Abstract
A new coronavirus (severe acute respiratory syndrome coronavirus [SARS-CoV]) has been identified to be the etiological agent of severe acute respiratory syndrome. Given the highly contagious and acute nature of the disease, there is an urgent need for the development of diagnostic assays that can detect SARS-CoV infection. For determination of which of the viral proteins encoded by the SARS-CoV genome may be exploited as diagnostic antigens for serological assays, the viral proteins were expressed individually in mammalian and/or bacterial cells and tested for reactivity with sera from SARS-CoV-infected patients by Western blot analysis. A total of 81 sera, including 67 from convalescent patients and seven pairs from two time points of infection, were analyzed, and all showed immunoreactivity towards the nucleocapsid protein (N). Sera from some of the patients also showed immunoreactivity to U274 (59 of 81 [73%]), a protein that is unique to SARS-CoV. In addition, all of the convalescent-phase sera showed immunoreactivity to the spike (S) protein when analyzed by an immunofluorescence method utilizing mammalian cells stably expressing S. However, samples from the acute phase (2 to 9 days after the onset of illness) did not react with S, suggesting that antibodies to N may appear earlier than antibodies to S. Alternatively, this could be due to the difference in the sensitivities of the two methods. The immunoreactivities to these recombinant viral proteins are highly specific, as sera from 100 healthy donors did not react with any of them. These results suggest that recombinant N, S, and U274 proteins may be used as antigens for the development of serological assays for SARS-CoV.
The recent severe acute respiratory syndrome (SARS) epidemic, which affected over 30 countries, has profoundly disturbed social and economic activities regionally as well as globally. The high mortality rate of up to 15%, together with the highly contagious and acute nature of the disease, has imposed tremendous psychological and economic burden on the public. In Singapore and elsewhere, to reduce the risk of contact with people who may have been exposed to the SARS-causing virus, strict quarantine orders were served to those who had traveled to SARS-affected countries, those who had been in direct contact with SARS patients, and those with temperatures exceeding 38°C. Early diagnoses of the disease during the early phase of infection could avoid unnecessary quarantines, reduce the stress to those concerned, and help doctors to decide on appropriate medical action and/or treatment. It is therefore vital to identify SARS patients as early as possible, with certainty and accuracy. Given that no effective anti-SARS therapeutics are currently available, the first line of defense is to identify and isolate infected patients as early as possible. Hence, the need for the development of sensitive and highly specific diagnostic kits that can be used in the field is urgent and immediate.
A novel coronavirus was identified as the etiological agent of SARS (2, 3, 5, 10). Coronaviruses are enveloped viruses that contain a single-stranded, positive-sense RNA genome of 27.6 to 31 kb. Analyses of the nucleotide sequence of the novel SARS coronavirus (SARS-CoV) showed that the viral genome is nearly 30 kb in length (9, 11) and contains 14 potential open reading frames (ORFs) (9). With the identification of the SARS-CoV genome, several diagnostic tests based on the detection of viral RNA sequences by use of PCR have been designed and are now available. Such tests, although sensitive, have inherent problems: scientists and clinicians around the world are unsure what types of samples (respiratory samples, saliva, stool, blood, or conjunctival fluid) from patients give the most reproducible RNA preparations; RNA extraction protocols are not straightforward, and if not done well, may produce RNA preparations that are not useful for the reverse transcription step that converts viral RNA to DNA; and the whole process of extraction, reverse transcription, and PCR can be time-consuming if confirmatory tests have to be done with several pairs of primers. In addition, false positives are possible with amplification methods, as was observed in August 2003 in Canada, when some patients infected with other human coronaviruses initially tested positive for SARS by a PCR method (http://www.bccdc.org). Contamination in PCR laboratories is always a concern, which in the case of SARS could lead to unnecessary quarantines.
Another commonly used method for the detection of viral infections is a serological test that assays for the presence of antibodies against viral proteins. The sequence of SARS-CoV reveals ORFs for four structural proteins, i.e., spike (S), membrane (M), envelope (E), and nucleocapsid (N), which are present in all coronaviruses (6, 9, 11, 13). In addition, there are several ORFs predicted from the SARS-CoV sequence that encode proteins unique to SARS-CoV, as they show no significant sequence homology to viral proteins of other coronaviruses. In this study, we screened a panel of SARS-CoV ORFs expressed in mammalian and/or bacterial cells for reactivity toward convalescent-phase patient sera, as well as sera from acutely infected patients, to determine which antigen(s) would be most suitable as a diagnostic marker for the detection of SARS-CoV infection.
MATERIALS AND METHODS
Materials.
All reagents used in this study were purchased from Sigma (St. Louis, Mo.), unless otherwise stated. All cell lines were purchased from the American Type Culture Collection (Manassas, Va.) and were cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle medium containing 1 g of glucose/liter, 2 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, 0.1 mM nonessential amino acids, 0.1 mg of streptomycin/ml, 100 U of penicillin, and 5% fetal bovine serum (HyClone, South Logan, Utah).
Construction of plasmids.
For transient expression in mammalian cells, the vectors used were pXJ40HA, for tagging proteins at the N terminus with one hemagglutinin (HA) epitope (8), and pXJ40-3′HA, for tagging proteins with three HA epitopes at the C terminus (T. Leung, Institute of Molecular and Cell Biology, Singapore, Republic of Singapore, personal communication). For the expression of glutathione S-transferase (GST) fusion proteins in bacteria, genes were cloned in frame with GST in pGEX-4T-1 (Amersham Pharmacia Biotech, Uppsala, Sweden). For the stable transfection of S in CHO cells, a full-length S construct tagged at the C-terminal end with green fluorescent protein (GFP) was cloned into the pMMTC vector (14). All of the constructs used for this study are listed in Table 1.
TABLE 1.
Plasmids used for this study
| Plasmid | Nucleotide posi- tions in ORF | Amino acid positions | Total no. of amino acids |
|---|---|---|---|
| pXJ40HA-U274 | 400-822 | 134-274 | 141 |
| pXJ40-E-3′HA | 1-228 | 1-76 | 76 |
| pXJ40-M-3′HA | 1-663 | 1-221 | 221 |
| pXJ40-U122-3′HA | 1-366 | 1-122 | 122 |
| pXJ40HA-N | 203-1266 | 69-422 | 354 |
| pMMTC-S-GFP | 1-3765 | 1-1255 | 1,255 |
| pGEX-4T1-U274 | 400-822 | 134-274 | 141 |
| pGEX-4T1-U122 | 46-333 | 16-111 | 96 |
| pGEX-4T1-N | 357-1266 | 120-422 | 303 |
SARS-CoV 2003VA2774, an isolate from a SARS patient in Singapore, was used for this study. For the cloning of various genes, RNAs were extracted by use of a Qiagen viral RNA kit (Valencia, Calif.) from a SARS-CoV-infected Vero E6 cell culture supernatant harvested when the cultures showed at least 75% cytopathic effects. Reverse transcription was performed with Superscript II RNase H− reverse transcriptase (Gibco BRL, Gaithersburg, Md.) and an oligo(dT) primer. PCRs were performed with either HotStar polymerase (Qiagen), Titanium Taq DNA polymerase (Clontech Laboratories Inc., Palo Alto, Calif.), or High Fidelity Taq polymerase (Roche Molecular Biochemicals, Indianapolis, Ind.). In some cases, overlapping cDNAs provided by the Genome Institute of Singapore (isolate SIN2774; accession no. AY283798 [12]) were used as templates instead.
All of the primers used for this study were synthesized by Genset Singapore Biotech (Singapore). The sequences of all constructs used in this study were confirmed by DNA sequencing performed at the core facility at the Institute of Molecular and Cell Biology by the dideoxy method with a Taq DyeDeoxy terminator cycle sequencing kit and an automated DNA sequencer (model 373) from PE Applied Biosystems (Foster City, Calif.).
Transient transfection of mammalian cells.
Transient transfection experiments were performed with Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. Typically, ∼106 COS-7 cells were plated on a 6-cm-diameter dish and allowed to attach for at least 4 h. One to 2 μg of DNA was used per plate, and the cells were left for at least 14 h before the cells were washed with phosphate-buffered saline (PBS), lysed directly in Laemmli's sodium dodecyl sulfate (SDS) buffer, and used for Western blot analysis.
Expression of GST fusion proteins.
Exponentially growing cultures (optical density at 600 nm of ∼0.7) of Escherichia coli (BL21/DE3) cells harboring the pGEX-4T-1 expression constructs were induced to synthesize fusion proteins by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), after which the cells were allowed to grow for another 4 h at 37°C or 12 h at 30°C. Cells were harvested, resuspended in PBS containing 0.5% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride, and then sonicated with an ultrasonic processor (Misonix Inc., Farmingdale, N.Y.). GST fusion proteins were then purified from the lysate by use of glutathione (GSH)-Sepharose beads (Amersham Pharmacia Biotech). Proteins were eluted from the beads with 10 mM reduced GSH in 50 mM Tris-HCl, pH 9.2, and 0.1% SDS, and protein concentrations were determined by use of a Coomassie Plus assay kit (Pierce, Rockford, Ill.). Proteins were also separated in SDS-polyacrylamide gels and stained with 0.25% Coomassie brilliant blue R-250 (Bio-Rad, Hercules, Calif.) in 45% methanol and 10% acetic acid.
Western blot analysis.
For Western blot analysis, approximately 105 transfected COS-7 cells or 50 ng of GST fusion proteins was separated in SDS-polyacrylamide gels and transferred to nitrocellulose Hybond C membranes (Amersham Pharmacia Biotech). The membranes were blocked with 5% nonfat dry milk. For the detection of HA-tagged or GST fusion proteins, the membranes were incubated with either an anti-HA polyclonal or anti-GST monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) overnight at 4°C and washed extensively with PBST (PBS containing 0.05% Tween 20), followed by incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (Pierce) for 1 h at room temperature. Membranes were then washed extensively with PBST, and the detection of signals by an enhanced chemiluminescence method (Pierce) was performed. For patient sera, each sample was first treated with 0.5% Triton X-100 and 0.1 mg of RNase (Sigma)/ml and then diluted 1:150 to 1:500 with PBST containing 1% nonfat dry milk. After incubation for 1 to 3 h at room temperature or overnight at 4°C, the membranes were incubated with an anti-human HRP-conjugated immunoglobulin G (IgG) (Santa Cruz Biochemicals), IgA, or IgM (Zymed Laboratories Inc., San Francisco, Calif.) antibody for 1 h at room temperature, followed by detection as described above. All secondary antibodies were used at a 1:2,000 dilution. Due to the limited amount of patient sera from the seven pairs of sera, Western blots of a mixture of three GST fusion proteins, N, U274, and U122, and GST were run in a single-slot SDS-10% polyacrylamide gel electrophoresis (PAGE) gel and transferred onto nitrocellulose membranes. The membranes were cut into strips of about 0.8 mm wide, and each strip was probed with 300 to 400 μl of diluted sera. For the late time point sera, the strips were incubated with the diluted sera for 1.5 h at room temperature. For the early time point sera, the strips had to be incubated overnight with the sera, at room temperature, to detect any signal. The secondary antibodies were incubated for 1 h at room temperature. This method was also used when large numbers of samples were being screened.
Expression of S-GFP fusion protein in CHO cells and immunofluorescence analysis to determine anti-S immunoreactivity in patient sera.
CHO cells were transfected with pMMTC-S-GFP by use of DMRIE-C reagent (Gibco BRL) according to the manufacturer's protocol. Transfected cells were selected in Geneticin (Gibco BRL) for >1 week. Cells were then analyzed under a Zeiss microscope (Carl Zeiss Vision GmbH, Hallbergmoss, Germany), and the clone with the strongest expression signal was picked and grown in medium containing 100 μM ZnSO4. Zn2+ ions increase the expression of genes by the pMMTC vector (14).
Cells were dislodged from the plates with 0.04% EDTA, seeded onto black Teflon Menzel diagnostic slides (Merck), and blown dry, followed by fixing in acetone at −20°C for 1 h. Then the cells were incubated with sera at dilutions of 1:20, 1:40, 1:80, and 1:160 (in PBS) for 1.5 h at 37°C, followed by incubation with a fluorescein isothiocyanate (FITC)- or rhodamine (Rh)-conjugated anti-human IgG (Sigma) at a 1:20 dilution for 1.5 h at 37°C. When Rh-conjugated anti-human IgG was used, it was diluted in PBS, and when FITC-conjugated anti-human IgG, IgM (Sigma), or IgA (Dako A/S, Glostrup, Denmark) was used, it was diluted in 0.05% Evans Blue solution (Fluka, Buchs, Switerland), which blocks GFP fluorescence from transfected cells. Slides were then viewed under a Zeiss microscope (Carl Zeiss Vision GmbH) and scored as follows: +++, very strong staining; ++, moderate staining; and +, weak staining.
Collection of sera from SARS-CoV-infected patients.
The first six serum samples (from patients 1 to 6), except for that from patient 3, were obtained by plasmapheresis. Plasmapheresis was performed on patients who (i) had previous documented SARS-CoV infection according to World Health Organization (WHO) criteria, (ii) had been in the convalescent phase for at least 6 weeks, and (iii) were symptom-free and willing to participate in the study. Blood was taken from patient 3, and the serum was separated out for the experiments. All participants have given written consent, and approval from the Tan Tock Seng Hospital Ethics Committee has been granted.
The second set of sera consisted of seven pairs of samples that were collected from patients admitted to Tan Tock Seng Hospital or Singapore General Hospital. These patients were admitted upon the onset of illness, as defined by WHO criteria, and the dates of admission and dates of collection of blood are shown in Table 2. The first samples (set A) were collected 2 to 11 days after the onset of illness, and the second set of samples (set B) were collected 16 to 54 days after the onset of illness.
TABLE 2.
Description of sera collected from seven SARS-CoV-infected patients at two different time points
| Patient | Sample | Date of onset of illness | Date of sample collection | No. of days after onset of illness |
|---|---|---|---|---|
| D2 | 2A | 18 March 2003 | 20 March 2003 | 2 |
| 2B | 18 March 2003 | 15 April 2003 | 28 | |
| D3 | 3A | 17 March 2003 | 20 March 2003 | 3 |
| 3B | 17 March 2003 | 17 April 2003 | 31 | |
| D4 | 4A | 16 March 2003 | 20 March 2003 | 4 |
| 4B | 16 March 2003 | 31 May 2003 | 48 | |
| D5 | 5A | 2 April 2003 | 7 April 2003 | 5 |
| 5B | 2 April 2003 | 29 April 2003 | 27 | |
| D8 | 8A | 19 March 2003 | 27 March 2003 | 8 |
| 8B | 19 March 2003 | 4 April 2003 | 16 | |
| D9 | 9A | 9 March 2003 | 18 March 2003 | 9 |
| 9B | 9 March 2003 | 2 May 2003 | 54 | |
| D11 | 11A | 24 February 2003 | 7 March 2003 | 11 |
| 11B | 24 February 2003 | 3 April 2003 | 38 |
The last set of sera was obtained from 61 probable SARS patients who were discharged from Tan Tock Seng Hospital. For some of these patients, sera were taken upon discharge (3 to 4 weeks after the onset of illness), while most of the patients were recruited >3 weeks after discharge (>6 weeks after the onset of illness) (see Table 3 for details). For one patient (patient P3L), a serum sample was taken 111 days (3 1/2 months) after the onset of illness. Samples from two other patients (P7 and P8) were obtained by plasmapheresis.
TABLE 3.
Reactivities of serum samples collected from 74 probable SARS patients 16 to 111 days after the onset of illness against N, U274, and S by Western blot analysis. Samples from two healthy contacts (Patient I.D. 868 and 873) were also examined.
| Patient no. | Reactivity to N proteina | Reactivity to U274 proteina | Reactivity to S proteinb | No. of days after onset of illnessc | Patient no. | Reactivity to N proteina | Reactivity to U274 proteina | Reactivity to S proteinb | No. of days after onset of illnessc | |
|---|---|---|---|---|---|---|---|---|---|---|
| 174 | +++ | ++ | + | 49 | ||||||
| 318 | +++ | − | + | 53 | ||||||
| 350 | +++ | ++ | + | 62 | ||||||
| 358 | +++ | + | + | 60 | ||||||
| 377 | +++ | + | + | 61 | ||||||
| 387 | +++ | − | + | 63 | ||||||
| 432 | +++ | ++ | +++ | 42 | ||||||
| 442 | +++ | + | ++ | 63 | ||||||
| 487 | +++ | + | ++ | 54 | ||||||
| 492 | +++ | + | + | 31 | ||||||
| 526 | +++ | + | + | 63 | ||||||
| 541 | +++ | + | ++ | 58 | ||||||
| 546 | +++ | ++ | ++ | 62 | ||||||
| 561 | +++ | ++ | + | 57 | ||||||
| 566 | +++ | + | ++ | 59 | ||||||
| 571 | +++ | + | + | 57 | ||||||
| 576 | +++ | + | ++ | 63 | ||||||
| 581 | +++ | − | ++ | 53 | ||||||
| 586 | +++ | ++ | ++ | 38 | ||||||
| 596 | +++ | − | + | 39 | ||||||
| 603 | +++ | ++ | ++ | 67 | ||||||
| 621 | +++ | ++ | + | 50 | ||||||
| 633 | +++ | − | +++ | 39 | ||||||
| 638 | +++ | + | + | 60 | ||||||
| 644 | +++ | + | ++ | 41 | ||||||
| 672 | +++ | + | + | 53 | ||||||
| 677 | +++ | ++ | ++ | 59 | ||||||
| 682 | +++ | − | + | 58 | ||||||
| 687 | +++ | + | ++ | 61 | ||||||
| 696 | +++ | − | + | 41 | ||||||
| 701 | +++ | ++ | ++ | 40 | ||||||
| 706 | +++ | + | ++ | 39 | ||||||
| 711 | +++ | − | + | 54 | ||||||
| 716 | +++ | − | ++ | 40 | ||||||
| 726 | +++ | + | ++ | 34 | ||||||
| 734 | +++ | − | + | 41 | ||||||
| 739 | +++ | − | ++ | 49 | ||||||
| 744 | +++ | + | ++ | 68 | ||||||
| 749 | +++ | +++ | ++ | 59 | ||||||
| 759 | +++ | + | + | 42 | ||||||
| 764 | +++ | + | ++ | 43 | ||||||
| 769 | +++ | + | ++ | 64 | ||||||
| 774 | +++ | + | ++ | 62 | ||||||
| 784 | +++ | − | +++ | 38 | ||||||
| 789 | +++ | − | ++ | 55 | ||||||
| 793 | +++ | + | + | 48 | ||||||
| 798 | +++ | − | ++ | 44 | ||||||
| 803 | +++ | − | ++ | 40 | ||||||
| 823 | +++ | + | ++ | 37 | ||||||
| 840 | +++ | ++ | +++ | 43 | ||||||
| 845 | +++ | ++ | ++ | 36 | ||||||
| 859 | +++ | +++ | ++ | 55 | ||||||
| 868d | − | − | − | Not applicable | ||||||
| 873d | − | − | − | Not applicable | ||||||
| 878 | +++ | ++ | +++ | 58 | ||||||
| 883 | +++ | + | ++ | 56 | ||||||
| 888 | +++ | + | ++ | 33 | ||||||
| 893 | +++ | ++ | +++ | 41 | ||||||
| 4153 | +++ | − | ++ | 41 | ||||||
| 4209 | +++ | − | ++ | 21 | ||||||
| P3L | +++ | ++ | + | 111 | ||||||
| P1e | +++ | ++ | ++ | >42 | ||||||
| P2e | +++ | − | + | >42 | ||||||
| P3 | +++ | ++ | + | >42 | ||||||
| P4e | +++ | ++ | + | >42 | ||||||
| P5e | +++ | ++ | + | >42 | ||||||
| P6e | +++ | ++ | + | >42 | ||||||
| P7e | +++ | + | + | >42 | ||||||
| P8e | +++ | ++ | ++ | >42 | ||||||
| 2Bf | +++ | + | ++ | 28 | ||||||
| 3Bf | +++ | ++ | +++ | 31 | ||||||
| 4Bf | +++ | + | + | 48 | ||||||
| 5Bf | +++ | + | +++ | 27 | ||||||
| 8Bf | +++ | + | +++ | 16 | ||||||
| 9Bf | +++ | + | ++ | 54 | ||||||
| 11Bf | +++ | ++ | ++ | 38 |
Determined by Western blot analysis. +++, ++, +, very strong, moderate, and weak signals, respectively, were observed on the autoradiograph; −, no signal was observed.
Determined by an immunofluorescence method in which CHO cells stably expressing the S protein were incubated with a diluted patient serum (1:40) followed by a FITC-conjugated anti-human IgG antibody. Slides were then viewed under a Zeiss microscope and scored as follows: +++, very strong staining; ++, moderate staining; +, weak staining. One hundred sera from healthy volunteers were examined at the same dilution, and none of them showed any staining.
Samples were obtained at the stated number of days after the onset of illness.
Samples from two healthy contacts (patients 868 and 873) were also examined.
Samples obtained by plasmapheresis.
Late time point from one of the seven pairs of sera described in Table 2.
A control serum was purchased from Sigma, and 99 serum samples were obtained from healthy donors who have given informed consent. These healthy donors (i) did not have a diagnosis of SARS, suspect SARS, or have contact with a person who was served a home quarantine order for SARS; (ii) did not have a fever, symptoms of influenza, runny nose, or sore throat within the last week at the time of donation; (iii) had not been on immunosuppressants; (iv) had no significant medical illnesses; and (v) had no history of travel to SARS-affected areas since November 2002.
RESULTS
Detection of IgG antibodies against viral proteins encoded by the SARS-CoV genome in the sera of probable SARS patients.
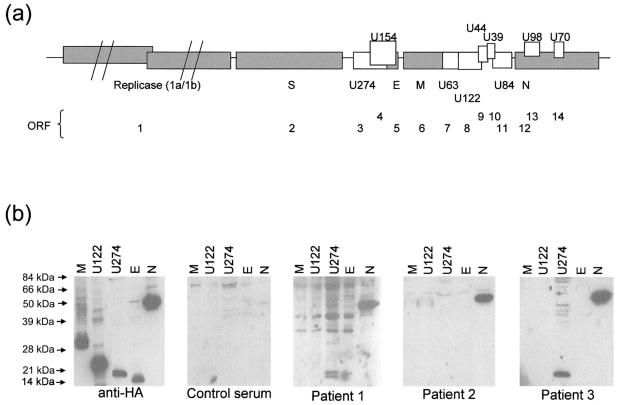
In order to assess which of the viral proteins encoded by the SARS-CoV genome may be exploited as diagnostic antigens for the development of a serological assay to detect SARS-CoV infection, we cloned three of the four structural proteins (E, M, and N) into expression vectors. Due to the large size (∼4.7 kbp) and heavy glycosylation of the spike (S) protein, it was analyzed separately by an immunofluorescence method (see below). In addition, two of the SARS-CoV unique proteins (U274 and U122) were also included in this study, and they will be referred to hereafter as UX, where “X” stands for the number of predicted amino acids of the proteins. In this study, we used only the C-terminal hydrophilic region of U274, as it has three potential transmembrane domains at its N-terminal end and these hydrophobic regions may affect the solubility of a recombinant U274 protein. The positions of the structural and unique proteins and their corresponding ORF numbers, as designated by Marra and coworkers (9), are shown in Fig. 1a.
FIG. 1.
Structural organization and expression of ORFs of SARS-CoV. (a) ORFs corresponding to structural proteins S, M, N, and E and unique proteins (UX) and corresponding annotated ORFs (1 to 14) are indicated. “X” denotes the number of amino acids encoded by the respective ORF. (b) Western blot analysis to determine the presence of antibodies against the various SARS-CoV viral proteins in patient sera. HA-tagged proteins were expressed in mammalian cells and probed with anti-HA antibody, antibodies in a control serum, or antibodies in three convalescent-phase sera (patients 1 to 3).
This panel of HA-tagged proteins was expressed in COS-7 cells by transient transfection, and total protein lysates were analyzed by Western blot analysis with sera from three convalescent patients (Fig. 1b, patients 1 to 3) to determine if these sera had any antibodies against the expressed proteins. Plasmapheresis was performed on patients 1 and 2, and the plasmapheresis products were used in Western blot analysis, while for patient 3, serum was used for the analysis. All three patients had antibodies against the N (amino acids [aa] 69 to 422) protein, but not against the other structural proteins, M and E. Interestingly, the C-terminal hydrophilic region of U274 (aa 134 to 274) was also detected by the sera of patients 1 and 3 but not by the serum of patient 2 (Fig. 1b). A control serum did not detect N, U274, or any other proteins. U122 was also not detected by any of the three sera, suggesting that it may not be expressed, it is not a structural protein, or it is not sufficiently antigenic.
As bacterially expressed proteins are easier and cheaper to produce on a large scale, we next expressed N and U274 as GST fusion proteins and tested them for reactivity to patient sera. Coomassie staining showed that GST-N (aa 120 to 422) and GST-U274 (aa 134 to 274) were of >90% purity after a one-step purification with GSH-Sepharose beads (Fig. 2a). The GST-N (aa 120 to 422) protein used here lacked the N-terminal part of the N protein, which contains a highly conserved motif (FYYLGTGP; aa 111 to 118 of SARS-CoV N) found in all coronaviruses (11), so there would be less chance of cross-reactivity with antibodies against other coronaviruses. From a 400-ml bacterial culture, ∼10 mg of GST-N and ∼2 mg of GST-U274 could be obtained, respectively. GST-U122 (aa 16 to 111) (lacking the signal peptide at the N terminus and the hydrophobic C terminus) and GST were also expressed and used as controls. The GST fusion proteins were probed with the same three patient sera and one control serum, as for the mammalian expressed proteins (Fig. 1b). Consistent with the proteins expressed in COS-7 cells, GST-N was clearly detected by all three patient sera and GST-U274 was detected only by the sera from patients 1 and 3 (Fig. 2b). Neither GST-U122 nor GST showed any background with all three patient sera, and there was no nonspecific binding of the control serum to any of the proteins (Fig. 2b). Sera from another three convalescent patients (patients 4 to 6) were also tested, and they were reactive specifically towards both GST-N and GST-U274, but not to GST-U122 or GST (Fig. 2b).
FIG. 2.
Expression and detection of bacterially expressed viral proteins with convalescent-phase patient sera. (a) GST-U274, GST-U122, GST-N, and GST proteins were expressed in bacteria and purified with GSH-Sepharose beads. Proteins were analyzed by SDS-PAGE and stained with Coomassie brilliant blue R-250 (Bio-Rad). Bovine serum albumin (BSA) standards were also run in the same gel. (b) These proteins (approximately 150 ng of each) were separated by SDS-PAGE and subjected to Western blot analysis with an anti-GST antibody, antibodies in a control serum, or antibodies in six convalescent-phase sera (patients 1 to 6).
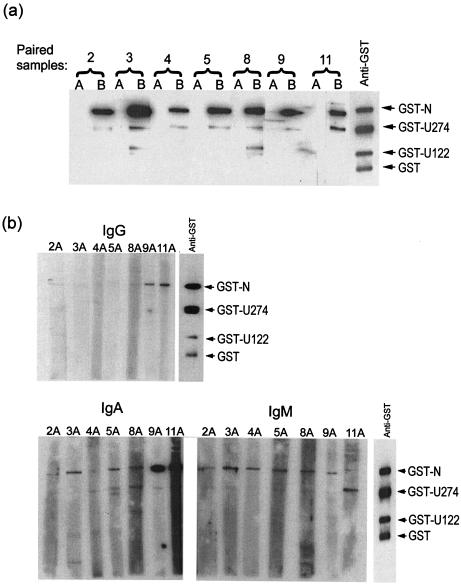
Profile of IgG, IgM, and IgA antibodies against N and U274 proteins in seven pairs of sera obtained at two time points of infection.
A set of paired sera from seven patients were also examined for reactivity against GST-N and GST-U274. These were obtained at two time points of infection, one at 2 to 11 days after the onset of illness (set A) and another at 16 to 54 days after the onset of illness (set B) (Table 2). For all seven cases, the second time point samples were taken at least 8 days after the first time point samples. As shown in Fig. 3a, anti-IgG antibodies against GST-N protein were present in all seven patients' sera at the later time point (set B), but not at the early time points (set A). For both sets of sera, the membranes were incubated with the diluted sera (1:150) for 1.5 h at room temperature, followed by incubation with the secondary antibody (HRP-conjugated anti-human IgG) for 1 h at room temperature. In addition, all seven of the late time point sera also contained antibodies against GST-U274, albeit at a lower level than that for GST-N (Fig. 3a, set B).
FIG. 3.
Detection of N and U274 with sera from patients during early and late phases of infection. (a) Reactivity to GST-N and GST-U274 was determined with sera from seven pairs of samples. Set A represents samples from an early time point (2 to 11 days after the onset of illness) and set B represents samples collected at a later time point (16 to 54 days after the onset of illness). GST fusion proteins were transferred to nitrocellulose membranes, and the membranes were incubated with diluted sera for 1.5 h at room temperature, followed by incubation with an HRP-conjugated anti-human IgG secondary antibody for 1 h at room temperature. (b) Samples from set A were reexamined by using three different secondary antibodies, namely anti-human IgG, anti-human IgA, or anti-human IgM antibody. The membranes were incubated with diluted sera overnight at room temperature followed by incubation with the respective secondary antibody for 1 h at room temperature.
We repeated the experiment for the early time point sera (set A) but incubated the blots with diluted sera overnight at room temperature instead of for 1.5 h and used three different secondary antibodies, anti-IgG, anti-IgM, and anti-IgA, and the results for each of them are shown in Fig. 3b. For IgG, only samples 9A and 11A showed some reactivity to GST-N. For IgA, though, all seven samples showed reactivity to GST-N, and the signals were particularly strong for samples 9A and 11A. For IgM, all seven samples showed a low level of reactivity to GST-N. As the early time point sample for each patient was collected a different number of days after the onset of illness, our results suggest that by as early as 2 days after the onset of illness (patient D2, sample 2A), IgM and IgA antibodies against the N protein were present. By 9 days after the onset of illness, very high levels of IgA antibodies against N were present and IgM and IgG antibodies were also detectable (Fig. 3b, samples 9A and 11A). As for GST-U274, some reactivities were observed for IgA (samples 5A, 8A, 9A, and 11A) and for IgM (sample 11A) from 5 to 11 days after the onset of illness. Interestingly, quite a strong signal for GST-U274 was observed for sample 11A (11 days after the onset of illness) when IgM was used, and for the corresponding early sample, 11B, the signal for GST-U274 was also strong (Fig. 3).
Determination of the sensitivity and specificity of immunoreactivity (IgG) against N and U274 proteins.
In order to determine the sensitivity and specificity of the immunoreactivity against N and U274, we obtained another 61 samples from probable SARS patients who were discharged from hospitals and tested them for the presence of IgG antibodies against N and U274 by Western blot analysis. As shown in Table 3, the samples were taken from 31 to 111 days after the onset of illness. Plasmapheresis products from 2 of the patients (P7 and P8) and sera from the other 59 patients were used. All of the samples showed strong immunoreactivity against the N protein (100%), and 44 of the 61 samples (72%) were positive for U274 (Table 3). In general, the signals observed for the U274 protein were much lower than those for the N protein. The exceptions were samples from patients 749 and 859, for which the signals for the U274 and N proteins were equally strong. Another 99 samples from healthy donors were also tested, and none of them showed any immunoreactivity to N or U274. In addition, samples from two patients (patients 868 and 873) who had close contact with probable SARS patients but never developed any of the clinical symptoms also did not show any immunoreactivity to N or U274 (Table 3). Therefore, the immunoreactivity against the N protein is highly sensitive (found in 100% of a total of 81 samples from probable SARS patients) and specific.
Detection of IgG antibodies against SARS-CoV S protein in convalescent-phase sera by immunofluorescence.
Due to the large size and heavy glycosylation of the S protein, it is advantageous to express this protein in mammalian cells instead of bacteria, as it is then possible to detect antibodies that may be dependent on conformation and/or glycosylation of the S protein. CHO cells were stably transfected with a full-length S construct tagged at the C-terminal end with GFP. After selection with antibiotics, a pool of clones expressing significant levels of S protein, as indicated by GFP fluorescence, were obtained (Fig. 4). These CHO cells were used for an immunofluorescence staining method to determine if there were IgG antibodies against the SARS-CoV S protein in convalescent-phase sera. Briefly, the cells were fixed with acetone and then incubated with diluted patient sera, followed by incubation with an Rh-conjugated anti-human IgG antibody. As shown in Fig. 4, sera from patients 1 to 6 all contained IgG antibodies against S, as detected by immunofluorescence, and no signal was observed when a control serum was used.
FIG. 4.
Detection of anti-S antibodies in patient sera by an immunofluorescence method utilizing mammalian cells stably expressing the S protein. Pools of CHO cells which were stably transfected with a full-length S construct tagged at the C-terminal end with GFP showed GFP fluorescence (GFP panel). These cells were fixed and incubated with control or patient sera (diluted 1:40), followed by an Rh-conjugated anti-human IgG antibody. Cells incubated with all patient sera (patients 1 to 6) showed strong staining, but no staining was seen when a control serum was used.
All remaining samples were tested similarly, except that the Rh-conjugated anti-human IgG antibody was replaced with a FITC-conjugated one in the presence of a high concentration of Evans Blue solution (0.05%). This is because of the greater ease of judging cells stained with FITC than those stained with Rh, and this concentration of Evans Blue solution was sufficient to block out all of the GFP fluorescence in the transfected cells (data not shown). As shown in Table 3, 100% of the convalescent-phase sera (74 samples) showed immunoreactivity against S at a dilution of 1:40, and 100 samples from healthy donors did not show any signal at the same dilution. The lowest dilution tested was 1:20, but a few of the samples from healthy donors showed weak signals at this concentration; therefore, the results from the 1:40 dilution were used for comparison. At higher dilutions (1:80 and 1:160), weaker signals were observed for most of the patient sera, as would be expected. In particular, for samples 432, 633, 784, 840, 878, 893, 3B, 5B, and 8B, for which very strong signals (+++) were observed at a 1:40 dilution, the signals decreased gradually at higher dilutions, i.e., moderate signals (++) were observed at 1:80 and weak signals (+) were observed at 1:160.
The seven early time point samples (Table 2, set A) were also analyzed in the same manner, but with IgG, IgM, and IgA secondary antibodies separately, but none of them showed any reactivity (at a 1:40 dilution) (data not shown).
DISCUSSION
The sequence of SARS-CoV reveals ORFs for four structural proteins, i.e., S, M, E, and N, which are present in all coronaviruses (6, 9, 11, 13). The S protein plays essential roles in mediating receptor binding and internalization of the virus and is one of the major antigens of the virus. The M and E proteins are essential for virion assembly, and the N protein binds to the viral genome to form the nucleocapsid. Besides these four common structural proteins, there are several unique ORFs predicted from the SARS-CoV sequence. Whether these ORFs are expressed or not and whether the expressed proteins serve a function in the viral replication cycle are yet to be determined.
We first screened a panel of ORFs expressed in mammalian and bacterial cells for reactivity towards convalescent-phase patient sera. As shown in Fig. 1b and 2b, the N protein was found to have strong immunoreactivity (IgG) with six of the convalescent-phase sera tested. Further tests using seven pairs of sera from patients also showed that high levels of IgG antibodies against the N protein were present at later time points (16 to 54 days) (Table 2) for all samples (Fig. 3a, set B). For the early time points (set A), there were lower levels of IgG and IgM antibodies against N, but for two of the patients, D9 and D11, the IgA antibodies against N were present at high levels (Fig. 3b, samples 9A and 11A). As the early time point serum from patient D9 (sample 9A) was taken at day 9 after the onset of illness, the data suggest that a high level of IgA antibodies against N can be present by this time. Taken together, all seven early samples (set A) showed immunoreactivities to the N protein, and one of the samples was taken as early as 2 days after the onset of illness (sample 2A). Finally, a third set of 61 samples from convalescent patients was 100% positive for IgG antibodies against N (Table 3). These samples were collected from 31 to 111 days after the onset of illness, and the high level of IgG antibodies against N observed here is consistent with a recent report that IgG antibodies against SARS-CoV in patients persisted for >13 weeks after the onset of symptoms (7). The presence of a strong immunoreactivity towards N suggests that it may be released from the virus or infected cells into the circulation at some stage of infection or that it may be presented by antigen-presenting cells for cytotoxic killing of infected cells. We therefore speculate that IgG antibody against N may contribute to the humoral immune response protecting patients against SARS, as has been observed for other coronaviruses (13). Future experiments will aim to address the mechanism by which N is presented as an antigen and its role in viral infection and/or replication. Our results also complement the preliminary mass spectrometric identification of N as the most immunogenic viral antigen (4). Interestingly, unlike some coronaviruses in which the M protein is most abundant, N appears to be the most abundant protein in SARS-CoV (11).
As may be expected, 100% (74 of 74) of the convalescent-phase sera also showed reactivity to the large S glycoprotein, which is responsible for the petal-shaped spikes found on the surfaces of coronaviruses (6, 13). In this case, we used full-length S expressed in mammalian cells (CHO) to achieve proper folding and glycosylation of the protein and immunofluorescence techniques for analysis. This has some advantages over immunofluorescence using SARS-CoV-infected Vero cells, as the latter would require a biosafety level 3 facility. However, this method still needs specialized expertise and is labor-intensive and thus not suited to mass testing. Therefore, in future studies, it is important to determine the epitope(s) on the S protein that is recognized by the antibodies in patient sera, as this epitope(s) may be easier to express than the full-length S protein and may allow the development of chromatographic or enzyme-linked immunosorbent assay (ELISA)-based diagnostic assays.
In addition, we also found that the majority of the patients (59 of 81 [73%]) had antibodies against the C-terminal end of U274 (corresponding to ORF 3, as annotated in reference 9, and X1 in reference 11), a protein unique to SARS-CoV. The immunoreactivity of U274 indicates that this novel and unique viral protein is expressed and is likely to be a protein involved in the biogenesis of SARS-CoV. Moreover, a potential transcription regulating sequence was found upstream of the U274 coding sequence, suggesting that it is the first ORF for one of the major subgenomic RNAs of SARS-CoV-infected cells (9, 11). U274 does not share any significant homology with any protein in the database, but it appears to have a similar topology to that of M, with three transmembrane regions and a large internal C-terminal domain. It would be interesting to unravel the function of U274. The patients in this study showed differential response to N and U274, and it is important to determine in future studies if this difference correlates with any significant clinical parameters.
With the identification of S, N, and U274 as antigenic proteins, diagnostic tests can be developed to detect the presence of antibodies against these proteins in suspected patients. Because of the fact that early-phase sera of all seven patients showed immunoreactivity to N, IgA and/or IgM offers a powerful and reliable system for the diagnosis of early infection. In addition, the presence of high levels of IgG antibodies against these proteins, in particular the N protein, in late infection or convalescent-phase sera suggests that this system would also be suitable for epidemiological studies. Further studies of sequentially bled patients in early stages of infection will be needed to characterize the development of these antibodies over time, and consequently, their utility in early infection. The N and U274 proteins can easily be expressed in bacteria and may be purified to a high degree, which might provide a better sensitivity and specificity over the current use of coarse viral lysates for serological assays, as the presence of antibodies against cellular components in viral lysates can result in false positivity. Therefore, the use of these recombinant viral antigens in a rapid chromatographic or ELISA-based kit can be expected to yield a superior diagnostic test. Due to the difficulty of expressing the S protein, the N protein appears to be the best candidate for the development of serological assays for the rapid detection of SARS-CoV infection. Indeed, the recombinant proteins S and U274 described here have been shown to work well in an ELISA format (3a).
It is still not clear whether SARS-CoV undergoes a high rate of genetic mutation, although some differences have been observed between the viral genomes sequenced so far (for examples, see references 1, 12, and 15). However, the S protein is predicted to contain hypervariable regions, as these have been found in other coronaviruses (6, 13), and this might pose another problem for the use of S in diagnostic assays. For the N protein, when we compared the sequences of 18 SARS isolates that were deposited in the GenBank database, we found that only two of them showed a difference, at one amino acid, from that of the Singapore isolate (SIN2774) that we used for this study (data not shown; also see reference 12). Therefore, the N protein does not appear to undergo rapid mutation, which is another advantage for its use for serological assay development. For U274, there is a slightly larger variance between the different reported sequences, with five sequences showing one amino acid substitution (when compared to SIN2774) and one sequence showing two amino acid substitutions (1). This may be one reason why fewer of the patient sera (73% compared to 100% for the N protein) reacted with the recombinant U274 protein used here.
This study is the first to dissect the range of antibody responses against SARS-CoV during infection. In summary, we have shown that antibodies against the S and N proteins are present in convalescent-phase sera from patients fulfilling the WHO case definition of probable SARS but not in sera from healthy contacts or volunteers. This observation is consistent with the finding that SARS-CoV is the causative agent of SARS. The analysis of seven pairs of sera showed that lower levels of antibodies (mainly IgM and IgA) against N were present during the acute phase of infection and that high levels of IgG antibodies were detected after 16 days or more. To date, serological diagnosis for SARS has been the most reliable method, but it requires convalescent-phase serum collected 28 days after the onset of illness to be the most accurate (http://www.cdc.gov/ncidod/sars/testresultsc.htm) and is thus retrospective. Our data showed that IgM and/or IgA assays could help in early diagnosis. Although Li and coworkers (7) reported that patients were positive for IgM only from week 2 of illness (when assayed with coarse viral lysates), the use of recombinant N protein may increase the sensitivity of ELISA assays and allow for earlier detection.
Acknowledgments
We thank the Genome Institute of Singapore for providing overlapping cDNAs from the SARS-CoV isolate SIN2774 and acknowledge the professional assistance provided by the Centre for Transfusional Medicine, Singapore General Hospital, for the plasmapheresis of convalescent patient donors.
This work was supported by grants from the Agency for Science, Technology and Research (A*STAR), Singapore.
REFERENCES
- 1.Chen, L. L., H. Y. Ou, R. Zhang, and C. T. Zhang. 2003. ZCURVE_CoV: a new system to recognize protein coding genes in coronavirus genomes, and its applications in analyzing SARS-CoV genomes. Biochem. Biophys. Res. Commun. 307:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Guan, M., H. Y. Chen, S. Y. Foo, Y.-J. Tan, P.-Y. Goh, and S. H. Wee. 2004. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 11:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krokhin, O., Y. Li, A. Andonov, H. Feldmann, R. Flick, S. Jones, U. Stroeher, N. Bastien, K. V. Dasuri, K. Cheng, J. N. Simonsen, H. Perreault, J. Wilkins, W. Ens, F. Plummer, and K. G. Standing. 2003. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics 2:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 6.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviruses, p. 1163-1185. In D. M. Knipe et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 7.Li, G., X. Chen, and A. Xu. 2003. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 349:508-509. [DOI] [PubMed] [Google Scholar]
- 8.Manser, E., H. Y. Huang, T. H. Loo, X. Q. Chen, J. M. Dong, T. Leung, and L. Lim. 1997. Expression of constitutively active PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17:1129-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 10.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, and SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 12.Ruan, Y. J., C. L. Wei, A. L. Ee, V. B. Vega, H. Thoreau, S. T. Su, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, T. Zhang, C. K. Peng, E. O. Lin, N. M. Lee, S. L. Yee, L. F. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siddell, S. G. 1995. The Coronaviridae. Plenum Press, New York, N.Y.
- 14.Tan, Y. H., and W. J. Hong. 1999. Gene expression in mammalian cells. UK patent GB2314332B.
- 15.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]