Abstract
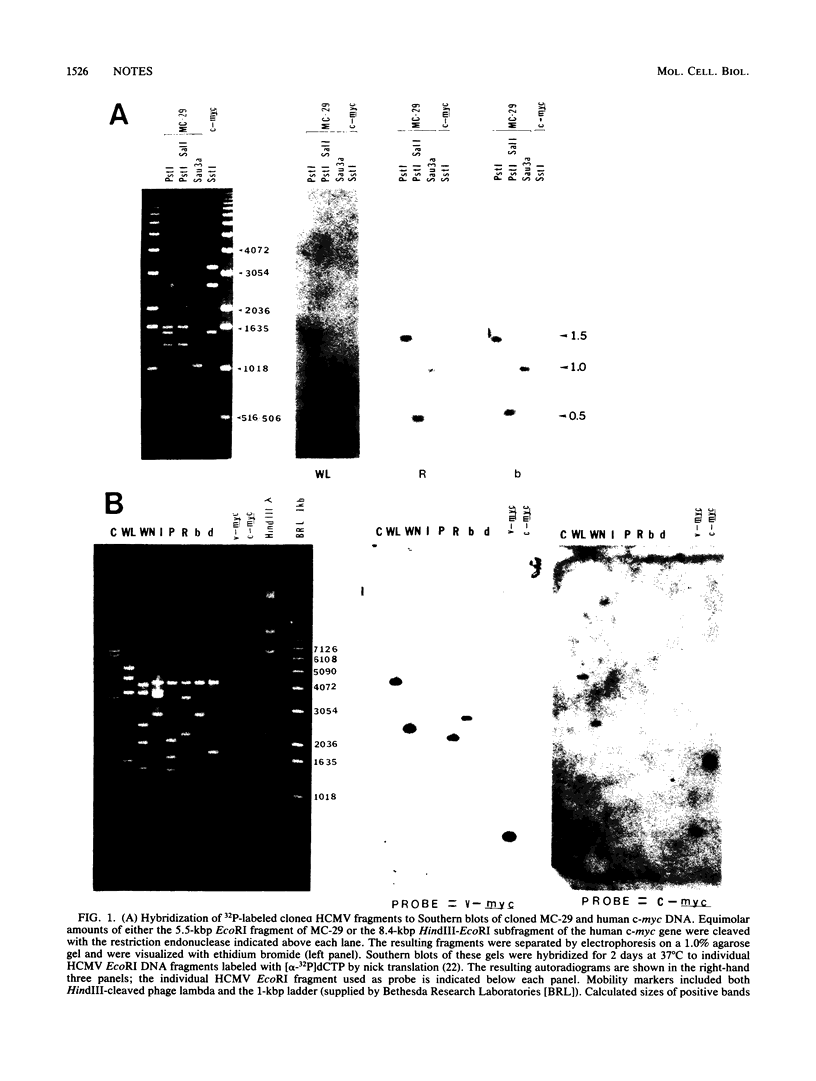
The degree of relatedness between previously identified cross-hybridizing regions within human cytomegalovirus strain AD169 and the avian retrovirus oncogene v-myc were investigated by nucleotide sequence comparison. We found that the homologous regions between the human cytomegalovirus genome and v-myc are limited to short G + C-rich regions in each genome and that the human cytomegalovirus genome shares little or no homology with the human c-myc gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alitalo K., Bishop J. M., Smith D. H., Chen E. Y., Colby W. W., Levinson A. D. Nucleotide sequence to the v-myc oncogene of avian retrovirus MC29. Proc Natl Acad Sci U S A. 1983 Jan;80(1):100–104. doi: 10.1073/pnas.80.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clanton D. J., Jariwalla R. J., Kress C., Rosenthal L. J. Neoplastic transformation by a cloned human cytomegalovirus DNA fragment uniquely homologous to one of the transforming regions of herpes simplex virus type 2. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3826–3830. doi: 10.1073/pnas.80.12.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby W. W., Chen E. Y., Smith D. H., Levinson A. D. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature. 1983 Feb 24;301(5902):722–725. doi: 10.1038/301722a0. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R., Gelmann E. P., Martinotti S., Franchini G., Papas T. S., Gallo R. C., Wong-Staal F. Cloning and characterization of different human sequences related to the onc gene (v-myc) of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1982 Nov;79(21):6497–6501. doi: 10.1073/pnas.79.21.6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Geelen J. L., Walig C., Wertheim P., van der Noordaa J. Human cytomegalovirus DNA. I. Molecular weight and infectivity. J Virol. 1978 Jun;26(3):813–816. doi: 10.1128/jvi.26.3.813-816.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmann E. P., Clanton D. J., Jariwalla R. J., Rosenthal L. J. Characterization and location of myc homologous sequences in human cytomegalovirus DNA. Proc Natl Acad Sci U S A. 1983 Aug;80(16):5107–5111. doi: 10.1073/pnas.80.16.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goad W. B., Kanehisa M. I. Pattern recognition in nucleic acid sequences. I. A general method for finding local homologies and symmetries. Nucleic Acids Res. 1982 Jan 11;10(1):247–263. doi: 10.1093/nar/10.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. I. Los Alamos sequence analysis package for nucleic acids and proteins. Nucleic Acids Res. 1982 Jan 11;10(1):183–196. doi: 10.1093/nar/10.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakeman A. D., Osborn J. E. Size of infectious DNA from human and murine cytomegaloviruses. J Virol. 1979 Apr;30(1):414–416. doi: 10.1128/jvi.30.1.414-416.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nelson J. A., Fleckenstein B., Galloway D. A., McDougall J. K. Transformation of NIH 3T3 cells with cloned fragments of human cytomegalovirus strain AD169. J Virol. 1982 Jul;43(1):83–91. doi: 10.1128/jvi.43.1.83-91.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer G., Goodheart C. R., Henson D., Bowling C. P. A comparative study of the DNA density and behavior in tissue cultures of fourteen different herpesviruses. Virology. 1969 Sep;39(1):134–137. doi: 10.1016/0042-6822(69)90355-9. [DOI] [PubMed] [Google Scholar]
- Reddy E. P., Reynolds R. K., Watson D. K., Schultz R. A., Lautenberger J., Papas T. S. Nucleotide sequence analysis of the proviral genome of avian myelocytomatosis virus (MC29). Proc Natl Acad Sci U S A. 1983 May;80(9):2500–2504. doi: 10.1073/pnas.80.9.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spector D. H., Hock L., Tamashiro J. C. Cleavage maps for human cytomegalovirus DNA strain AD169 for restriction endonucleases EcoRI, BglII, and HindIII. J Virol. 1982 May;42(2):558–582. doi: 10.1128/jvi.42.2.558-582.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D. H., Spector S. A. The oncogenic potential of human cytomegalovirus. Prog Med Virol. 1984;29:45–89. [PubMed] [Google Scholar]
- Spector D. H., Vacquier J. P. Human cytomegalovirus (strain AD169) contains sequences related to the avian retrovirus oncogene v-myc. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3889–3893. doi: 10.1073/pnas.80.13.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro J. C., Hock L. J., Spector D. H. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J Virol. 1982 May;42(2):547–557. doi: 10.1128/jvi.42.2.547-557.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman J. S., McCarthy B. J. The relationship between mismatched base pairs and the thermal stability of DNA duplexes. II. Effects of deamination of cytosine. Biochim Biophys Acta. 1973 Feb 4;294(1):416–424. doi: 10.1016/0005-2787(73)90096-8. [DOI] [PubMed] [Google Scholar]
- Vennström B., Moscovici C., Goodman H. M., Bishop J. M. Molecular cloning of the avian myelocytomatosis virus genome and recovery of infectious virus by transfection of chicken cells. J Virol. 1981 Aug;39(2):625–631. doi: 10.1128/jvi.39.2.625-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Watson D. K., Psallidopoulos M. C., Samuel K. P., Dalla-Favera R., Papas T. S. Nucleotide sequence analysis of human c-myc locus, chicken homologue, and myelocytomatosis virus MC29 transforming gene reveals a highly conserved gene product. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3642–3645. doi: 10.1073/pnas.80.12.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt R., Stanton L. W., Marcu K. B., Gallo R. C., Croce C. M., Rovera G. Nucleotide sequence of cloned cDNA of human c-myc oncogene. Nature. 1983 Jun 23;303(5919):725–728. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]