Abstract
Nicastrin (NCSTN) is a component of the γ-secretase complex and therefore potentially a candidate risk gene for Alzheimer's disease. Here, we have developed a novel functional genomics methodology to express common locus haplotypes to assess functional differences. DNA recombination was used to engineer 5 bacterial artificial chromosomes (BACs) to each express a different haplotype of the NCSTN locus. Each NCSTN-BAC was delivered to knockout nicastrin (Ncstn−/−) cells and clonal NCSTN-BAC+/Ncstn−/− cell lines were created for functional analyses. We showed that all NCSTN-BAC haplotypes expressed nicastrin protein and rescued γ-secretase activity and amyloid beta (Aβ) production in NCSTN-BAC+/Ncstn−/− lines. We then showed that genetic variation at the NCSTN locus affected alternative splicing in human postmortem brain tissue. However, there was no robust functional difference between clonal cell lines rescued by each of the 5 different haplotypes. Finally, there was no statistically significant association of NCSTN with disease risk in the 4 cohorts. We therefore conclude that it is unlikely that common variation at the NCSTN locus is a risk factor for Alzheimer's disease.
Keywords: Nicastrin, Haplotype variation, Functional genomics, Alzheimer's disease, γ-Secretase complex
1. Introduction
Alzheimer's disease (AD) is the most common cause of neurodegeneration. Mutations in 3 genes (amyloid precursor protein [APP], presenilin 1 [PS1], and presenilin 2 [PS2]) are known to cause rare familial early onset AD (EOAD). However, until recently the genetic factors which influenced the common sporadic form of AD, late onset AD (LOAD), were unclear with the ε4 allele of the apolipoprotein E (APOE) gene being for many years the only well-replicated genetic susceptibility factor for LOAD. Recent genome-wide association studies (GWAS) have now confirmed the association with the APOE ε4 allele and shown that susceptibility to LOAD is influenced by multiple genetic factors of small effect. At least 9 candidate susceptibility genes for LOAD have been identified: ATP-binding cassette, sub-family A, member 7 (ABCA7), bridging integrator 1 (BIN1), CD2-associated protein (CD2AP), CD33 molecule (CD33), clusterin (CLU), complement receptor 1 (CR1), EPH receptor A1 (EPHA1), membrane-spanning 4-domains subfamily A member 4 (MS4A4)/membrane-spanning 4-domains subfamily A member 6E (MS4A6E), and phosphatidylinositol binding clathrin assembly protein (PICALM) (Harold et al., 2009; Hollingworth et al., 2011; Lambert et al., 2009; Naj et al., 2011). It is predicted that variation in these genes, including APOE, account for approximately 32% of AD genetic risk (Fagan, 2011). However, despite the consistency achieved by recent GWAS studies, there has been limited success in the identification of functional variants (Brouwers et al., 2012).
Nicastrin (NCSTN, MIM 605254) is a protein critical to the function of the γ-secretase complex (Yu et al., 2000). However, evidence for a role of NCSTN in AD susceptibility remains unclear. A recent study has shown that individuals carrying a rare NCSTN variant show an increased risk of AD (Lupton et al., 2011). In addition, common variation at the NCSTN locus may affect risk for AD. Four main haplotypes have been identified at the NCSTN locus; HapA, HapB, HapC, and HapD (Dermaut et al., 2002). In addition, we have previously defined a further haplotype, HapE (Deary et al., 2005). Of these, HapB was shown to increase risk of developing familial early onset Alzheimer's disease in Dutch individuals, particularly those without an APOE ε4 allele (Dermaut et al., 2002). This was replicated in both a Sardinian (Piscopo et al., 2006) and Finnish study (Helisalmi et al., 2004), although the latter result was nonsignificant. However, it was not replicated in an Italian (Confaloni et al., 2003) or French study (Cousin et al., 2003). Overall, the evidence indicates HapB may be a risk factor for AD, while we have shown that HapB may have an effect on general cognitive ability (Deary et al., 2005).
Although the genetic data are conflicting, there is evidence that protein sequence changes at the NCSTN locus affect protein function (Yu et al., 2000). In addition to NCSTN, the γ-secretase complex consists of 3 additional cofactors: the presenilins (PS1/PS2), PS enhancer 2 (PEN2), and anterior pharynx defective (APH)-1. NCSTN has important roles in the assembly and function of γ-secretase. It has been implicated in the control of intracellular trafficking of the γ-secretase (Spasic et al., 2007; Zhang, Y.W. et al., 2005), which is critical to its function. The transmembrane domain (TMD) of NCSTN is required in the interaction with the C terminus of PS1 (Kaether et al., 2004), and the transmembrane domain and juxtamembrane region are required in the interaction of NCSTN with APH-1 (Walker et al., 2006). A recent molecular study identified an alternatively spliced transcript of NCSTN that lacked exon 16, and therefore lacked this juxtamembrane region. Transcripts lacking exon 16 showed a trend for association with AD in patients with an APOE ε4 allele (Mitsuda et al., 2006). Another important region of NCSTN is the ectodomain. A conformational change in the NCSTN ectodomain is associated with γ-secretase activity (Shirotani et al., 2003). In addition, this domain contains a DYIGS and peptidase homologous region (DAP) domain. Although it has not retained its peptidase activity (Fagan et al., 2001; Fergani et al., 2001), this domain has retained its ability for substrate recognition (Shah et al., 2005). Point mutations within the DAP domain appear to augment amyloid precursor protein (APP) processing, whereas deletions disrupt APP processing, probably by preventing the exit of NCSTN from the endoplasmic reticulum (ER) (Chen et al., 2001; Yu et al., 2000).
Given that previous genetic studies have been inconclusive as to the role of NCSTN in AD susceptibility, we have developed a novel functional genomics approach based on the expression of different haplotypes. In this study we have used bacterial artificial chromosome (BAC) vectors expressing different NCSTN haplotypes to evaluate whether haplotypic variation at the NCSTN locus contributes to functional variation. We first identified a BAC carrying the NCSTN HapA variant and then used serial homologous recombination to create 4 further BACs with NCSTN HapsB–E. Subsequent analyses of clonal cell lines expressing these haplotypes demonstrated that all haplotypes were capable of expressing NCSTN to restore γ-secretase activity and amyloid beta (Aβ) production to nicastrin (Ncstn−/−) knockout cells. However, we were unable to demonstrate any robust functional difference between cell lines expressing different haplotypes. Analysis of postmortem human brain tissue indicated that haplotypic variation may affect splicing of NCSTN in specific brain regions, although we were unable to demonstrate any difference in the alternative splicing of NCSTN exon 16 in our cell culture models. Finally, analysis of GWAS data from the NCSTN locus did not reveal any statistically significant association with disease. Overall, our thorough approach combining studies in functional genomics, genetic association, and gene expression and splicing demonstrates that NCSTN is unlikely to be a susceptibility gene in AD.
2. Methods
2.1. Genotyping
All primers were designed using Primer3 online (Rozen and Skaletsky, 2000) and genomic fragments were amplified using standard polymerase chain reaction (PCR) procedures. Genotyping was carried out by restriction digestion and/or sequencing (Big Dye Version 3.1, Applied Biosystems, Carlsbad, CA, USA).
2.2. BAC characterization
BAC clones spanning the NCSTN genomic locus were identified using the University of California at Santa Cruz (UCSC) genome browser (Kent et al., 2002). BACs were obtained from Invitrogen or the BACPAC Resource Centre (BPRC), and confirmed by exon amplification and restriction digestion (Supplementary Table 1). Each BAC was genotyped to determine the NCSTN haplotype expressed at each locus (Supplementary Table 2).
2.3. Haplotype characterization
NCSTN haplotypes are characterized by the 4 single nucleotide polymorphisms (SNPs) originally described by Dermaut et al. (2002). These SNPs may not be functional and may be proxies for the true untyped functional variant in the haplotype region. To address this, we sequenced the haplotype region. We obtained genomic DNA from individuals expressing different NCSTN haplotypes (gifts from I. Deary and S. Harris, University of Edinburgh, UK). Specifically, these individuals expressed haplotypes AD, AE, BB, and CC respectively. Disease status with respect to Alzheimer's disease was unknown. DNA representing each haplotype was amplified using a forward primer containing a SalI site at the 5' end (5'-TTTTTTGTCGACTCCTATGGTGGTTCTAGGATAACT-3') and a reverse primer containing the NotI site at the 5' end (5'-TTTTTTGCGGCCGCCCTTTTATGTCCTTCCCAGTCA-3'). PCR amplified DNA for each haplotype was cloned into pBluescript (pBS) and delivered to ElectroMAX DH10B cells (Invitrogen) for sequence analysis. pBS-NCSTN plasmids were sequenced in both directions with 21 primer pairs that spanned the haplotype region (Supplementary Table 3). Polymorphisms were counted as true if present in both directions and in more than 1 clone for each haplotype.
2.4. BAC modification using Red/ET recombination
2.4.1. NCSTN
HapA BACs were modified to create NCSTN haplotypes B, C, D, and E in a sequential manner using a 2-step recombination protocol based on Red/ET recombination (Muyrers et al., 2000, 2001; Zhang Y.W. et al., 2000). All modifications were carried out in ElectroMAX DH10B cells.
In brief, a plasmid containing RpsL-neo was linearized and amplified using forward (5'-GGCCTGGTGATGATGGCGGGATC-'3) and reverse (5'-TCAGAAGAACTCGTCAAGAAGG-3') primers with 5' homology arms that matched specific genomic NCSTN DNA insertion sites (Supplementary Table 4). Amplified products were added to recombination-induced cells containing the Red/ET plasmid pSC101-BAD-gbaA. The DNA fragment containing the replacement polymorphism was amplified from the corresponding pBS-NCSTN clone (Supplementary Table 5), and the product added to cells containing correct recombinants from the first round. Correct recombinants were confirmed by restriction digestion and sequencing.
2.5. BAC modification using Cre/LoxP recombination
pEHHG-NCSTN was obtained by incubating pEHHG and NCSTN BAC with Cre enzyme (Merck, Darmstadt, Germany).
2.6. Tissue culture
Three mouse embryonic fibroblast cell lines, Ncstn wild type (Ncstn+/+), Ncstn heterozygotes (Ncstn+/−) and Ncstn knockout (Ncstn−/−) (gifts from P. Wong, Johns Hopkins University School of Medicine, USA), were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS), P/S, and L-Glut. pEHHG-NCSTN stable clones were generated by transfecting pEHHG-NCSTN into Ncstn−/− cells using Lipofectamine 2000 (Invitrogen). Transfected cells were diluted after 48–72 hours in medium containing 225 μg/mL hygromycin (Invitrogen) and single colonies isolated after 10–15 days. All cells were cultured at 37 °C in 5% (vol/vol) CO2.
2.7. RT-PCR
RNA was prepared using TRIzol (Invitrogen). RNA was resuspended in diethylpyrocarbonate (DEPC) water. cDNA was made from 2 μg RNA using the first Strand cDNA Synthesis Kit for RT-PCR (AMV, Roche, West Sussex, UK). Human-specific primers amplified exon 9 to exon 11; forward (5'-CAGCTCGAGGATGGTCTACG-3'), reverse (5'-GAGAGGCTGGGACTGATTG-3'), product 250 base pair (bp).
2.8. Western blot analysis
Protein was prepared using lysis buffer (50 mM Tris pH 8, 150 mM NaCl, 1% Triton X200, protease inhibitor). Protein was reduced with β-mercaptoethanol or dithiothreitol (DTT) and 5–10 μg was resolved on either a 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (Invitrogen) or an 8% SDS-PAGE gel (Pierce). Following transfer to a positively charged nylon membrane, Hybond N+ (GE Healthcare, Buckinghamshire, UK), protein samples were incubated with antibody; anti-nicastrin (1:1000) (Sigma-Aldrich, Dorset, UK), anti-β-actin (1:10,000) (Abcam, Cambridge, UK), polyclonal rabbit anti-mouse horseradish peroxidase (HRP) (1:5000) (Dako, Glostrup, Denmark), and polyclonal goat anti-rabbit (1:4000) (Dako). Samples were visualized using ECL plus (GE Healthcare) or SuperSignal West Dura Extended Duration Substrate (Thermo, Fisher Scientific, Rockford, IL, USA). Each protein sample was analyzed in triplicate. β-actin expression was used to normalize for equal loading between samples. Total protein from Ncstn+/+ cells was used to compare gels for NCSTN. Gels were quantified using GeneSnap (Syngene, Cambridgeshire, UK).
2.9. Copy number determination
NCSTN transgene copy number of each clonal cell line was determined using human-specific quantitative real-time PCR (RT-PCR). Primers were designed within intron 2 (IN2) of NCSTN and 2 control primer sets targeting mouse sequences were used: D3MIT12 and D14MIT5 (Supplementary Table 6). A positive control construct for each primer set was generated by the amplification of human and mouse genomic DNA with the NCSTN IN2 and D3MIT12 and D14MIT5 primer pairs respectively. Amplified products were cloned into pCR2.1-TOPO (Invitrogen), introduced into DH5α cells (One Shot MAX Efficiency, Invitrogen) and confirmed by sequence analysis. For real-time detection a MyiQ Single-Color Real-Time PCR machine (Bio-Rad, Hertfordshire, UK) was used using a standard PCR program with amplification at 65 °C and data collection at 72 °C (NCSTN IN2, D14MIT5), or amplification at 68 °C with data collection at 78 °C (D3MIT5). Plasmid DNA was titrated and amplified with the DyNAmo Flash SYBR Green qPCR kit (NEB, Ipswich, MA, USA), using 0.25 μM (NCSTN IN2, D14MIT5) or 1-μM primer (D3MIT12) primer. All reactions had an efficiency of 90%−110%. A standard curve for each primer pair was generated by plotting the log of the copy number (x-axis) versus CT values (y-axis).
To determine the copy number of each NCSTN clone, genomic DNA was amplified in separate reactions with all 3 primer pairs. Specifically, 10 ng genomic DNA was amplified using optimized conditions and each reaction was set up in triplicate. Experimental CT values were converted to log copy number values using the appropriate standard curve and absolute copy number for each clonal cell line was determined by normalization to the average value obtained from D3MIT12 and D14MIT5.
2.10. Notch activity assay
Cells were seeded in a 24-well plate at a density of 1 × 105 in media with no antibiotics. Cells were transfected with 444-ng firefly luciferase Notch-CBF-1 reporter, 4xwtCBF-1-luc (a gift from Dr. G. Weinmaster, UCLA Medical School, USA) and 500 ng of the S2 cleaved form of human Notch-1, ▵EN1 (a gift from Dr. J. Aster, Brigham and Women's Hospital, USA) using Lipofectamine 2000. To control for transfection efficiency, 54 ng phTKRenilla luciferase (Promega, Madison, WI, USA) was used. Empty vector (pcDNA3.1) was included where necessary to maintain constant DNA concentrations. A constitutively active luciferase plasmid, pGL4.13 (Promega), was used as a between plates control. Transfected samples were analyzed for firefly and renilla luciferase activities after 24 hours using the Dual-Glo Luciferase Assay System (Promega) and measured using a Synergy 2 luminescence Microplate Reader (Bio-Tek, Winooski, VT, USA). All values were normalized to wells containing reagent but no cells. Data were obtained from 3 independent experiments of 6 repeats.
2.11. Aβ enzyme-linked immunosorbent assay (ELISA)
Cells were seeded in a 24-well plate at a density of 1.5 × 105 in media containing no hygromycin. Cells were transfected with 250 ng of a hAPPswe plasmid (Mudher et al., 2001) and 250 ng β-galactosidase (Promega) using Lipofectamine 2000 (Invitrogen). Media was removed 4 hours posttransfection media and 250-μL fresh media added. Conditioned media was removed after 20 hours and cleared by centrifugation. Supernatant for Aβ40 assays was diluted 1:2 using dilution buffer containing 1 mM AEBSF, while supernatant for Aβ42 assays was used neat with 1 mM AEBSF. Supernatants were stored at –20 °C until measured. Cell lysates were prepared and assayed for β-galactosidase expression using the β-galactoside Enzyme Assay System (Promega). Secreted Aβ40 and Aβ42 levels were determined using colorimetric ELISA kits (Aβ40, Invitrogen; Aβ42, Millipore, Billerica, MA, USA). Data were obtained from 3 independent experiments of 6 repeats. Aβ values were normalized using β-galactosidase results.
2.12. Expression of alternative transcripts from postmortem brain tissue
Brain tissue was obtained from healthy control individuals from 3 UK brain banks; the Oxford Project to Investigate Ageing and Memory (OPTIMA), the MRC London Brain Bank for Neurodegenerative Diseases and the South West Dementia Brain Bank (SWDBB). RNA was extracted from brain tissue using Trizol and an RNase Easy Column (Qiagen, West Sussex, UK). cDNA was made as before.
Expression levels of NCSTN transcripts containing and lacking exon 16 were determined using quantitative RT-PCR. Primers were designed that uniquely amplified transcripts that contained exon 16 (full length transcript, FL Tc, forward: 5'-AGTGAAAACAAGGATCTGTATGAGT-3', reverse: 5'CAGGGTGATCAACTCAAGCTC-3') or lacked exon 16 (▵Ex16, forward: 5'-CAAGGATTTGATCACCCTGA-3', reverse: 5'-GCTGGGGTCCTCCTCAGTAT-3'). Three human housekeeping genes (PGBD, GNB2L1, ABL1) were selected from an initial panel of 10 (Zhang, X. et al., 2005). For real-time detection a MyiQ Single-Color Real-Time PCR machine (Bio-Rad) was used. cDNA was amplified using 0.5−1-μM primer with the DyNAmo Flash SYBR Green qPCR kit (NEB) and using a standard PCR program with amplification at 68 °C and data collection at 78 °C. Each cDNA sample was amplified in triplicate with all primer pairs. Experimental CT values for each sample were compared and sample representing CT minimum for each primer pair was identified. The equation where raw data = 1 + ECtmin-Ct was used to determine relative expression levels for each sample (Livak and Schmittgen, 2001; Pfaffl, 2001). Raw values obtained for FL Tc and ΔEx16 were normalized using the geomean of the 3 housekeeping genes. Standard residual scores were calculated for each data point to incorporate age and gender, using linear regression in SPSS, v14.0. Results are presented as a normalized ratio of FL Tc to ΔEx16 transcript. Statistical analysis was carried out using a 1-way ANOVA (SPSS, v14.0).
2.13. Expression of alternative transcripts from clonal cell lines
cDNA was made from clonal cell lines as previously described. Expression levels of NCSTN transcripts containing and lacking exon 16 were determined using quantitative real-time PCR (RT-PCR) as described in 2.12. Two mouse housekeeping genes (RPL6 and OAZ1) were selected from an initial panel of 5 (de Jonge et al., 2007).
2.14. Analysis of NCSTN genotype data
AD case and control genotype data were obtained from 4 consortia; the Genetic and Environmental Risk for Alzheimer's disease (GERAD1) Consortium (Harold et al., 2009) (Supplementary Fig. 1), the Translational Genomics Research Institute (TGen) (Corneveaux et al., 2010) (Supplementary Fig. 2), the European Alzheimer Disease Initiative (EADI) (Lambert et al., 2009) (Supplementary Fig. 3), and the Mayo (MAYO) Clinic (Carrasquillo et al., 2009) (Supplementary Fig. 4) (Supplementary Table 7).
Power calculations were carried out using CaTS (Skol et al., 2006). All calculations were carried out with a disease prevalence of 0.3 and a type I error rate of 0.01. Directly genotyped and imputed data were available for 16 NCSTN SNPs (Supplementary Table 8). Four SNPs, rs2274185, rs6664438, rs6677637, and rs17370539, overlap with haplotype SNPs identified in our sequence analysis. Statistical analysis was carried out using PLINK v1.07 (Purcell et al., 2007). SNPs were examined for association with disease singly or by specified haplotype analysis. Two stratified data sets, with or without the APOE ε4 allele, were analyzed similarly. METAL, a tool for the meta-analysis of genome-wide association scans, was used to analyze the combined SNP and haplotype data from the 4 cognitive studies (Willer et al., 2010). A significance threshold of p ≤ 0.05 was applied to the meta-analysis.
3. Results
3.1. BAC characterization
The genomic locus of NCSTN is 15.7 kb in length. We identified 16 BACs that contained the NCSTN gene and each was genotyped at 4 SNP locations (rs12239747, rs2274185, rs7528638, rs17370539). All BACs contained the most common haplotype, HapA. One BAC, CTC635J11, was selected for all further experiments (Fig. 1A). CTC635J11 is approximately 121.1 kb in size and carried by the vector pBeloBAC11. In addition to NCSTN, CTC635J11 carries the full length Nescient helix loop helix 1 (NHLM1) and Vang-like 2 (VANGL2) genes. NHLM1 belongs to a putative family of transcription factors and VANGL2 is involved in planar cell polarity. It also contains the 5′ end of the coatomer protein complex, subunit alpha (COPA) gene.
Fig. 1.
Genomic structure of the nicastrin (NCSTN) locus. (A) Bacterial artificial chromosome (BAC) CTC635J11 encompasses the NCSTN, Nascient helix loop helix 1 (NHLH1) and Vang-like 2 (VANGL2) genes and the 5′ end of the coatomer protein complex, subunit alpha (COPA) gene. The genomic sequence of NCSTN spans 15.7 kb and is composed of 17 exons. The haplotype of interest, marked in gray, spans exon 6 to intron 16 and covers 7 kb of genomic sequence. The polymorphisms of interest within the haplotype are marked on the first row. Polymorphisms on the second row indicate single nucleotide polymorphisms (SNPs) that were genotyped in the Genetic and Environmental Risk for Alzheimer's disease (GERAD1) study, SNPs in common are highlighted in bold. The transmembrane domain is marked in red. An alternative transcript lacking exon 16 is marked. Scale bars indicate 5 kb. (B) Use of loxP sites on the pEHHG vector and the BAC allowed sequence-specific integration of these 2 vectors, and (C) pulse field gel of NotI digested constructs before (−) and after (+) the addition of a single copy of pEHHG after Cre/loxP recombineering in the BAC. The asterisk (*) indicates the 12.6 kb size change in the BAC library vector corresponding to pEHHG.
3.2. Haplotype characterization
The original SNPs representing the haplotype may not themselves be functional and may be proxies for the true untyped functional variant in the haplotype region. The NCSTN haplotype spans a genomic region of approximately 7000 bp (Dermaut et al., 2002). To identify unknown potential functional variants, we obtained DNA from 4 individuals expressing different NCSTN haplotypes (S. Harris and I. Deary, University of Edinburgh). These were sub-cloned into pBS. Two pBS-NCSTN clones per haplotype were sequenced across a 7638-bp region. Sequence results confirmed the 4 original SNPs and identified an additional 10 SNPs that differed between the haplotypes (Table 1, Fig. 1A). HapC and HapE were most similar to HapA and differed at 1 SNP position each (rs2274185, HapC; rs12239747, HapE). HapD was found to differ at 6 SNP positions (rs1041066, rs78110462, rs6427515, rs45503695, rs17370539, rs41266887). HapB was most dissimilar to HapA, differing at 8 SNP positions (rs12240253, rs12239747, rs6664438, rs6664627, IVS8+316G→A, rs7528638, rs6677637, rs17370539). Two of the 14 SNPs were located in exons (rs12239747, rs6664627), but are synonymous changes not expected to affect amino acid sequence.
Table 1.
The 5 nicastrin (NCSTN) haplotypes differ at up to 14 locations
| rs12240253 | rs12239747 | rs2274185 | rs6664438 | rs6664627 | rs1041066 | IVS8+316G→A | rs7528638 | rs78110462 | rs6677637 | rs6427515 | rs455036955 | rs17370539 | rs41266887 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hap A | C | A | C | C | C | G | G | C | G | C | C | T | G | C |
| Hap B | T | G | C | T | T | G | A | G | G | T | C | T | C | C |
| Hap C | C | A | G | C | C | G | G | C | G | C | C | T | G | C |
| Hap D | C | A | C | C | C | A | G | C | A | C | T | C | C | T |
| Hap E | C | G | C | C | C | G | G | C | G | C | C | T | G | C |
| Location | Intron 5 | Exon 6 | Intron 6 | Intron 6 | Exon 7 | Intron 7 | Intron 7 | Intron 10 | Intron 11 | Intron 11 | Intron 11 | Intron 13 | Intron 16 | Intron 16 |
Sequence analysis of the 7 kb haplotype (Hap) region identified 14 SNPs that differed between nicastrin (NCSTN) haplotypes A–E. SNPs previously shown to tag each haplotype are marked in bold and underlined.
We compared the sequencing results to information deposited in dbSNP (National Centre for Biotechnology Information) and the 1000 Genomes Project (1000 Genomes Project Consortium, 2010). On comparison with dbSNP, our study has identified 13 of the 26 SNPs located in the haplotype region, plus 1 novel SNP. Compared with the 1000 Genomes Project we identified 5 SNPs of the 9 located in the haplotype region. SNPs not identified in our study were either very rare or were heterozygous between individuals with the same haplotype, suggesting they were not haplotype-specific SNPs.
3.3. BAC modification
Sequential rounds of Red/ET recombination, using a positive/negative selection strategy, were used to introduce each polymorphism identified in our sequencing analysis to the HapA BAC, CTC635J11, to generate an additional 4 vectors; BAC-NCSTN HapB, HapC, HapD, and HapE. Following these modifications, BAC-NCSTN Haps A–E were further modified by the addition of pEHHG (Fig. 1B). Successful addition of pEHHG by cre-LoxP recombination was confirmed by pulse field gel electrophoresis (Fig. 1C).
3.4. Characterization of clonal cell lines
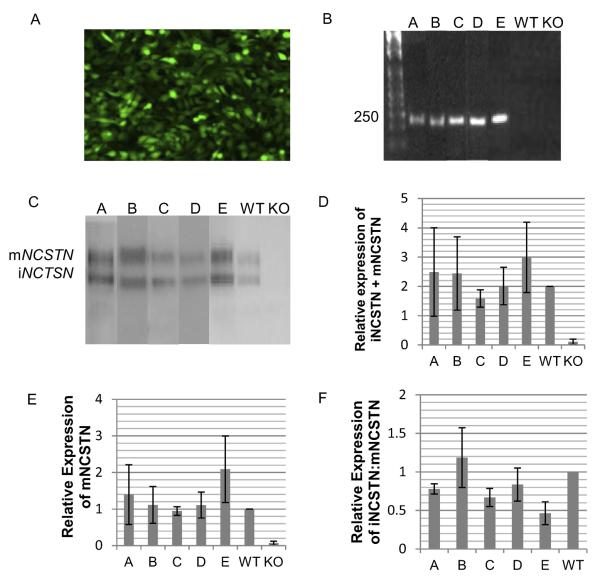
The successful delivery of pEHHG-NCSTN Haps A–E to Ncstn−/− cells was visualized by the expression of green fluorescent protein (GFP) (Fig. 2A). Thirty-five clonal cell lines were initially established; 6 for HapA, 8 for HapB, 7 for HapC, 6 for HapD, and 8 for HapE. Subsequently, each clonal cell line was characterized to confirm expression of NCSTN at both the RNA and protein level. cDNA was amplified using human-specific primers that amplified a 250 bp DNA fragment spanning exon 9 to exon 11. The NCSTN antibody used here binds to the N-terminal and binds both immature, nonfully glycosylated, NCSTN, approximately 110 kDa (immature nicastrin [iNCSTN]) and mature, fully glycosylated, NCSTN, approximately 130 kDa (mature nicastrin [mNCSTN]). Of the 35 cell lines originally picked, 32 were shown to express correctly spliced mRNA (Fig. 2B) and protein (Fig. 2C). Each NCSTN haplotype returned the level of total NCSTN (Fig. 2D), mNCSTN (Fig. 2E), and the ratio of iNCSTN:mNCSTN (Fig. 2F) to broadly the same level observed in wild type Ncstn cells.
Fig. 2.
Characterization of Ncstn−/− cell lines expressing nicastrin (NCSTN) haplotypes. NCSTN cell lines express: (A) green fluorescent protein (GFP); (B) NCSTN RNA; and (C) NCSTN protein; pictures representative. Quantitative immunoblot analyses show that compared with knockout (KO) cell lines, NCSTN cell lines have restored: (D) total NCSTN levels (sum of immature nicastrin [iNCSTN] and mature nicastrin [mNCSTN]); (E) mNCSTN levels; and (F) iNCSTN:mNCSTN ratio. Haplotype results are the average of results obtained with specific clones; A, n = 6; B, n = 6; C, n = 3; D, n = 9; E, n = 8; wild type (WT), n = 3; and KO, n = 3. Error bars are the mean ± standard deviations. Human genomic DNA positive control (+).
NCSTN cell lines were analyzed for copy number. Absolute copy number was determined by normalizing to the average copy number of 2 diploid mouse control genes (D3MIT12, D14MIT5). The majority of clones had fewer than 10 copies of NCSTN, however 4 had 50 to 60 copies. Twelve clonal cell lines with a similar NCSTN copy number and comparable protein expression were selected for functional analyses; 2 HapA clones, 2 HapB clones, 1 HapC clone, 4 HapD clones, and 3 HapE clones.
3.5. Functional analysis of NCSTN clones
Twelve clonal cell lines were used to evaluate the ability of each NCSTN haplotype to rescue γ-secretase function and Aβ production in the Ncstn−/− cells.
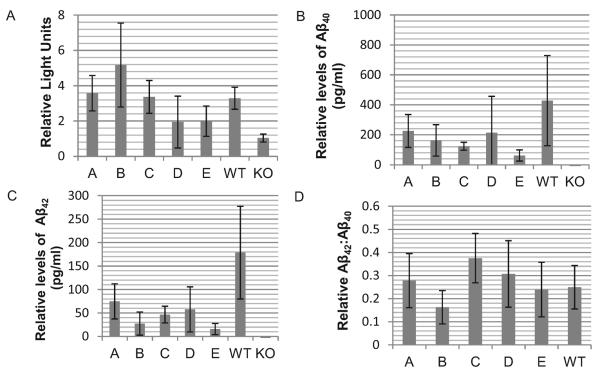
First, we compared γ-secretase activity by assessing luciferase levels from a Notch reporter gene assay transfected into each cell line (Fig. 3A). Membrane bound Notch is processed by functional γ-secretase generating an intracellular fragment that activates gene transcription. We show here that each NCSTN haplotype restored γ-secretase activity toward the level observed in wild type Ncstn cells (Fig. 3A). There was no correlation between γ-secretase activity (Fig. 3A) and the expression level of mNCSTN (Fig. 2E).
Fig. 3.
Functional rescue of γ-secretase activity in Ncstn−/− cells by nicastrin (NCSTN) haplotypes. (A) Cells expressing each NCSTN haplotype recover the ability to process Notch, compared with knockout (KO) cells. Haplotype results are the average obtained with specific clones; A, n = 36; B, n = 36; C, n = 18; D, n = 72; E, n = 54; wild type (WT), n = 12; and KO, n = 18. Cells expressing each NCSTN haplotype have increased levels of both (B) secreted Aβ40, (C) Aβ42, and (D) secreted Aβ42:Aβ40, compared with KO cells. Haplotype results are the average obtained with specific clones; A, n = 35, 25, 25; B, n = 33, 30, 30; C, all n = 12; D, all n = 46; E, all n = 10; WT, n = 14, 9, 8; and KO, both n = 18. Error bars are the mean ± standard deviations. Abbreviation: mNCSTN, mature nicastrin.
Next, to determine the ability of each NCSTN haplotype to restore Aβ production to Ncstn−/− cells, we quantified levels of secreted Aβ40 and Aβ42 in each cell line following transfection of the hAPPswe plasmid. Again, we showed that levels of secreted Aβ40 (Fig. 3B) and secreted Aβ42 (Fig. 3C) are above levels observed in Ncstn−/− cells. Notably, the ratio of secreted Aβ42 to Aβ40 (Aβ42:Aβ40) was broadly similar to the ratio observed in wild type Ncstn cells (Fig. 3D).
3.6. Expression analysis of NCSTN in postmortem human brain samples
To evaluate the existence of tissue-specific haplotype-related splicing variation, we carried out expression analysis of postmortem brain tissue (Fig. 4D). Twenty-seven individuals were genotyped and brain tissue was obtained from those expressing specific NCSTN: HapAA (n = 8), HapAB (n = 10), and HapAC (n = 9). Frontal cortex, entorhinal cortex, and hippocampus was obtained where available (Table 2).
Fig. 4.
Alternative splicing of nicastrin (NCSTN) exon 16. Quantitative real time polymerase chain reaction (PCR) results for brain samples from healthy individuals. Brain regions denoted: frontal cortex (FC), hippocampus (Hip), and entorhinal cortex (EC). (A) and (B) An average normalized expression ratio of standardized scores was generated for each haplotype and brain region for statistical comparison. (C) Averaged quantitative real time PCR results for cell lines expressing each haplotype. (D) Transcripts were amplified using exon-specific primers and transcript expression is presented as a ratio of transcripts containing exon 16 (FL Tc) over transcripts lacking exon 16 (ΔEx16). Significance values, * < 0.05, ** < 0.01, and *** < 0.001. Abbreviation: gDNA, genomic DNA.
Table 2.
Brain tissue samples were collected from individuals expressing different haplotypes of nicastrin (NCSTN)
| FC | EC | Hip | |
|---|---|---|---|
| A/A | 8 | 5 | 8 |
| A/B | 10 | 8 | 2 |
| A/C | 9 | 9 | 5 |
Tissue samples were collected, where possible, from the frontal cortex (FC), entorhinal cortex (EC), and hippocampus (Hip) regions.
Our results show that not only is there variation in the ratio of FL Tc:ΔEx16 transcripts in different brain regions from the same individual, but that there is also variation between individuals with the same NCSTN haplotype. Analysis of the haplotype combined data showed that the most significant difference observed was an increased level of FL Tc in entorhinal cortex and frontal cortex from individuals with HapAA compared with those with HapAB (p < 0.05 and p < 0.01) (Fig. 4B). Within individuals carrying HapAA, we observed that there was a highly significantly increased level of FL Tc in the entorhinal cortex compared with frontal cortex and hippocampus (both p < 0.05) (Fig. 4A). Additionally, within individuals carrying HapAB, we observed a nominally significant increase in FL Tc in the hippocampus compared with frontal cortex (p < 0.001) and entorhinal cortex (p < 0.05) (Fig. 4A).
3.7. Expression analysis of NCSTN in clonal cell lines
An alternatively spliced NCSTN variant, lacking exon 16, has been reported. To determine whether haplotypespecific sequence variation affects splicing we quantified expression levels of each transcript in each clonal cell line using RT-PCR. We determined the expression level of full length NCSTN transcript (FL Tc) and transcript lacking exon 16 (ΔEx16) from each sample (Fig. 4D) and found there was no statistically significant difference between haplotypes (p = 0.18) (Fig. 4C).
3.8. Genetic analysis of NCSTN genotype data
To evaluate genetic variation at the NCSTN locus for association with AD, we have analyzed genotype data from 16 SNPs in 4 cohorts. Power calculations show that this study had 100% power to detect effect sizes of odds ratio (OR) 1.2 and greater with a minor allele frequency of at least 0.05 (data not shown).
No individual SNP was associated with disease risk in the overall or APOE stratified sample in our meta-analysis at p ≤ 0.05 (Supplementary Table 9).
Four SNPs, rs2274185, rs6664438, rs6677637, and rs17370539, overlapped with SNPs that were engineered as part of the haplotype (Fig. 1A); genotype data for these SNPs were available from the TGen and EADI cohorts. No haplotype was associated with disease risk in the overall or APOE stratified sample in our meta-analysis at p ≤ 0.05 (Supplementary Table 10). Indeed, haplotype analyses indicate that SNP rs17370539 is not a haplotype tagging SNP as originally reported; HapA is the most common haplotype and was reported to be tagged by the major allele of rs17370539 (G), however the most common haplotype in the TGen cohort at this loci expresses the minor allele of rs17370539 (A).
4. Discussion
NCSTN is a critical component of the γ-secretase complex and there is some previous evidence to suggest that it is a genetic risk factor for Alzheimer's disease. In this study, we undertook a novel functional genomics approach to investigate the functional impact of common haplotypic variation on NCSTN function and also sought to replicate previous genetic findings in a large meta-analysis.
Through sequencing analysis we have determined that the putative NCSTN risk haplotype, HapB, was most different from the common haplotype, HapA: differing at 8 SNPs throughout the haplotype region. HapD differed at 6 SNPs, while HapC and HapE were most similar to HapA, differing at 1 SNP each. However, we cannot exclude the effect of very rare variants, which may contribute to disease. The NCSTN haplotype region spans a number of functional domains; the DAP domain is coded by NCSTN exons 7–13 and is likely to be important in substrate recognition, while the transmembrane region of NCSTN is required in the interaction with the C terminus of PS1. The SNPs that we identified in our sequencing analysis either lie outside coding sequences or do not change the amino acids encoded. Here we tested the hypothesis that noncoding genetic variation in the NCSTN locus could affect NCSTN function in other ways, for example by regulating expression or splicing.
Our data are the first example of the functional analyses of NCSTN haplotypic variation. We have demonstrated that that all 5 NCSTN haplotypes restore γ-secretase activity and Aβ production to an Ncstn−/− cell line and that Notch activity and secreted levels of Aβ42:Aβ40 are broadly similar across all 5 haplotypes. Overall, we were not able to detect any robust functional differences between the haplotypes. Furthermore, we did not detect any statistically significant genetic association of NCSTN polymorphisms with disease risk in the meta-analysis of 4 large consortia, however, again, this does not exclude a functional role for rare variation.
However, we do demonstrate a significant difference in NCSTN splicing from postmortem brain tissue of a small number of healthy individuals expressing different haplotypes, indicating a potential effect on neuronal-specific splicing. Here, we show that there was a highly significantly increase in full length NCSTN transcript expressed from HapAA individuals than from HapAB individuals in entorhinal cortex and frontal cortex tissue. Further, within HapAA individuals the level of full length transcript in entorhinal cortex tissue was significantly increased compared with that observed in the hippocampus and frontal cortex. Alternatively spliced exon 16 encodes residues important in stabilizing the interaction of NCSTN and APH-1 (Walker et al., 2006). This result suggested that there could be an increased association of NCSTN with APH-1 in the entorhinal cortex of HapAA individuals. Due to the competitive binding of APH-1 and Rer1p to NCSTN, one may hypothesize that NCSTN molecules lacking exon 16 are likely to remain in the early endosomal compartments and not participate in a functional γ-secretase complex. HapA is the most common haplotype of NCSTN (f > 0.8), which may indicate a positive selection pressure; however, this study was very small and replication is required in a larger sample. We did not detect any differences in the alternative splicing of exon 16 in our clonal cell lines, although this could be due to inherent differences in the in vivo tissue analyses and in vitro cell culture work. Further, it was necessary to use a mouse Ncstn−/− knockout cell line so as to be able assay NCSTN transgene function in this study and we cannot exclude any species-specific effects on NCSTN splicing; nor can we exclude the potential effect of trans-factors that may have haplotype-specific effects.
The functional effect of noncoding variants, particularly for a late-onset disease, is likely to be very subtle and may be tissue-specific. Here, we demonstrate that splicing differences are specific to only certain regions of the brain, emphasizing the difficulty in modeling this effect in vitro.
4.1. Conclusions
The NCSTN gene has been implicated as a risk factor for AD in numerous genetic studies. In addition, there is evidence to indicate that genetic variation at this locus may affect function. To clarify the role of NCSTN polymorphisms in AD we have carried out a study with 3 strands using genomic DNA expression vectors, analysis of human postmortem tissue, and genetic association. Using clonal cell lines, we were able to functionally rescue an Ncstn−/− cell line with 5 haplotypes of NCSTN; however, the function of each NCSTN haplotype was broadly similar and we did not detect any robust haplotype-specific functional differences. Further, we detected no difference in splicing between clonal cell lines expressing different haplotypes. In addition, we did not detect any statistically significant genetic association of NCSTN polymorphisms with disease risk. Our study is limited to the common variants that we have identified; and we cannot exclude the contribution of additional rare variants that we have not been able to evaluate. However, in postmortem brain tissue, we show that genetic variation at the NCSTN locus may participate in the tissue-specific control of alternative splicing. The possibility remains that genetic variation at the NCSTN locus may affect disease risk by influencing splicing control, however, we believe that taken together, our results conclude that NCSTN is probably not a risk factor for Alzheimer's disease. This is the first time that the functional importance of genomic variation to Alzheimer's disease has been assessed in this way.
Supplementary Material
Acknowledgements
The authors thank Dr. T. Caffrey, Dr. M. Lufino, and Dr. C. Grünewald for helpful advice and suggestions. DNA was kindly provided by Dr. S. Harris and Professor I.J. Deary, University of Edinburgh, UK; Ncstn cell lines by Professor P. Wong, Johns Hopkins University School of Medicine, USA; the pEHHG plasmid from Professor E.A. Chiocca, Massachusetts General Hospital, USA; the Notch-CBF-1 reporter from Dr. G. Weinmaster, UCLA Medical School, USA; and the ΔEN1 plasmid from Dr. J. Aster, Brigham and Women's Hospital, USA. Brain tissue was provided by MRC, London Brain Bank for Neurodegenerative Diseases (Dr. C. Troakes), Oxford Project to Investigate Memory in Ageing (C. Joachim), and the South West Dementia Brain Bank (R. Barber). We are very grateful to the GERAD1, MAYO, TGen, and EADI consortia for sharing their data. A full list of contributors and research support for these is listed in Supplementary Figs. 1–4. This work was supported by a Fellowship award to G.H. from the Alzheimer's Research Trust. G.H. was an Alzheimer's Society and Alzheimer Scotland Fellow. R.W.M. was a Wellcome Trust Research Career Development Fellow. Financial support for the work carried out in this manuscript was provided by Alzheimer's Research UK.
Footnotes
Disclosure statement The authors disclose no conflicts of interest.
Appendix: A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.neurobiolaging.2012.02.005.
References
- Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert JC, Bettens K, Le Bastard N, Pasquier F, Montoya AG, Peeters K, Mattheijssens M, Vandenberghe R, De Deyn PP, Cruts M, Amouyel P, Sleegers K, Van Broeckhoven C. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing c3b/c4b binding sites. Mol. Psychiatry. 2012;17:223–233. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, Ertekin-Taner N, Crook JE, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG. Genetic variation in pcdh11× is associated with susceptibility to late-onset alzheimer's disease. Nat. Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Yu G, Arawaka S, Nishimura M, Kawarai T, Yu H, Tandon A, Supala A, Song YQ, Rogaeva E, Milman P, Sato C, Yu C, Janus C, Lee J, Song L, Zhang L, Fraser PE, St George-Hyslop PH. Nicastrin binds to membrane-tethered notch. Nat. Cell Biol. 2001;3:751–754. doi: 10.1038/35087069. [DOI] [PubMed] [Google Scholar]
- Confaloni A, Terreni L, Piscopo P, Crestini A, Campeggi LM, Frigerio CS, Blotta I, Perri M, Di Natale M, Maletta R, Marcon G, Franceschi M, Bruni AC, Forloni G, Cantafora A. Nicastrin gene in familial and sporadic alzheimer's disease. Neurosci. Lett. 2003;353:61–65. doi: 10.1016/j.neulet.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O'Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren-Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ. Association of cr1, clu and picalm with alzheimer's disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum. Mol. Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin E, Hannequin D, Macé S, Dubois B, Ricard S, Génin E, Brun C, Chansac C, Pradier L, Frebourg T, Brice A, Campion D, Deleuze JF. No replication of the association between the nicastrin gene and familial early-onset alzheimer's disease. Neurosci. Lett. 2003;353:153–155. doi: 10.1016/s0304-3940(03)01105-4. [DOI] [PubMed] [Google Scholar]
- de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Hamilton G, Hayward C, Whalley LJ, Powell J, Starr JM, Lovestone S. Nicastrin gene polymorphisms, cognitive ability level and cognitive ageing. Neurosci. Lett. 2005;373:110–114. doi: 10.1016/j.neulet.2004.09.073. [DOI] [PubMed] [Google Scholar]
- Dermaut B, Theuns J, Sleegers K, Hasegawa H, Van den Broeck M, Vennekens K, Corsmit E, St George-Hyslop P, Cruts M, van Duijn CM, Van Broeckhoven C. The gene encoding nicastrin, a major gamma-secretase component, modifies risk for familial early-onset alzheimer disease in a dutch population-based sample. Am. J. Hum. Genet. 2002;70:1568–1574. doi: 10.1086/340732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium A map of human genome variation from population-scale sequencing [erratum in: 2011;473:544] Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan R, Swindells M, Overington J, Weir M. Nicastrin, a presenilin-interacting protein, contains an aminopeptidase/transferrin receptor superfamily domain. Trends Biochem. Sci. 2001;26:213–214. doi: 10.1016/s0968-0004(01)01789-3. [DOI] [PubMed] [Google Scholar]
- Fagan T. Large genetic analysis pays off with new ad risk genes. 2011 Available at: www.alzforum.org/new/detail.asp?id=2747.
- Fergani A, Yu G, St George-Hyslop P, Checler F. Wild-type and mutated nicastrins do not display aminopeptidase m- and b-like activities. Biochem. Biophys. Res. Commun. 2001;289:678–680. doi: 10.1006/bbrc.2001.6030. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at clu and picalm associated with Alzheimer's disease. Nat. Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helisalmi S, Dermaut B, Hiltunen M, Mannermaa A, Van den Broeck M, Lehtovirta M, Koivisto AM, Iivonen S, Cruts M, Soininen H, Van Broeckhoven C. Possible association of nicastrin polymorphisms and alzheimer disease in the finnish population. Neurology. 2004;63:173–175. doi: 10.1212/01.wnl.0000133153.98139.4e. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein M, Lawlor B, Lynch A, Brown KS, Passmore D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wil-cock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate JS, Kauwe C, Cruchaga P, Nowotny JC, Morris K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn MM, Breteler MA, Ikram AL, Destefano AL, Fitzpatrick O, Lopez LJ, Launer S, Seshadri C, Berr D, Campion J, Epelbaum JF, Dartigues C, Tzourio A, Alperovitch M, Lathrop TM, Feulner P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans T, Jonsson M, Riemen-schneider K, Younkin SG, Owen MJ, O'Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaether C, Capell A, Edbauer D, Winkler E, Novak B, Steiner H, Haass C. The presenilin c-terminus is required for er-retention, nicastrin-binding and gamma-secretase activity. EMBO J. 2004;23:4738–4748. doi: 10.1038/sj.emboj.7600478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, European Alzheimer's Disease Initiative Investigators. de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lupton MK, Proitsi P, Danillidou M, Tsolaki M, Hamilton G, Wroe R, Pritchard M, Lord K, Martin BM, Kloszewska I, Soininen H, Mecocci P, Vellas B, Harold D, Hollingworth P, Lovestone S, Powell JF. Deep sequencing of the nicastrin gene in pooled DNA, the identification of genetic variants that affect risk of alzheimer's disease. PLoS One. 2011;6:e17298. doi: 10.1371/journal.pone.0017298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Yamagata HD, Zhong W, Aoto M, Akatsu H, Uekawa N, Kamino K, Taguchi K, Yamamoto T, Maruyama M, Kosaka K, Takeda M, Kondo I, Miki T. A novel alternative splice variant of nicastrin and its implication in alzheimer disease. Life Sci. 2006;78:2444–2448. doi: 10.1016/j.lfs.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Mudher A, Chapman S, Richardson J, Asuni A, Gibb G, Pollard C, Killick R, Iqbal T, Raymond L, Varndell I, Sheppard P, Makoff A, Gower E, Soden PE, Lewis P, Murphy M, Golde TE, Rupniak HT, Anderton BH, Lovestone S. Dishevelled regulates the metabolism of amyloid precursor protein via protein kinase C/mitogen-activated protein kinase and c-Jun terminal kinase. J. Neurosci. 2001;21:4987–4995. doi: 10.1523/JNEUROSCI.21-14-04987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Benes V, Testa G, Ansorge W, Stewart AF. Point mutation of bacterial artificial chromosomes by et recombination. EMBO Rep. 2000;1:239–243. doi: 10.1093/embo-reports/kvd049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem. Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Huentelman J, Myers MJ, Barmada AJ, Demirci MM, Baldwin FY, Green RC, Rogaeva E, George-Hyslop PS, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui CHC, lark DG, Corneveaux J, Cotman CW, Cummings JL, Decarli C, Dekosky T, Diaz-Arrastia R, Dickson M, Ellis DW, Faber WG, Fallon KM, Farlow KB, Ferris MR, Frosch S, Galasko MP, Ganguli DR, Gearing M, Geschwind M, Ghetti DH, Gilbert B, Gilman JR, Giordani S, Glass B, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Karlawish N, Karydas J, Kaye A, Kim JA, Koo R, Kowall EH, Lah NW, Levey JJ, Lieberman AI, Lopez AP, Mack OL, Marson WJ, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piscopo P, Manfredi A, Malvezzi-Campeggi L, Crestini A, Spadoni O, Cherchi R, Deiana E, Piras MR, Confaloni A. Genetic study of sardinian patients with alzheimer's disease. Neurosci. Lett. 2006;398:124–128. doi: 10.1016/j.neulet.2005.12.063. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, 3rd, Südhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Shirotani K, Edbauer D, Capell A, Schmitz J, Steiner H, Haass C. Gamma-secretase activity is associated with a conformational change of nicastrin. J. Biol. Chem. 2003;278:16474–16477. doi: 10.1074/jbc.C300095200. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Spasic D, Raemaekers T, Dillen K, Declerck I, Baert V, Serneels L, Füllekrug J, Annaert W. Rer1p competes with aph-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J. Cell Biol. 2007;176:629–640. doi: 10.1083/jcb.200609180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ES, Martinez M, Wang J, Goate A. Conserved residues in juxtamembrane region of the extracellular domain of nicastrin are essential for gamma-secretase complex formation. J. Neurochem. 2006;98:300–309. doi: 10.1111/j.1471-4159.2006.03881.x. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Nishimura M, Arawaka S, Levitan D, Zhang L, Tandon A, Song YQ, Rogaeva E, Chen F, Kawarai T, Supala A, Levesque L, Yu H, Yang DS, Holmes E, Milman P, Liang Y, Zhang DM, Xu DH, Sato C, Rogaev E, Smith M, Janus C, Zhang Y, Aebersold R, Farrer LS, Sorbi S, Bruni A, Fraser P, St George-Hyslop P. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ding L, Sandford AJ. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol. Biol. 2005;6:4. doi: 10.1186/1471-2199-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in escherichia coli. Nat. Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Luo WJ, Wang H, Lin P, Vetrivel KS, Liao F, Li F, Wong PC, Farquhar MG, Thinakaran G, Xu H. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J. Biol. Chem. 2005;280:17020–17026. doi: 10.1074/jbc.M409467200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.