may bring multiple copies of A20’s ubiquitin modifying activities, as well as potential binding partners, to ubiquitinated complexes.
Table 1. A20 OTU domains form conserved dimers in crystal structures.
Structure-based alignments among the three A20 dimers in the PDB coordinates 3DKB and the two A20 dimers in the PDB coordinates 2VFJ are shown.
| 3DKB, chains C and F | |
| 3DKB, chains A and D | 704 aligned Cα, 0.40 Å |
| 3DKB, chains B and E | 704 aligned Cα, 0.44 Å |
| 2VFJ, chains A and D | 623 aligned Cα, 1.1 Å |
| 2VFJ, chains B and C | 623 aligned Cα, 0.75 Å |
Discussion
Our studies of Tnfaip3ZF4/ZF4, Tnfaip3OTU/OTU, and Tnfaip3OTU/ZF4 compound mutant mice reveal several facets of A20’s regulation of TNF signaling. We have discovered a role for A20’s ZF4 motif in recruiting A20 proteins to ubiquitinated RIP1 during TNF signaling. We have found that A20’s C103 and ZF4 mutant proteins complement each other in cells. This complementation is facilitated by dimerization of A20 proteins and we have defined a dimerization interface in A20. These motifs perform distinct biochemical functions in regulating TNF signals.
While prior studies suggested that A20’s N-terminal OTU domain is not required for A20’s ability to restrict TNF signals, our current experiments demonstrate that A20’s C103 based DUB activity restricts TNF induced signals (Song et al, 1996, Heyninck and Beyaert, 1999). Our serial TNFR and RIP1 IP experiments with Tnfaip3OTU/OTU cells revealed that RIP1 proteins bear increased amounts of both K48- and K63-linked ubiquitin chains in Tnfaip3OTU/OTU cells, suggesting that A20’s DUB activity removes both types of chains from RIP1 in cells. These chains might be removed together if they are present in mixed K48- and K63-linked ubiquitin chains. Distinguishing these physiological chain conformations will require more biochemically detailed analyses of physiological signaling complexes.
A20’s C103 may also support degradation of Ubc5hc and Ubc13 proteins approximately 4-6 hours after TNF stimulation (Shembade et al, 2010). This function of A20’s C103 appears temporally distinct from the more acute differences in RIP1 ubiquitination we have observed 10-15 minutes after TNF stimulation. We have not observed differences in expression of these E2 enzymes in Tnfaip3OTU/OTU cells at acute time points (e.g., 10-15 minutes) (data not shown). Nor have we observed acute recruitment defects of A20OTU proteins to TNFR signaling complexes. Hence, A20’s C103 acute functions regulating RIP1 ubiquitination appear distinct from apparently later functions regulating E2 enzyme stability. The net physiological function of A20’s C103 vis a vis RIP1 is to limit both K48 and K63 ubiquitination of this protein. Failure to perform this function leads to increased IKK activation and NF-κB signaling.
A20’s ZF4 is a complex motif that has been shown to bind ubiquitin, build K48 ubiquitin chains on RIP1, and support degradation of E2 enzymes (Wertz et al, 2004, Bosanac et al, 2010, Shembade et al, 2010). Our results reveal that A20’s ZF4 is critical for mediating A20’s recruitment to ubiquitinated RIP1 in TNFR signaling complexes. Thus, A20’s ZF4 may support several functions for A20. One potential mechanism by which this motif might support several functions would be to collaborate with other A20 motifs, including other A20 zinc fingers, to bind different ubiquitinated molecules. For example, A20’s ZF1 and ZF2 appear to support binding to RIP1, while A20’s ZF7 binds linear and K63 linked polyubiquitin chains (Skaug et al, 2011, Tokunaga et al, 2012, Verhelst et al, 2012). While the principles by which ubiquitin binding proteins recognize distinct substrates are poorly understood, recent studies suggest that these proteins can recognize distinct conformations of ubiquitin chains via multiple ubiquitin binding motifs (Sims and Cohen, 2009; Sims et al, 2009). Thus, A20’s ZF4 motif may contribute to A20’s binding to ubiquitinated E2, ubiquitinated RIP1, and potentially other ubiquitinated species.
Our observation of increased K48 ubiquitinated RIP1 in A20ZF4/ZF4 cells--even in the presence of proteasome inhibition--was an unexpected finding given A20’s ZF4 mediated support of E3 ligase function building K48 chains. The most straightforward interpretation of our findings is that A20ZF4 proteins fail to bind ubiquitinated RIP1 in TNFR signaling complexes and thus fail to deubiquitinate ubiquitinated RIP1 in these complexes. As the net effect of A20’s ZF4 mutation on RIP1 ubiquitination is increased—rather than diminished--ubiquitination, the predominant physiological function of this motif is to limit RIP1 ubiquitination during TNF signaling. In addition, our studies indicate that other E3 ligases such as cIAPs or TRAFs likely build ubiquitin chains on RIP1 during TNF signaling. Future studies may unveil greater complexities in types of chains and ubiquitination sites on RIP1. A combination of recruitment and E3 ligase functions may contribute to A20’s ZF4’s roles in regulating RIP1 ubiquitination. Overall, our studies unveil a critical role for A20’s ZF4 in recruiting A20 to ubiquitinated RIP1 independently of its role in supporting E3 ligase activity.
Our studies of compound mutant Tnfaip3OTU/ZF4 cells reveal that A20 proteins dimerize in vivo. Successful recruitment of A20OTU and A20ZF4 proteins to the TNFR signaling complex in these cells indicates that A20 proteins require neither C103 nor ZF4 motifs to dimerize, and these dimers require only a single intact ZF4 motif to be recruited to TNFR signaling complexes. These findings suggest that coordination between A20’s deubiquitinating, E3 ligase and ubiquitin binding functions occur in higher order complexes rather than within a single A20 molecule. Oligomerization of signaling complexes has emerged as an important principle in propagating activating signals (Krappmann and Scheidereit, 2005). Our studies indicate that oligomerization of negative regulatory enzymes may also be a general theme.
Our observations of A20 oligomers in cells led us to discover that A20 proteins form dimers with extensive interfaces. Biochemical identification of A20’s oligomerization and recruitment motifs provides additional opportunities for the regulation of A20’s functions. Dimerization of the ubiquitin hydrolase UCH-L1 influences its ability to function as a ligase, so dimerization of A20 may also regulate its enzymatic functions (Liu et al, 2002). Further studies of A20 proteins should reveal important insights into how A20 coordinates its ubiquitin modifying functions.
A20’s C103 and ZF4 motifs have been associated with A20’s repression of TNF induced NF-κB signals (Wertz et al, 2004, Shembade et al, 2010). Thus, Tnfaip3OTU and A20ZF4 mice might be expected to resemble Tnfaip3−/− mice (Lee et al, 2000). However, Tnfaip3−/− mice develop spontaneous multi-organ inflammation and perinatal lethality, while both Tnfaip3OTU and Tnfaip3ZF4 mice exhibit little spontaneous disease. As these mice have all been analyzed on inbred C57BL/6J backgrounds in the same facility, these differences are unlikely to be strain or environment related. One potential explanation for this difference is that A20’s DUB and E3 ligase activities partly compensate for each other in vivo, so that mice expressing double mutant A20 proteins (i.e., OTU and ZF4 mutations within the same protein) would more closely resemble A20−/− mice. As A20’s C103 and ZF4 motifs are obviously tightly linked genetically, additional gene-targeting studies will be necessary to investigate these possibilities. Another explanation could be that other motifs of A20 perform critical functions that are important for regulating NF-κB signals and preserving immune homeostasis. For example, A20’s ZF7 has recently been described to restrict NF-κB signaling at the IKKγ complex, and this function involves binding of ZF7 to linear ubiquitin chains (Skaug et al, 2011, Tokunaga et al, 2012, Verhelst et al, 2012). In sum, the distinct functions of A20’s C103 and ZF4 motifs imply that they may impart distinct immune perturbations and disease susceptibilities. These functions may provide important insight into how A20 regulates diverse NF-κB signals (D. Baltimore 2011, Malynn and Ma, 2012). Given the variety of coding and non-coding A20 mutations that have been described in human diseases, understanding A20’s functions will be crucial for deciphering the pathophysiology of these diseases.
Materials and Methods
Generation of Tnfaip3OTU and Tnfaip3ZF4 mice
To generate gene A20OTU and A20ZF4 mice, we utilized two distinct bacterial artificial chromosomes (BACs) bearing the A20 gene from the C57BL/6J strain. Site directed mutagenesis was used to change the catalytic cysteine at amino acid 103 to an alanine to generate the OTU mutant gene targeting construct. In a separate construct, two cysteines (C609, C612) in A20’s ZF4 were mutated to alanines to generate the ZF4 gene targeting construct. These targeting constructs were transfected into PRXB6T (C57BL/6J) embryonic stem (ES) cells. Properly targeted ES cell clones from both constructs were identified by Southern analysis and transiently transfected with a plasmid expressing Cre enzyme to delete the floxed neomycin cassettes. Blastocyst injections of targeted ES cells were performed by the UCSF Transgenic Core. Chimeric mice were bred with C57BL/6J mice to obtain Tnfaip3ZF4 and Tnfaip3OTU mice on in inbred C57BL/6J background. All mouse handling was done according to the UCSF’s institutional guidelines.
Flow cytometry and ELlSA
Cell preparations, flow cytometric and ELISA analyses were performed as previously described (Tavares et al, 2010). All antibodies were purchased from BD Biosciences. Cells were analyzed by flow cytometry using LSRII (BD Biosciences) and Flowjo software (Tree Star).
Cell signaling assays
MEFs were derived from Tnfaip3OTU/OTU, Tnfaip3ZF4/AF4 and Tnfaip3OTU/ZF4 embryos as previously described (Oshima et al, 2009). MEFs were stimulated with 10ng/ml TNF, and lysed in lysis buffer (20mM Tris HCl pH 7.4, 150mM NaCl, 10% glycerol, 0.2% NP-40 supplemented with Roche protease inhibitors, phosphatase inhibitors (1mM NaV, 5mM NaF, 20mM ß-glycerol phosphate) and 10mM N-ethylmaleimide. Cells were lysed on ice for 20 minutes, and cleared by centrifugation at 14000 rpm for 20 minutes. For immunoprecipitation of the TNF receptor complex, cells were stimulated and lysed as above. Supernatants were immunoprecipitated with anti-TNFR antibody (R and D) and Protein G Dynabeads.
For the IKK kinase assay of TNF-treated MEFs, total cell lysates from repeatedly TNF-treated MEFs were immunoprecipitated with an anti-IKKγ antibody, and kinase activity was assessed using a GST-IκBα substrate. Comparable IKKβ protein in immunoprecipitated samples was confirmed by western blot.
Serial IP analyses of RIP1 ubiquitination
For sequential immunoprecipitation, cells were lysed in lysis buffer supplemented with 10μM MG-132 as above. For the primary immunoprecipitation, TNFR complexes were immunoprecipitated and washed twice with lysis buffer, twice with lysis buffer supplemented with 1M NaCl, and washed twice with lysis buffer. TNFR complexes were denatured and eluted with lysis buffer containing 6M Urea. Eluates were diluted 1:25 and immunoprecipitated with an anti-RIP antibody overnight at 4°C. Immune complexes were collected with protein G dynabeads, extensively washed, and analyzed by western blot. Antibodies used included: anti-RIP1 (BD 610459, Cell Signal 3493), anti-A20 (Cell Signal 5630), anti-TNFR (R and D AF-425-PB, Abcam 19139), anti-pIkba (Cell Signal 9246), anti-Ikba (cell signal 9242), anti-K63 ubiquitin (Millipore 05-1307), anti-K48 ubiquitin (Millipore 05-1308), anti-Ub (P4D1, SCBT).
Ubiquitin binding assays
Ubiquitin binding of A20 proteins were performed by incubating recombinant GST-A20 proteins with recombinant ubiquitin chains followed by immunoblotting analyses. Binding studies of A20 to ubiquitinated RIP1 were perfomed by incubating whole cell lysates from TNF stimulated A20−/− MEFs with recombinant GST-A20 proteins.
DSS colitis
Sex-matched wildtype, Tnfaip3OTU/OTU, and Tnfaip3ZF4/AF4 mice between 2-3 months of age were co-housed and exposed to drinking water with 3% DSS (MP Biomedicals) for 5 days. Tissues samples were taken 6 days after removal of DSS for histological and mRNA analysis. The degree of inflammation in the colon was graded according to a previously described grading system that evaluates inflammatory cell infiltration and tissue damage (Onizawa et al., 2009). Briefly, the scoring for inflammatory cell infiltration is as follows: (0) occasional inflammatory cells in the lamina propia; (1) increased numbers of inflammatory cells in the lamina propria; (2) confluence of inflammatory cells, extending into the submucosa; (3) transmural extension of the infiltrate. Tissue damage was scored as follows: (0) no mucosal damage; (1) discrete lymphoepithelial lesions; (2) surface mucosal erosion or focal ulceration; (3) extensive mucosal damage and extension into deeper structures of the bowel wall. The combined histological score ranged from 0 (no changes) to 6 (extensive cell infiltration and tissue damage).
RNA analyses
RNA was isolated from stimulated cells and reverse transcribed (Applied Biosystems). Taqman gene expression master mix and Taqman gene expression assay primers from Applied Biosystems were used for quantitative real time PCR on an ABI 7300 (Applied Biosystems). Relative mRNA units were calculated as 2^-(CT gene of interest − CT actin).
Multi-angle Light Scattering (MALS) Analyses
The molar mass of A20 protein complexes (residues x-y, or 1-370) was determined by MALS. Protein sample was injected into a Superdex 200 (10/300 GL) gel filtration column (GE Healthcare) equilibrated in a buffer containing 20 mM Tris at pH 8.0 and 150 mM NaCl. The chromatography system was coupled to a three-angle light scattering detector (mini-DAWN TRISTAR) and a refractive index detector (Optilab DSP) (Wyatt Technology). Data were collected every 0.5 s with a flow rate of 0.2 mL/min. Data analysis was carried out using ASTRA V.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adrianto I, Wen F, Templeton A, Wiley G, King JB, Lessard CJ, Bates JS, Hu Y, Kelly JA, Kaufman KM, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltimore D. NF-kappaB is 25. Nat Immunol. 2011;12:683–685. doi: 10.1038/ni.2072. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, et al. IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, Burtt NP, Guiducci C, Parkin M, Gates C, Plenge RM, Behrens TW, Wither JE, Rioux JD, Fortin PR, Graham DC, Wong AK, Vyse TJ, Daly MJ, Altshuler D, Moser KL, Gaffney PM. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, Reizis B, DeFranco A, Criswell LA, Nakamura MC, Ma A. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nat Immunol. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyninck K, Beyaert R. The cytokine-inducible zinc finger protein A20 inhibits IL-1-induced NF-κB activation at the level of TRAF6. FEBS Lett. 1999;442:147–150. doi: 10.1016/s0014-5793(98)01645-7. [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S, Mori S, Ishikawa Y, Okamoto K, Tobinai K, Nakagama H, Nakahata T, Yoshino T, Kobayashi Y, Ogawa S. Frequent inactivation of A20 in B-cell lymphomas. Nature. 2009;459:712–716. doi: 10.1038/nature07969. [DOI] [PubMed] [Google Scholar]
- Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- Krappman D, Scheidereit C. A pervasive role of ubiquitin conjugation in activation and termination of IkB kinase pathways. EMBO Rep. 2005;6:321–326. doi: 10.1038/sj.embor.7400380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM. Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT. The UCH-L1 gene encodes two opposing enzymatic activities that affect α-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Ma A. A20 takes on tumors: tumor suppression by an ubiquitin-editing enzyme. J Exp Med. 2010;206:977–980. doi: 10.1084/jem.20090765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin (Ub) and functions as a Ub ligase through an atypical Ub-interacting motif and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- Musone SL, Taylor KE, Lu TT, Nititham J, Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, Seldin MF, Gregersen PK, Behrens TW, Ma A, Kwok PY, Criswell LA. Multiple polymorphisms in the TNFAIP3 region are independently associated with systemic lupus erythematosus. Nat Genet. 2008;40:1062–64. doi: 10.1038/ng.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise CA, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR, Collaborative Association Study of Psoriasis Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;21:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizawa M, Nagaishi T, Kanai T, Nagano K, Oshima S, Nemoto Y, Yoshioka A, Totsuka T, Okamoto R, Nakamura T, Sakamoto N, Tsuchiya K, Aoki K, Ohya K, Yagita H, Watanabe M. Signaling pathway via TNF-alpha/NF-kappaB in intestinal epithelial cells may be directly involved in colitis-associated carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G850–859. doi: 10.1152/ajpgi.00071.2008. [DOI] [PubMed] [Google Scholar]
- Opipari AW, Jr., Boguski MS, Dixit VM. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J Biol Chem. 1990;265:14705–14708. [PubMed] [Google Scholar]
- Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Yen TS, Woo T, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe’er I, Burtt NP, Blumenstiel B, DeFelice M, Parkin M, Barry R, Winslow W, Healy C, Graham RR, Neale BM, Izmailova E, Roubenoff R, Parker AN, Glass R, Karlson EW, Maher N, Hafler DA, Lee DM, Seldin MF, Remmers EF, Lee AT, Padyukov L, Alfredsson L, Coblyn J, Weinblatt ME, Gabriel SB, Purcell S, Klareskog L, Gregersen PK, Shadick NA, Daly MJ, Altshuler D. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genetics. 2007;39:1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Shembade N, Ma A, Harhaj EW. Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science. 2010;327:1135–1139. doi: 10.1126/science.1182364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ. Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NF-kB activation. Proc. Natl. Acad. Sci. USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, Donn R, Symmons D, Hider S, Bruce IN, Wellcome Trust Case Control Consortium. Wilson AG, Marinou I, Morgan A, Emery P, YEAR Consortium. Carter A, Steer S, Hocking L, Reid DM, Wordsworth P, Harrison P, Strachan D, Worthington J. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Nishimasu H, Ishitani R, Goto E, Noguchi T, Mio K, Kamei K, Ma A, Iwai K, Nureki O. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst K, Carpentier I, Kreike M, Meloni L, Verstrepen L, Kensche T, Dikic I, Beyaert R. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–55. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O’Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.