Abstract
There are seven members of the APOBEC3 family in humans (APOBEC3A through APOBEC3H) that have antiviral activity against retroviruses and/or retroelements. To determine whether variants in APOBEC3 genes in human populations have altered antiviral activity, we identified and functionally tested novel single nucleotide variants (SNVs) in APOBEC3 genes present in the 1000 Genome Project dataset. We found that common variants minor allele frequency (>1%) of APOBEC3A, C, F, and G do not affect protein function. However, we found that two common novel polymorphisms in APOBEC3D decrease antiviral activity against HIV-1, and one polymorphism decreases activity against Alu retrotransposons. We characterized the diversity of APOBEC3 genes in three human populations and find significant evidence that APOBEC3D has evolved under purifying selection in recent human history. These data suggest that the antiviral activity of APOBEC3D has been maintained in human populations for a cellular function in host defense.
Keywords: Apobec3 locus, common polymorphisms, HIV, Alu elements, Apobec3D, human evolution
Introduction
The APOBEC3 family of retroviral restriction factors contains seven paralogs in humans (APOBEC3A through APOBEC3H) (Jarmuz et al., 2002; OhAinle et al., 2006). APOBEC3s are cytidine deaminases that inhibit the replication of a diverse array of viruses and retrotransposons (Chen et al., 2006; Esnault et al., 2005; Mangeat et al., 2003; Okeoma et al., 2007; Sheehy et al., 2002; Turelli et al., 2004). Most APOBEC3 proteins are constitutively expressed in viral target cells, though some APOBEC3 proteins are induced by interferon (Koning et al., 2009; Refsland et al., 2010). APOBECSs inhibit viral replication during reverse transcription by inducing hypermutation of viral genomes and by deamination-independent methods (Harris et al., 2003; Macmillan et al., 2013; Newman et al., 2005; Zhang et al., 2003).
Most lentiviral genomes contain an antagonist of APOBEC3s, which is encoded by the vif gene in HIV-1, to overcome the restriction imposed by APOBEC3s(Mariani et al., 2003; Sheehy et al., 2003). This has led to an evolutionary ‘arms race’ between hosts and viruses that has left a signature of positive selection, or rapid evolution, in host and viral genes (Compton and Emerman, 2013). Many APOBEC3 genes, including APOBEC3G, APOBEC3H, and APOBEC3D, which was called “APOBEC3DE” in earlier papers, have evolved under positive selection for millions of years in primates (Duggal et al., 2011; OhAinle et al., 2008; Sawyer et al., 2004). At least one APOBEC3 gene within an Old World monkey species has acquired population-specific polymorphisms in recent history that allow host evasion from lentiviral infection (Compton et al., 2012), suggesting that APOBEC3s may have rapidly evolved in recent primate history as well.
Within humans, several APOBEC3s are known to have common polymorphisms that render them defective. For example, two variants of APOBEC3H have decreased protein expression (OhAinle et al., 2008), and deletions of APOBEC3B are common in some human populations (Kidd et al., 2007). Polymorphisms in APOBEC3F and APOBEC3G have also been described (An et al., 2004; Mulder et al., 2010; Reddy et al., 2010). However, whether variants that have functional consequences are present in other APOBEC3 genes is unknown. Moreover, the only allele of the human APOBEC3D gene that has been functionally tested for antiviral activity is less active against lentiviruses than the orthologous chimpanzee APOBEC3D gene (Duggal et al., 2011). Because of widespread interest in understanding the contributions of human genetics to viral susceptibility and disease, we sought to comprehensively identify and functionally characterize variants of APOBEC3 genes in human populations.
Using the 1000 Genome Project dataset, we identify 21 amino acid-altering mutations in the APOBEC3 locus, of which 9 are reported for the first time. We find that 6 common (minor allele frequency, MAF >1%) single nucleotide variants (SNVs) in APOBEC3A, C, F, and G have no effect on antiviral activity. However, two SNVs in APOBEC3D decrease antiviral activity against HIV-1 and Alu retrotransposons. To better understand the evolutionary pressures acting on the APOBEC3 locus, we perform neutrality tests and find that APOBEC3D is subject to purifying selection in humans. These results highlight the conserved role of APOBEC3D in host defense and suggest that APOBEC3D variants may be only slightly deleterious.
Results
Common polymorphisms in the APOBEC3 locus
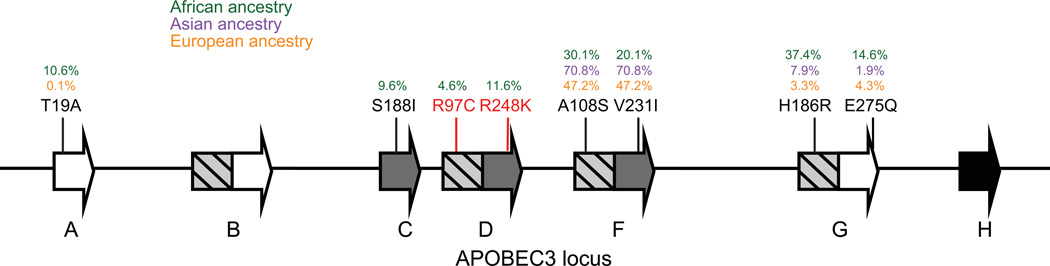
To identify single nucleotide variants in the APOBEC3 locus, we accessed the 1000 Genome Project Phase I genotypes for the coding regions of each APOBEC3 gene, which consists of genotypes from 913 geographically diverse individuals of African, Asian, and European ancestries. We also obtained insertions and deletions for 911 of the same individuals from the 1000 Genome Project Integrated Phase 1 release. A summary of the most common (MAF > 1%) codon-altering variants found in APOBEC3A through H is found in Fig. 1 and Table 1.
Fig. 1. Variants in the APOBEC3 locus tested in this study.
The human APOBEC3 locus with polymorphisms tested in this study. Polymorphisms in red have functional effects. Shading of APOBEC3 genes represents regions of homology. Percentages in green represent the derived allele frequency in individuals of African ancestry, purple represents individuals of Asian ancestry, and orange represents individuals of European ancestry. Diagram is not to scale.
Table 1.
Derived allele frequency of common variants in APOBEC3 genes.
| APOBEC3 | Variant | ID | African (na = 246) |
Asian (n = 286) |
European (n = 381) |
Combined (n = 913) |
Citation |
|---|---|---|---|---|---|---|---|
| A | T19A | rs17000556 | 10.6 | 0 | 0.1 | 2.9 | |
| B | K62E | rs2076109 | 68.1 | 63.6 | 62.2 | 64.2 | |
| B | P98L | rs59708943 | 16.3 | 0 | 5.4 | 6.6 | |
| B | T146K | rs5995649 | 5.9 | 2.1 | 3.9 | 3.9 | |
| B | L189P | rs146055882 | 10.6 | 0 | 0 | 2.8 | |
| B | A254V | rs138093253 | 5.7 | 0 | 0.9 | 1.9 | |
| B | CNVb | CNV_36030 | 6.1 | 31.1 | 7.8c | 14.6d | (Kidd et al., 2007) |
| C | S188I | rs112120857 | 9.6 | 0 | 0 | 2.6 | |
| D | R97C | rs75858538 | 4.7 | 0 | 0 | 1.3 | |
| D | R248K | rs61748819 | 11.6 | 0 | 0 | 3.1 | |
| F | A108S | rs2020390 | 30.1 | 70.8 | 47.2 | 50.0 | (Mulder et al., 2010) |
| F | V231I | rs2076101 | 20.1 | 70.8 | 47.2 | 47.3 | (Mulder et al., 2010) |
| F | Y307C | rs12157816 | 3.7 | 0 | 1.1 | 1.4 | (Mulder et al., 2010) |
| G | H186R | rs8177832 | 37.4 | 7.9 | 3.3 | 13.9 | (An et al., 2004) |
| G | E275Q | rs17496046 | 14.6 | 1.9 | 4.3 | 6.4 | (Reddy et al., 2010) |
| H | R18L | rs139293 | 6.1 | 16.1 | 31.1 | 19.7 | (OhAinle et al., 2008) |
| H | R105G | rs139297 | 15.9 | 67.7 | 52.2 | 47.3 | (OhAinle et al., 2008) |
| H | E121K | rs139298 | 13.8 | 68.0 | 51.8 | 46.7 | (OhAinle et al., 2008) |
| H | E121D | rs139299 | 86.8 | 32.3 | 48.4 | 53.7 | (OhAinle et al., 2008) |
| H | E178D | rs139302 | 15.4 | 67.5 | 52.2 | 47.1 | (OhAinle et al., 2008) |
| H | Dele 15 | rs59165009 | 33.1 | 24.8 | 35.2c | 31.3d | (OhAinle et al., 2008) |
n = number of individuals.
CNV = copy number variant.
n = 379.
n = 911.
Del = deletion.
The 1000 Genome Project dataset contains 12 previously reported common variants in the APOBEC3 locus, including the deletion of APOBEC3B (Kidd et al., 2007), the deletion of residue 15 in APOBEC3H (OhAinle et al., 2008), and SNVs in APOBEC3F (A108S, V231I, Y307C) (Mulder et al., 2010), APOBEC3G (H186R, Q275E) (An et al., 2004; Reddy et al., 2010), and APOBEC3H (R18L, R105G, E121K/D, D178E) (OhAinle et al., 2008). These polymorphisms were previously identified using smaller datasets because of their very high frequency. In addition to these previously reported variants, 9 additional common variants can also be found in this dataset, including SNVs in APOBEC3A (T19A), APOBEC3B (K62E, P98L, T146K, L189P, A254V), APOBEC3C (S188I), and APOBEC3D (R97C, R248K) (Figure 1 and Table 1). Using the 1000 Genome Project data, we have now identified all common polymorphisms in the APOBEC3 locus by eliminating an ascertainment bias.
Consistent with previous reports of APOBEC3 variants in HapMap populations, the deletion of APOBEC3B, the polymorphisms in APOBEC3F at codons 108 and 231, and the polymorphisms in APOBEC3H at codons 15, 105, 121, and 178 are found at their highest frequencies in populations with Asian or European ancestries (Figure 1). In contrast, SNVs in APOBEC3A, APOBEC3C, APOBEC3D, and APOBEC3G are found at their highest frequencies in populations of African ancestry. Thus, all APOBEC3 paralogs contain at least one common non-synonymous polymorphism in a human population. In addition, most variants in the APOBEC3 locus have population-specific distributions, indicative of demographic events or directional selection acting in a population-specific manner.
Common polymorphisms in APOBEC3A, C, F, and G do not affect antiviral activity
All common variants in APOBEC3H have been well described functionally in previous studies: the deletion of residue 15 decreases protein stability; the R105G mutation decreases protein stability, alters cellular localization, and reduces RNA binding; the D121K mutation decreases sensitivity to HIV-1 Vif; and the R18L and D178E mutations have no effect on protein function (Harari et al., 2009; Li et al., 2010; OhAinle et al., 2008; Tan et al., 2009; Wang et al., 2011; Zhen et al., 2012; Zhen et al., 2010). The loss of APOBEC3B in many individuals suggests that APOBEC3B is not essential for human health, and the insensitivity of APOBEC3B to HIV-1 Vif (Doehle et al., 2005) suggests it is not relevant to HIV-1 infection. Therefore, for the rest of our functional analyses of SNVs in APOBEC3 genes, we focused on those found in APOBEC3A, C, DE, F, and G.
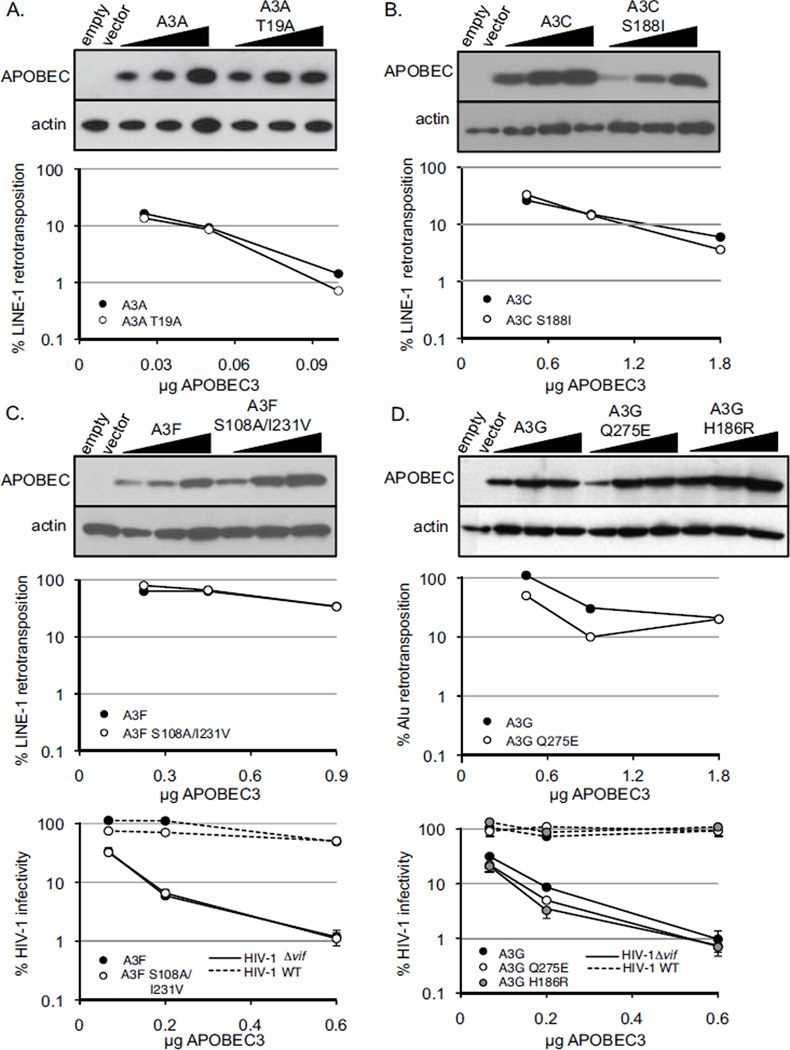
Human APOBEC3A and APOBEC3C inhibit the replication of LINE-1 (Bogerd et al., 2006; Muckenfuss et al., 2006), with the activity of APOBEC3A more potent than APOBEC3C. APOBEC3A T19A and APOBEC3C S1881 mutations are present at frequencies near 10% in African populations and <1% in other populations (Table 1). As no SNVs in either paralog have been previously described, we tested whether these APOBEC3A or APOBEC3C variants affect their activity against a retroelement, LINE-1. Each of these mutations was introduced into a plasmid containing epitope-tagged APOBEC3A or APOBEC3C. We co-transfected increasing amounts of APOBEC3 plasmids with a plasmid containing the non-LTR human retrotransposon LINE-1, which expresses neomycin resistance after retrotransposition. Because APOBEC3A is a stronger inhibitor of LINE-1 (Niewiadomska et al., 2007), we used 10 times less APOBEC3A plasmid DNA compared to APOBEC3C in these assays. After selection in G418, neomycin-resistant colonies were counted as a measure of LINE-1 activity. Although APOBEC3A T19A and APOBEC3C S188I had slightly decreased protein expression compared to wild-type APOBEC3A or APOBEC3C (Fig. 2A and 2B, upper panels), these polymorphisms did not affect the ability of APOBEC3A or APOBEC3C to restrict the activity of LINE-1 in a dose-dependent manner (Fig. 2A and 2B, lower panels). These data suggest that the SNVs in APOBEC3A and APOBEC3C do not alter their capacity to restrict LINE-1.
Fig. 2. Common variants in APOBEC3A, C, F, and G do not affect antiviral activity.
The antiviral activity of APOBEC3 mutants against HIV-1, Alu, or LINE-1 elements was compared to the antiviral activity of wild-type APOBEC3. Infectivity and retrotransposition are represented as a percentage of virus infectivity or retrotransposon activity in the absence of APOBEC3, which was normalized to 100%. Infections were performed with dilutions of APOBEC3, and APOBEC3 levels are increasing from left to right. Cellular APOBEC3 protein was detected by Western blot using an anti-HA antibody, and actin was used as a loading control. (A) APOBEC3A T19A. (Upper panel) Western blot analysis of cellular APOBEC3A (A3A) protein levels. (Lower panel) LINE-1 retrotransposition. Filled circles are wild-type APOBEC3A, and open circles are APOBEC3A T19A. (B) APOBEC3C S1881. (Upper panel) Western blot analysis of cellular APOBEC3C (A3C) protein levels. (Lower panel) LINE-1 retrotransposition. Filled circles are wild-type APOBEC3C, and open circles are APOBEC3C S188I. (C) APOBEC3F S108A/I231V. (Upper panel) Western blot analysis of cellular APOBEC3F (A3F) protein levels. (Middle panel) HIV-1 infectivity. Filled circles are wild-type APOBEC3F, and open circles are APOBEC3F S108A/I231V. Dashed lines are infections with HIV-1 containing Vif (HIV-1 WT), and solid lines are infections with HIV-1 lacking Vif (HIV-1Δvif). (Lower panel) LINE-1 retrotransposition. (D) APOBEC3G Q275E and APOBEC3G H186R. (Upper panel) Western blot analysis of cellular APOBEC3G (A3G) protein levels. (Middle panel) Alu retrotransposition. (Lower panel) HIV-1 infectivity. Filled circles are wild-type APOBEC3G, open circles are APOBEC3G Q275E, and gray circles are APOBEC3G H186R. Dashed lines are infections with HIV-1 containing Vif (HIV-1 WT), and solid lines are infections with HIV-1 lacking Vif (HIV-1Δvif). Experiments were performed at least 3 times, and results from a representative experiment are shown. Error bars represent the standard deviation of experimental triplicates.
Human APOBEC3F has potent antiviral activity against HIV-1Δvif and is sensitive to HIV-1 Vif (Bishop et al., 2004; Liddament et al., 2004; Wiegand et al., 2004; Zheng et al., 2004), but reports vary in its ability to inhibit LINE-1 (Bogerd et al., 2006; Hulme et al., 2007; Stenglein and Harris, 2006). In this dataset, APOBEC3F Y307C is present at a low frequency in African and European populations (MAF <5%) and is absent in Asian populations. Previous reports have shown that APOBEC3F Y307C has reduced antiviral activity (Dang et al., 2011) and increased sensitivity to HIV-1 Vif (Mulder et al., 2010). In addition, APOBEC3F A108S and V231I are present at intermediate to high frequencies (MAF 20 – 70%) in all human populations, with the highest frequencies found in individuals outside of Africa. We hypothesized that these high frequency variants of APOBEC3F may explain the discrepancy in reported anti-LINE activity. Therefore, we tested these two SNVs in APOBEC3F for antiviral activity. As these two polymorphisms are in strong linkage disequilibrium (LD) in all populations (r2 = 0.91), we introduced the mutations into the same APOBEC3F plasmid and functionally assayed them together. Plasmids containing APOBEC3F were co-transfected at multiple concentrations with HIV-1 proviral constructs containing a luciferase gene and VSV-G to produce pseudotyped virions. Equivalent amounts of virions were used to infect SupT1 cells, and viral infectivity was determined by luciferase activity of infected cells. While these mutations slightly increased protein expression of transfected APOBEC3F (Fig. 2C, upper panel), they did not alter the ability of APOBEC3F to restrict HIV-1Δvif (Fig. 2C, lower panel, solid lines) or HIV-2Δvif (data not shown), and they did not alter their sensitivity to degradation by HIV-1 Vif (Fig. 2C, lower panel, dashed lines) or HIV-2 Vif (data not shown). We next asked if wild-type or mutant APOBEC3F could inhibit LINE-1 retrotransposition. However, neither wild-type nor mutant APOBEC3F restricted LINE-1 by more than 3-fold, even at the highest concentration (Fig. 2C, middle panel). From these data, we conclude that APOBEC3F does not inhibit LINE-1 and that the common SNVs in APOBEC3F do not alter its anti-HIV activity.
Human APOBEC3G has the most potent antiviral activity against HIV-1Δvif, is highly sensitive to HIV-1 Vif (Refsland et al., 2012), and restricts Alu elements (Esnault et al., 2005; Hulme et al., 2007). In this dataset, APOBEC3G H186R and Q275E variants are at their highest frequency in African individuals (MAF 37% and 15%, respectively), compared to lower frequencies (MAF <10% or <5%, respectively) in other populations (Table 1). APOBEC3G H186R has been shown to restrict HIV-1Δvif and be sensitive to HIV-1 Vif (An et al., 2004), but APOBEC3G Q275E has not been described functionally. As these SNVs are in linkage equilibrium (r2 < 0.01), we introduced these mutations into separate plasmids containing epitopetagged APOBEC3G. When transfected into 293T cells, APOBEC3G H186R and APOBEC3G Q275E had similar protein expression levels as compared to wild-type APOBEC3G (Fig. 2D, upper panel). We assayed the anti-HIV activity of the H186R and Q275E mutants compared to wild-type APOBEC3G and found that all APOBEC3G variants restricted HIV-1Δvif infectivity to similar levels in a dose-dependent manner (Fig. 2D, lower panel, solid lines). APOBEC3G H186R and APOBEC3G Q275E were also highly sensitive to HIV-1 Vif, as infectivity was recovered to 90–100% in the presence of Vif at all concentrations of APOBEC3G (Fig. 2D, lower panel, dashed lines). Furthermore, we tested the ability of APOBEC3G Q275E to restrict Alu elements. We found that APOBEC3G Q275E restricted Alu elements similarly to wild-type APOBEC3G (Fig. 2D, middle panel). These data suggest that variants of APOBEC3G do not have altered antiviral activity against HIV or Alu. Thus, we have not found differences in activity for common SNVs in APOBEC3A, APOBEC3C, APOBEC3F, and APOBEC3G
Two polymorphisms in APOBEC3D decrease antiviral activity
Human APOBEC3D has weak antiviral activity against HIV-1 in the absence of Vif, is highly sensitive to degradation by HIV-1 Vif, and has strong anti-retroelement activity against Alu elements (Dang et al., 2006; Duggal et al., 2011; Tan et al., 2009). We wanted to identify SNVs of APOBEC3D in human populations to determine whether any individuals have an APOBEC3D allele with stronger anti-HIV activity. In the 1000 Genome Project dataset, we found that the most common non-synonymous SNVs in APOBEC3D were an arginine to cysteine change at position 97 (R97C) and an arginine to lysine change at position 248 (R248K), which exist at 1.3% and 3.1% frequencies, respectively, in the combined human populations. These non-synonymous SNVs are found almost exclusively in individuals with African ancestry at low frequencies (MAF 4.7% and 11.6%).
As these novel SNVs in APOBEC3D are in linkage equilibrium (r2 < 0.01), we introduced the R97C and R248K mutations into separate plasmids containing epitope-tagged APOBEC3D. These plasmids were transfected into 293T cells at multiple concentrations, and we compared wild-type and mutant APOBEC3D for protein expression by Western blot. We found that APOBEC3D R97C has similar protein levels as wild-type APOBEC3D, while APOBEC3D R248K has drastically lower (approximately 9-fold) protein expression than wild-type APOBEC3D (Fig. 3A). We next assessed the two APOBEC3D mutants for antiviral activity in a single-round HIV-1 infectivity assay. APOBEC3D R97C and APOBEC3D R248K restricted HIV-1Δvif by only 5- and 2-fold, which was significantly lower than wild-type APOBEC3D, which restricted HIV-1Δvif by nearly 30-fold (p<0.05, Fig. 3C, filled bars). The decreased antiviral activity of APOBEC3D R97C and APOBEC3D R248K was observed at all APOBEC3 concentrations tested (Fig. 3B). Similarly to wild-type APOBEC3D, APOBEC3D R97C and APOBEC3D R248K were highly sensitive to HIV-1 Vif, as infectivity was rescued to 100% in the presence of HIV-1 Vif, and were partially sensitive to HIV-2 Vif, as infectivity was rescued to 20 – 40% in the presence of HIV-2 Vif (Fig. 3C, open and gray bars). APOBEC3G was used as a control for Vif sensitivity, as infectivity was rescued to nearly 100% by Vif from HIV-1 and HIV-2. Thus, variants of APOBEC3D have significantly decreased antiviral activity against HIV-1 but similar sensitivities to Vif.
Fig. 3. Single nucleotide variants in APOBEC3D decrease antiviral activity.
The antiviral activity of APOBEC3D R97C and R248K mutants against HIV-1 was compared to the antiviral activity of wild-type APOBEC3D. Infectivity is represented as a percentage of virus infectivity in the absence of APOBEC3D (A3D), which was normalized to 100%. (A) Western blot analysis of cellular APOBEC3D protein levels. APOBEC3D was detected using an anti-HA antibody, and actin was used as a loading control. Transfections were performed with 3-fold dilutions of APOBEC3D, and APOBEC3D levels are increasing from left to right. (B) HIV-1 infectivity. Infections were performed with HIV-1 lacking Vif (HIV-1Δvif). Wild-type APOBEC3D is represented by filled circles, APOBEC3D R97C is represented by open circles, and APOBEC3D R248K is represented by gray circles. Transfections were performed with 3-fold dilutions of APOBEC3D, and APOBEC3D levels are increasing from left to right. (C) HIV-1 infectivity. Open bars are infections with HIV-1 lacking Vif (HIV-1Δvif), filled bars are infections with HIV-1 containing Vif (HIV-1 WT), and gray bars are infections with HIV-1 lacking Vif but containing HIV-2 Vif (HIV-1Δvif + HIV-2 Vif). Transfections were performed with 0.6µg APOBEC3D. (D) Distribution of deleterious APOBEC3D alleles in African populations. The proportion of deleterious APOBEC3D alleles (97C or 248K) is shown in filled bars, and the proportion of wild-type APOBEC3D alleles (97R and 248R) is shown in gray bars. YRI, Yoruba individuals from Ibadan, Nigeria; LWK, Luhya individuals in Webuye, Kenya; ASW, African ancestry individuals from southwest US. Experiments were performed at least 3 times, and results from a representative experiment are shown. Error bars represent the standard deviation of experimental triplicates.
While the R97C and R248K variants of APOBEC3D are independent mutations, they have the same functional effect of reducing the anti-HIV potential of APOBEC3D. By determining the combined frequency of alleles containing either the R97C or the R248K mutation, we find that 3.4% of APOBEC3D alleles in humans are deleterious with regards to blocking HIV replication. Within individuals of African ancestry, 16.2% of APOBEC3D alleles are deleterious (Fig. 3D), and 4.5% of individuals with African ancestry have two deleterious APOBEC3D alleles. Importantly, we did not find any common (MAF >1%) polymorphisms in human APOBEC3D with increased antiviral activity relative to the wild-type allele. Therefore, we conclude that the previously reported reduced antiviral activity of human APOBEC3D relative to chimpanzee APOBEC3D (Duggal et al., 2011) is not a reflection of ascertainment bias. In fact, common variants of APOBEC3D in human populations are less effective at restricting HIV-1 than wild-type APOBEC3D.
Genetic separation of APOBEC3D antiviral activities
While the antiviral activity of APOBEC3D against HIV-1 is moderate, APOBEC3D restricts Alu elements potently. To test whether the polymorphisms in APOBEC3D affect anti-Alu activity of APOBEC3D, we compared the anti-Alu activity of wild-type APOBEC3D to the anti-Alu activity of APOBEC3D carrying the R97C or R248K mutation. Interestingly, while the R97C and R248K mutations both decreased APOBEC3D antiviral activity against HIV-1Δvif, only the R248K mutation decreased APOBEC3D antiviral activity against Alu elements. At every dilution, APOBEC3D R248K (Fig. 4, gray circles) had 4- to 5-fold lower anti-Alu activity than wild-type APOBEC3D, where as APOBEC3D R97C (Fig. 4, open circles) restricted Alu similarly to wild-type APOBEC3D. These data suggest that the R248K mutation in APOBEC3D causes a global decrease in activity, likely due to its low protein expression, whereas the R97C mutation decreases HIV-1, but not Alu, restriction. Thus, the mechanism of human APOBEC3D viral restriction can be separated from its mechanism of retrotransposon restriction by a mutation at position 97.
Fig. 4. Effect of SNVs on APOBEC3D restriction of Alu elements.
The antiviral activity of APOBEC3D R97C and R248K mutants against Alu elements was compared to the antiviral activity of wild-type APOBEC3D. Retrotransposition is represented as a percentage of Alu activity in the absence of APOBEC3D (A3D), which was normalized to 100%. Wild-type APOBEC3D is represented by filled circles, APOBEC3D R97C is represented by open circles, and APOBEC3D R248K is represented by gray circles. Assays were performed with 2-fold dilutions of APOBEC3D, and APOBEC3D levels are increasing from left to right. Experiments were performed at least 3 times, and results from a representative experiment are shown. Error bars represent the standard deviation of experimental triplicates. p-values were calculated across experiments using a paired two-tailed Student’s t-test.
Molecular evolution of the APOBEC3 locus in human populations
The presence of common loss-of-function polymorphisms in APOBEC3D, APOBEC3F, APOBEC3H, and APOBEC3B in human populations is indicative of recent changes in demographic history or selective pressures acting on the APOBEC3 locus. By comparing the ratio of the rate of non-synonymous mutation (dN) to the rate of synonymous mutations (dS), previous studies have shown APOBEC3D, APOBEC3G, and APOBEC3H to be evolving under recurrent ancient positive selection in primates (Duggal et al., 2011; OhAinle et al., 2008; Sawyer et al., 2004). To determine whether the evolution of the APOBEC3 locus has changed in recent human history, we calculated the interspecies divergence and intraspecies nucleotide diversity of APOBEC3A, C, DE, F, G, and H for each human population (African, Asian, and European ancestries). We did not evaluate APOBEC3B because its copy number is highly variable in humans. We found that nucleotide diversity (π) varies between APOBEC3 genes and across each gene, but diversity is similar between populations. The APOBEC3 genes with the lowest average nucleotide diversity are APOBEC3C and APOBEC3D (Table 2). Using the Hudson Kreitman Aguade (HKA) test, we compared the polymorphic and fixed changes in APOBEC3 genes to the polymorphic and fixed changes in a non-coding (putatively neutral) region within the APOBEC3 locus. We found the nucleotide diversity in APOBEC3D in humans to be significantly lower than expected under neutrality (HKA χ2 = 4.31, p<0.05, Table 2). The trend is similar within each human population (African, Asian, European), suggesting that this is not a population-specific event (Table 2). This test is indicative of selective or demographic events acting on APOBEC3D to reduce diversity.
Table 2.
Neutrality test statistics for APOBEC3 genes.
| Sa | πb | Divergencec | Tajima's D | Fay and Wu’s H |
HKA χ 2d | ||
|---|---|---|---|---|---|---|---|
| APOBEC3A | African | 7 | 1.9x10−3 | 0.013 | 0.19 | −3.6 | 0.20 |
| 597 nt | Asian | 5 | 1.9x10−3 | 0.013 | 0.92 | −1.29 | 0.58 |
| European | 5 | 1.1x10−3 | 0.013 | −0.06 | −2.44 | 0.53 | |
| Combined | 8 | 1.6x10−3 | 0.013 | −0.05 | −3.80 | 0.01 | |
| APOBEC3C | African | 1 | 3.0x10−4 | 0.0089 | 0.15 | 0.15 | 0.87 |
| 570 nt | Asian | 2 | 2.0x10−5 | 0.0087 | −1.13 | −1.97 | 0.54 |
| European | 1 | 1.0x10−5 | 0.0087 | −0.83 | 0.00 | 0.16 | |
| Combined | 4 | 1.0x10−4 | 0.0088 | −1.24 | −1.94 | 0.06 | |
| APOBEC3D | African | 7 | 3.0x10−4 | 0.036 | −1.27 | 0.31 | 2.40 |
| 1159 nt | Asian | 3 | 2.0x10−5 | 0.036 | −1.34 | 0.02 | 2.87# |
| European | 4 | 1.0x10−5 | 0.036 | −1.47 | −1.98 | 2.87# | |
| Combined | 13 | 1.0x10−4 | 0.036 | −1.89 | −1.89 | 4.31* | |
| APOBEC3F | African | 10 | 1.4x10−3 | 0.016 | 0.15 | 0.71 | 0.01 |
| 1119 nt | Asian | 7 | 1.2x10−3 | 0.016 | 0.55 | −3.71 | 0.16 |
| European | 7 | 1.4x10−3 | 0.017 | 1.08 | 0.20 | 0.77 | |
| Combined | 14 | 1.5x10−3 | 0.017 | −0.13 | −1.97 | 0.24 | |
| APOBEC3G | African | 9 | 8.4x10−4 | 0.017 | −0.55 | 0.64 | 0.60 |
| 1152 nt | Asian | 5 | 5.0x10−4 | 0.017 | −0.34 | 0.45 | 0.19 |
| European | 3 | 5.5x10−4 | 0.017 | 0.74 | 0.25 | 0.06 | |
| Combined | 11 | 7.0x10−4 | 0.017 | −0.79 | 0.62 | 0.23 | |
| APOBEC3H | African | 11 | 2.7x10−3 | 0.020 | −0.18 | −1.61 | 0.78 |
| 549 nt | Asian | 8 | 4.5x10−3 | 0.021 | 2.24* | −0.74 | 0.46 |
| European | 6 | 5.3x10−3 | 0.021 | 4.39* | 0.15 | 0.01 | |
| Combined | 11 | 5.1x10−3 | 0.021 | 2.11 | 0.28 | 0.48 |
S = number of segregating sites.
π = nucleotide diversity.
Divergence = humanchimpanzee divergence.
HKA = Hudson Kreitman Aguade test, silent sites only.
p<0.05,
p<0.10
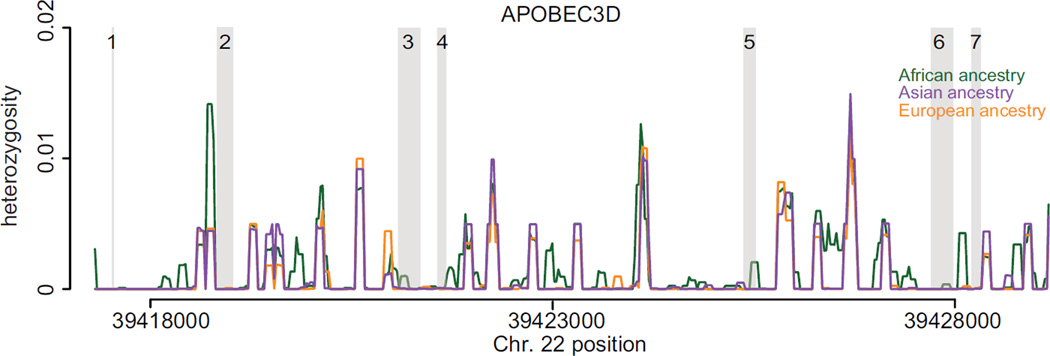
To determine whether a selective sweep has reduced the diversity across the APOBEC3D genomic locus in each population, we calculated the polymorphism and divergence across APOBEC3D exons and introns. A recent selective sweep would remove diversity across the entire APOBEC3D locus, including introns. However, we find that the nucleotide diversity in APOBEC3D exons is lower than the diversity within introns (Fig. 5, position of exons highlighted in gray). This is suggestive of purifying selection acting on the coding regions of APOBEC3D to reduce non-synonymous variation, rather than an advantageous haplotype sweeping through a population.
Fig. 5. Diversity across APOBEC3D within human populations.
Sliding window analysis of diversity across intronic and exonic regions of APOBEC3D, stratified by population. Heterozygosity (π) is plotted for each population. Green represents individuals of African ancestry, purple represents individuals of Asian ancestry, and orange represents individuals of European ancestry. Positions of exons are highlighted in gray.
From these population genetics analyses, we conclude that, while APOBEC3D, APOBEC3G, and APOBEC3H have previously been shown to evolve under positive selection during human-chimpanzee divergence, it is unlikely that APOBEC3D is still evolving under positive selection within humans. Importantly, APOBEC3D has evolved under purifying selection in recent human history, suggesting that a cellular function of APOBEC3D has been optimized and conserved due to continuous selective pressure.
Discussion
In this study, we characterized the level of diversity in the APOBEC3 locus in human populations and the effects of variants on APOBEC3 antiviral activity. We find that high frequency SNVs in APOBEC3A, C, F, and G genes do not affect antiviral activity. However, we found that two common variants in APOBEC3D (R97C and R248K), which are limited to African populations, decrease the antiviral activity of APOBEC3D against HIV-1, and one variant (R248K) also decreases anti-Alu activity. Further, we show that the pattern of diversity in the APOBEC3D is indicative of recent purifying selection in humans. Together, these data indicate that APOBEC3D plays an important role in host defense in humans.
Because the R248K mutation likely has decreased anti-Alu activity due to its low protein expression (Fig. 4), its deleterious effects may be compensated in vivo by another polymorphism that was not included in the plasmid used for transfection. For example, the R248K3 polymorphism is in tight linkage disequlibrium (r2 > 0.8) with two additional polymorphisms in non-coding regions near APOBEC3D that may compensate for the low in vitro expression level. It is also possible that the R248K polymorphism is only slightly deleterious and that the purifying selection acting on APOBEC3D is weak. This is supported by the relatively low frequency of the R248K polymorphism in the human population. Either way, we conclude that APOBEC3D is subject to purifying selection, and the anti-Alu function of APOBEC3D is conserved in most individuals. Thus, we suggest that anti-Alu activity is the critical cellular function that has driven purifying selection of APOBEC3D in humans. Because APOBEC3D has rapidly evolved in chimpanzees yet has not altered anti-Alu activity (Duggal et al., 2011), it is likely that this function is subject to purifying selection in chimpanzees as well. However, in the absence of an additional selective pressure, purifying selection is the main evolutionary force affecting APOBEC3D in humans. This is supported by previous studies showing that APOBEC3D mRNA is expressed in human embryonic stem cells (Wissing et al., 2011), the cell type where Alu elements can have the most deleterious effect on a population. In addition, because endogenous mobile elements do not trigger an interferon response, it is interesting that APOBEC3D mRNA is constitutively expressed with only moderate upregulation by interferon.
The observation of recent purifying selection in APOBEC3D (Fig. 5 and Table 2) is in contrast to the ancient adaptive evolution that has been implicated for many APOBEC3 family members. The arms race between restriction factors and retroviruses is generally thought to induce rapid evolution in immune-related genes and viruses (Duggal and Emerman, 2012). However, recent purifying selection suggests that the antiviral activity of the human repertoire of APOBEC3 genes was optimized against a selective pressure, and this antiviral activity has been maintained. While APOBEC3 restriction factors have an important role in host defense in humans, the consequences of purifying selective pressure are that some APOBEC3 genes, such as APOBEC3D, are not optimized to restrict the lentiviruses circulating today.
Based on other studies showing that endogenous APOBEC3D, APOBEC3F, and APOBEC3G expression decreases HIV-1 replication in HIV target cells (Refsland et al., 2012), it is possible that the novel variants in APOBEC3D described here may have a role in HIV-1 acquisition or disease progression. These polymorphisms will be very difficult to associate with disease outcomes due to their low frequency, though detecting the influence of rare genetic variants on disease susceptibility is becoming a possibility (Tennessen et al., 2012). However, this study supports investigating the functional consequences of low frequency alleles in host defense genes, and more studies evaluating the effects of mutations in immune-related genes are needed to fully understand the cumulative impact that variants could have on human health.
Materials and Methods
APOBEC3 genotype data
SNVs in the APOBEC3 locus (GRCh37, Chr22: 39347756 – 39501072) from 913 individuals were obtained through the 1000 Genome Project May 2011 phase 1 low-coverage phased genotype release (www.1000genomes.org). Insertions and deletions were obtained for 911 individuals from the 1000 Genome Project February 2012 integrated phase 1 release. The African population includes 61 African ancestry individuals from southwest US, 97 Luhya individuals in Webuye, Kenya, and 88 Yoruba individuals from Ibadan, Nigeria. The Asian population includes 97 Han Chinese individuals in Beijing, 100 Chinese individuals in Denver, and 89 Japanese individuals in Tokyo. The European population includes 87 individuals with European ancestry in Utah, 93 Finnish individuals from Finland, 89 British individuals from England and Scotland, 14 individuals from Iberian populations in Spain, and 98 Toscani in Italy. The ancestral state of each allele was determined by comparison to the chimpanzee genome.
APOBEC3 plasmids
Human APOBEC3A, APOBEC3C, APOBEC3D, and APOBEC3G were cloned into the expression vector pCS2, and human APOBEC3F was cloned into the expression vector pCDNA3.1. A hemagglutinin (HA) tag was added to the C-terminus of all APOBEC3 proteins. Mutations were introduced into APOBEC3 plasmids using overlapping PCR. Primer sequences are available upon request.
Cell lines, Transfections, and Western Blot analysis
SupT1 cells were maintained in RPMI/1% Pen/Strep/10% FBS at 37°C in a CO2 incubator. 293T, HeLa, and HeLa-HA cells were maintained similarly in DMEM/1% Pen/Strep/10% BGS. Transfections were performed with TransIT-LT1 transfection reagent (Mirus Bio) at a reagent:plasmid DNA ratio of 2:1. Transfected cells and viral lysates were harvested 48 hours after transfection. Cells were lysed in NP40-doc buffer (1% NP40, 0.2% sodiumdeoxycholate, 0.12M NaCl, 20mM Tris pH 8.0, 2.4mM DTT) with protease inhibitors (Roche) and spun for 1 minute at 16,000xg, and supernatants were quantified by Bradford assay. Lysates were resolved by 10% SDS-PAGE, transferred to PVDF membranes, and probed with anti-HA (Santa Cruz Biotech) or anti-actin (Sigma).
Retrotransposition assays
LINE-1 and Alu assays were performed as described previously (Dewannieux et al., 2003; OhAinle et al., 2008; Ribet et al., 2004; Wei et al., 2001) with some modifications. Cells were plated in 6-well plates at 8×104 cells/well, and, 24 hours later, cells were transfected. For L1 assays, 0.1 µg LINE-1 plasmid (JM101/L1.3 (Moran et al., 1999; Wei et al., 2001)) and 0.6 µg APOBEC or empty plasmid were transfected into HeLa cells. For Alu assays, 1 µg Alu plasmid (AluneoTET (Dewannieux et al., 2003) or Alu-Ya5-eab (Bennett et al., 2008)), 0.3 µg Orf2p (pCep5’UTRORF2Δneo (Alisch et al., 2006)), and 0.45 µg APOBEC or empty plasmid were transfected into HeLa-HA cells. Three days after transfection, cells were selected in G418 for 10–12 days. Colonies were stained with crystal violet and counted manually.
Viral infectivity assays
Single-round HIV-1 infectivity assays were performed as described (Yamashita and Emerman, 2004). To produce VSV-G-pseudotyped HIV-1, 3×106 293T cells were plated in a 6-well plate, and 24 hours later, co-transfected with 0.6 µg lentiviral vector (pLai3ΔenvLuc2 (Yamashita and Emerman, 2004), pLai3ΔenvLuc2Δvif (OhAinle et al., 2006), 0.2 µg L-VSV-G, and 0.2 µg APOBEC or empty plasmid. HIV-1 expressing Vif from HIV-2 was made using pLai3ΔenvLuc2ΔvifLk/vifROD9 (Li et al., 2010). Virus was harvested 48 hours after transfection and filtered through a 0.2µm filter. Virus was quantified by p24 gag ELISA (Advanced BioScience), and virus equivalent to two nanograms of p24 gag was used to infect 4×104 SupT1 cells in a 96-well plate in the presence of 20 ug/mL DEAE-dextran. Forty-eight hours after transfection, cells from triplicate infections were lysed in Bright-Glo Luciferase Assay Reagent (Promega) and read on a luminometer. Error bars are the standard deviation of experimental triplicates. p-values were calculated using a two-tailed Student’s t-test.
Population genetics analyses
Population genetics calculations were performed using DnaSP v5 (Librado and Rozas, 2009). The HKA test was performed using by comparing APOBEC3 genes to a 2 kb noncoding region of the APOBEC3 locus (Chr. 22: 39403000 – 39405000). Significance of the HKA test was determined using the χ2 value with 1 degree of freedom. Significance of Tajima’s D and Fay and Wu’s H tests were determined using 10,000 coalescent simulations based on the observed θ with no recombination. The outgroup used for all tests was the UCSC chimpanzee sequence, accessed through the SeattleSeq Annotation 134 webserver. Sliding windows were calculated with a window size of 100 nucleotides and an overlap of 25 nucleotides. Linkage disequilibrium (r2) was calculated using Haploview v4.2 (Barrett et al., 2005).
Highlights.
Common human variants in the coding genes of the APOBEC3 locus were identified.
Most common variants have no effect on APOBEC3 anti-retroelement function.
Two APOBEC3D variants decrease anti-HIV activity, but none increase it.
APOBEC3D has evolved under purifying selection in recent human evolution.
Acknowledgements
We thank Lily Wu for assistance and Mia Levine and Harmit Malik for comments and advice. This work was supported by R01 AI30937 (ME) and an NSF graduate fellowship to NKD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S, Goedert JJ, Phair J, Buchbinder S, O'Brien SJ, Telenti A, Winkler CA. APOBEC3G genetic variants and their influence on the progression to AIDS. Journal of virology. 2004;78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bennett EA, Keller H, Mills RE, Schmidt S, Moran JV, Weichenrieder O, Devine SE. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Current biology : CB. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Compton AA, Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 2013;9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Abudu A, Son S, Harjes E, Spearman P, Matsuo H, Zheng YH. Identification of a single amino acid required for APOBEC3 antiretroviral cytidine deaminase activity. J Virol. 2011;85:5691–5695. doi: 10.1128/JVI.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. Journal of virology. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Doehle BP, Schafer A, Cullen BR. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nature reviews. Immunology. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Malik HS, Emerman M. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. Journal of virology. 2011;85:11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Harari A, Ooms M, Mulder LC, Simon V. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J Virol. 2009;83:295–303. doi: 10.1128/JVI.01665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. Journal of virology. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wu LI, Emerman M. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol. 2010;84:88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Macmillan AL, Kohli RM, Ross SR. APOBEC3 Inhibition of Mouse Mammary Tumor Virus Infection: the Role of Cytidine Deamination versus Inhibition of Reverse Transcription. Journal of virology. 2013;87:4808–4817. doi: 10.1128/JVI.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Löwer J, Cichutek K, Flory E, Schumann GG, Münk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Mulder LC, Ooms M, Majdak S, Smedresman J, Linscheid C, Harari A, Kunz A, Simon V. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. Journal of virology. 2010;84:9613–9617. doi: 10.1128/JVI.02630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Current biology : CB. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. Journal of virology. 2007;81:9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell host & microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Reddy K, Winkler CA, Werner L, Mlisana K, Abdool Karim SS, Ndung'u T, Team CAIS. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. Aids. 2010;24:195–204. doi: 10.1097/QAD.0b013e3283353bba. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic acids research. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Dewannieux M, Heidmann T. An active murine transposon family pair: retrotransposition of "master" MusD copies and ETn trans-mobilization. Genome Res. 2004;14:2261–2267. doi: 10.1101/gr.2924904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS biology. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature medicine. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, GO B, GO S, Project obotNES. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012 doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. Journal of virology. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem. 2011;286:36427–36437. doi: 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Du J, Zhou X, Xiong Y, Yu XF. Reduced APOBEC3H variant anti-viral activities are associated with altered RNA binding activities. PloS one. 2012;7:e38771. doi: 10.1371/journal.pone.0038771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. Journal of virology. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]