Abstract
The dynamin-related GTPase Dnm1 controls mitochondrial morphology in yeast. Here we show that dnm1 mutations convert the mitochondrial compartment into a planar ‘net’ of interconnected tubules. We propose that this net morphology results from a defect in mitochondrial fission. Immunogold labelling localizes Dnm1 to the cytoplasmic face of constricted mitochondrial tubules that appear to be dividing and to the ends of mitochondrial tubules that appear to have recently completed division. The activity of Dnm1 is epistatic to that of Fzo1, a GTPase in the outer mitochondrial membrane that regulates mitochondrial fusion. dnm1 mutations prevent mitochondrial fragmentation in fzo1 mutant strains. These findings indicate that Dnm1 regulates mitochondrial fission, assembling on the cytoplasmic face of mitochondrial tubules at sites at which division will occur.
The mitochondrion is a complex organelle with a double membrane, its own genome and an independent protein-synthetic machinery. Although the role of mitochondria in metabolism and ATP production is more widely recognized, alterations in mitochondrial shape and abundance are also important for cellular function and differentiation. For example, mitochondrial morphology and copy number change in response to nutrient availability in yeast cells1, cell damage and apoptosis in mammalian cells2 and developmental cues in Xenopus and Drosophila3,4. Cytological studies indicate that the ‘steady-state’ mitochondrial morphology and copy number can vary dramatically in different cell types, ranging from multiple, spherical organelles to the branched, tubular networks found in budding yeast and some mammalian cells5–7. These differences in mitochondrial morphology and copy number are largely determined by the balance between ongoing mitochondrial fission and fusion events.
Little is known about molecules that regulate mitochondrial fission and fusion. The fuzzy onions (fzo) family of transmembrane GTPases was recently shown to control mitochondrial fusion in different organisms and cell types. In Drosophila, fzo is required for a developmentally regulated mitochondrial fusion event during spermatogenesis8. In budding yeast, mutations in fzo1 cause mitochondrial networks to fragment9,10 and prevent mitochondrial fusion during yeast mating9. Molecules required for mitochondrial fission have been more difficult to define, although candidates have been identified in budding yeast and mammalian cells. Dnm1, a GTPase of high relative molecular mass, regulates mitochondrial morphology in yeast11. Mutations in DNM1 cause mitochondrial membranes to collapse to one side of the cell but do not interfere with the morphology or distribution of other cytoplasmic organelles11. A Dnm1-like molecule called Drp1 also controls the distribution of mitochondria in mammalian cells12. Dnm1 is structurally related to the GTPase dynamin13, which is required for the scission and release of clathrin-coated vesicles from the plasma membrane during endocytosis (ref. 14 and references therein). On the basis of this structural similarity, we proposed that Dnm1 might play a part in the fission/division of mitochondrial tubules11. The localization of Dnm1 to punctate structures on the tips and sides of mitochondrial tubules was consistent with this model, but did not rule out alternative functions for Dnm1 in the maintenance of mitochondrial morphology11.
Here we provide new evidence that Dnm1 participates in mitochondrial fission. Mitochondrial membranes in dnm1Δ (dnm1-null) cells are organized as ‘nets’ of interconnected tubules that most likely result from a defect in tubule division. In immunogold labelling studies, Dnm1 appears to be associated with mitochondrial tubules at sites that are undergoing division and at sites that have recently completed division. Genetic studies indicate that the activities of the GTPases Dnm1 and Fzo1 are antagonistic: dnm1 mutations block the mitochondrial fission and fragmentation normally observed in the fzo1 mutant strain. Together, these results support our hypothesis that Dnm1 acts at the outer mitochondrial membrane to regulate mitochondrial fission.
Results
Mitochondrial membranes form nets in dnm1 mutant cells
We previously reported that mitochondrial membranes collapse to one side of the cell in a dnm1Δ mutant strain11. Transmission electron microscopy indicated that these collapsed membranes might be organized in an unusual structure11. To characterize this structure further, we studied mitochondrial morphology in dnm1Δ cells under several different conditions.
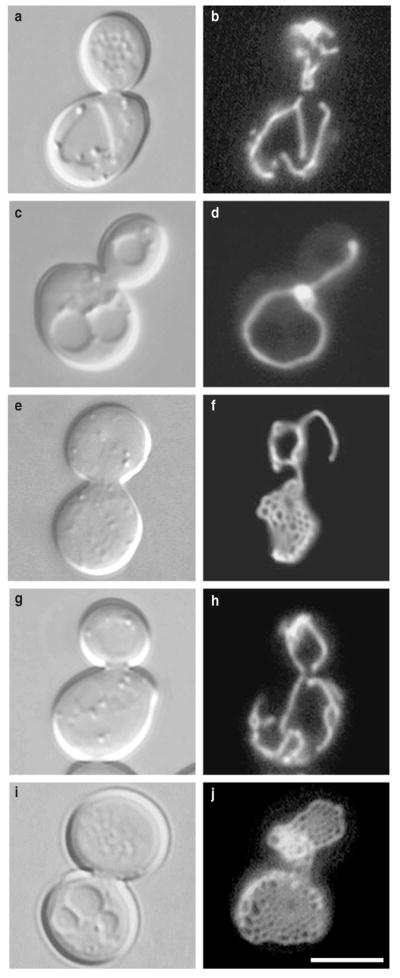
As reported previously, dnm1Δ mutants grown at 25 °C lack the highly branched mitochondrial network characteristic of wild-type cells (Figs 1a, b, 2; 98% of wild-type cells show such a network). Instead, mitochondrial membranes in the dnm1Δ strain collapse into one or more long tubular structures that contain fewer branches than in wild-type cells and remain near the cell cortex (Figs 1c, d, 2; 39%). These collapsed mitochondrial membranes often contain one or more expanded regions that appear as bright spots of variable size in the fluorescence microscope (Fig. 1d, 2; 49%). In a few cells, these bright regions expand further to reveal mitochondrial membranes organized as ‘nets’ (Figs 1e, f, 2; 11%). This net morphology is very unusual and has not been reported in other yeast mitochondrial-morphology mutants or cell types.
Figure 1. Mitochondrial membranes form nets in dnm1 mutant cells.
Morphology of GFP-labelled mitochondrial membranes (pDO12) in b, wild-type (strain JSY3096), d, f, dnm1Δ (JSY3097), h, mdm20Δ (JSY3094), and j, dnm1Δ mdm20Δ (JSY3095) cells at 25 °C. The corresponding differential interference contrast images are shown in a, c, e, g, i. The four strains are sister spores from a single tetrad. Scale bar represents 5 μm.
Figure 2.

Mitochondrial-membrane morphology in dnm1Δ and dnm1Δ mdm20Δ mutant cells.
Genetic and biochemical studies indicate that the distribution of mitochondrial membranes in yeast is regulated, in part, by interactions with actin cables (bundles of actin filaments) which extend from the base of the mother cell into the growing bud (reviewed in ref. 15). We reasoned that such organelle–actin interactions might prevent mitochondrial nets from spreading out along the cortex, impeding their visualization in many cells. To test this idea, we combined the dnm1Δ mutation with a mutation in MDM20 (mitochondrial distribution and morphology, ref. 16) that disrupts actin cables. We reported previously that loss of actin cables in mdm20Δ cells causes moderate mitochondrial-inheritance defects at 25 °C, which become more severe at 37 °C (ref. 16). However, mitochondrial morphology in the mdm20Δ mutant remains wild-type at the permissive temperature (Figs 1g, h, ***********2; 72%). Mitochondrial nets occupy a greater volume of the cell and are more easily visualized in the dnm1Δ mdm20Δ double mutant (Fig. 1i, j). Moreover, in asynchronously growing cultures, the percentage of cells with visible mitochondrial nets increases from 11% in the dnm1Δ mutant to 81% in the dnm1Δ mdm20Δ double mutant (Fig. 2). We used the mdm20Δ mutation as a tool to enhance visualization and analysis of the mitochondrial nets formed in dnm1 mutant strains.
Transmission electron microscopy confirmed that mitochondrial membranes are organized as nets in dnm1Δ mdm20Δ cells (Fig. 3). Previous studies indicated that mitochondrial profiles in wild-type cells are usually oval or tubular in shape and are distributed evenly at the cell periphery9,11. In contrast, mitochondrial membranes in dnm1Δ mdm20Δ cells are organized as flattened sheets perforated by a series of regularly spaced holes (Fig. 3a). Reconstruction of twelve serial sections containing these mitochondrial sheets reveals a curved mitochondrial net (Fig. 3b) resembling those visualized by fluorescence microscopy in the dnm1Δ and dnm1Δ mdm20Δ mutants (Fig. 1f, j).
Figure 3. Transmission electron microscopy and serial reconstruction of a mitochondrial net.
a, Section through a mitochondrial net (m) in a dnm1Δ mdm20Δ cell (strain JSY3095). Black arrowheads mark ‘holes’ in the mitochondrial compartment. b, Computer reconstruction of a mitochondrial net (grey and white) generated from 12 serial sections, including the image shown in a. Scale bars represent 1 μm.
The mitochondrial nets formed in dnm1 mutant strains could arise from a defect in mitochondrial fission. For example, the Dnm1 GTPase normally may function in vivo to ‘clip’ membranes that separate two adjacent holes in the mitochondrial net, releasing mitochondrial ends or tips and forming tubules.
Localization of Dnm1
Time-lapse imaging studies of wild-type mitochondrial networks revealed that fission events usually occur within a tubule (creating two ends) or at branchpoints in the network (creating one end and a tubule)17. If Dnm1 is required for mitochondrial fission, it should be associated with sites on the mitochondrial compartment that are preparing to divide (on the sides of tubules), are undergoing division (at constriction sites in tubules), or have just completed division (at the tips/ends of tubules). Our previous indirect immunofluorescence studies localized a haemagglutinin-tagged Dnm1 protein (Dnm1–HAcp) to punctate structures at the tips and sides of mitochondrial tubules and at branchpoints in the network11. Although this localization pattern was consistent with a role for Dnm1 in fission, indirect immunofluorescence techniques lack the resolution necessary to detect constriction sites in mitochondrial tubules. To examine sites of Dnm1 localization in more detail, we performed immunogold labelling experiments.
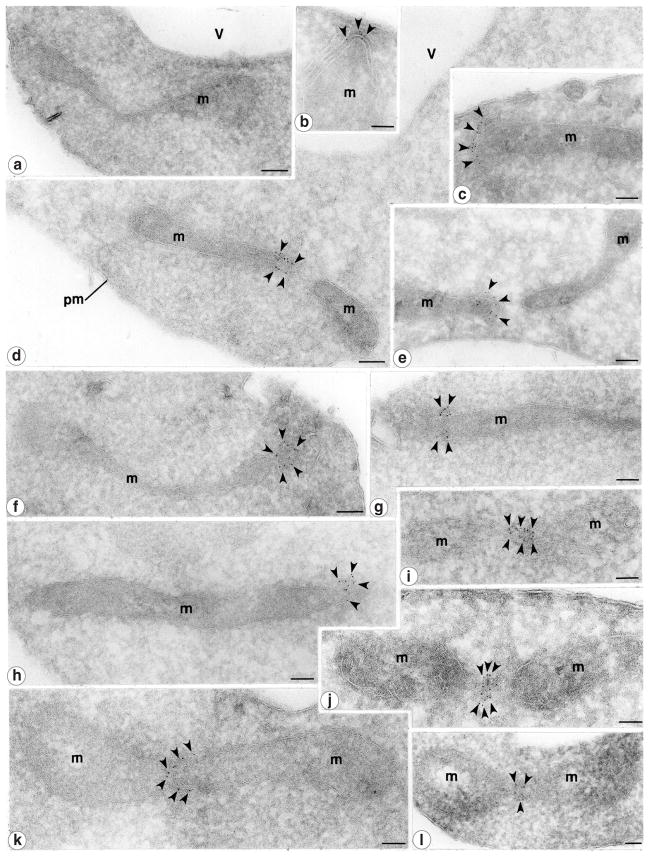
Immunogold labelling was carried out on ultrathin cryosections of cells expressing the Dnm1–HAcp protein (strain JSY1781; ref. 11). Consistent with previous immunofluorescence studies11, 87% of the 5-nm gold particles were found at the tips/ends of mitochondrial tubules (Fig. 4b–f, h, Table 1) and at discrete sites on the sides of mitochondrial tubules (Fig. 4g, i–l, Table 1). Interestingly, 34.6% of the gold particles observed were clustered at constriction sites on mitochondrial tubules, indicating that these tubules may have been undergoing division (Fig. 4i–l, Table 1). In some sections, label was detected on the end of a mitochondrial tubule in close proximity to another unlabelled mitochondrial end (Fig. 4d, e; there were 13 pairs of closely spaced tips with gold particles on only one of the two tips; 157 gold particles were counted). In cases in which two tips were juxtaposed, gold particles were never found on both tips. This labelling pattern could represent a recently completed division event. Together, these results support the idea that Dnm1 is required for mitochondrial fission.
Figure 4. Dnm1 localizes to mitochondrial constriction sites and mitochondrial tips.
The distribution of the Dnm1–HAcp protein (black arrowheads; strain JSY1781) was determined by immunogold labelling of ultrathin cryosections. Gold particles were found concentrated at the tips of mitochondrial tubules proximal to the plasma membrane (b, c, f), at the tips of mitochondrial tubules that may be newly divided (d, e, h), and at putative mitochondrial constriction/division sites (i–l). g, Gold particles are also detected on mitochondria at sites that are not constricted. a, No mitochondrial labelling was detected in cells lacking the Dnm1–HAcp protein (strain JSY3097). m, mitochondria; pm, plasma membrane; v, vacuole. Scale bars represent 0.1 μm.
Table 1.
Distribution of Dnm1–HAc protein in yeast
| Compartment | Number of gold particles | Percentage of total |
|---|---|---|
| Mitochondrial tips | 411 | 40.8 |
| Mitochondrial sides | 117 | 11.6 |
| Mitochondrial constriction sites | 348 | 34.6 |
| Mitochondrial other* | 13 | 1.3 |
| Mitochondrial total | 889 | 88.4 |
| Plasma membrane | 37 | 3.7 |
| Cytoplasm | 62 | 6.2 |
| Vacuole | 18 | 1.8 |
Mitochondrial boundary unclear
We also observed gold particles on the ends of mitochondrial tubules in close proximity to the plasma membrane (Fig. 4b, c; 9 tips with 76 gold particles). Thus, Dnm1 may also localize to sites at which the outer mitochondrial membrane is anchored at the cell cortex. This localization is consistent with our previous finding that disruption of DNM1 interferes with the cortical distribution of mitochondrial membranes11. Although the localization of the yeast mitochondrial network at the cell periphery is well documented (reviewed in ref. 15), the role of Dnm1 in mitochondrial anchoring and cortical distribution is unclear.
dnm1 mutations block mitochondrial fragmentation in fzo1-1 cells
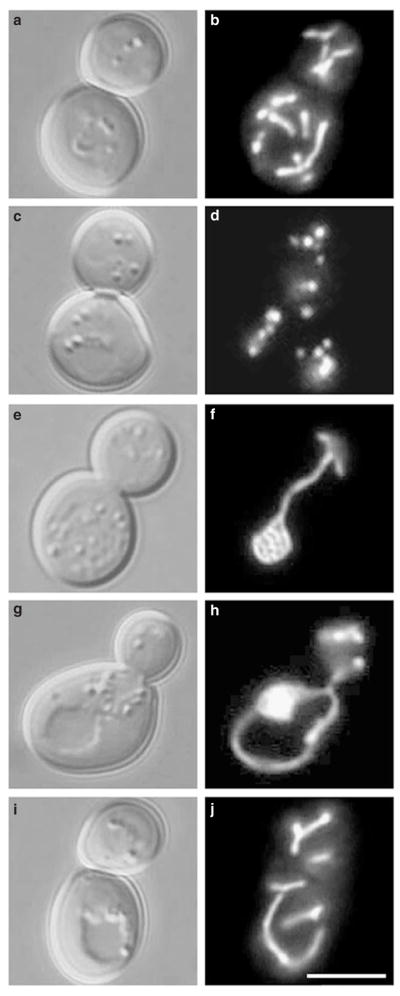
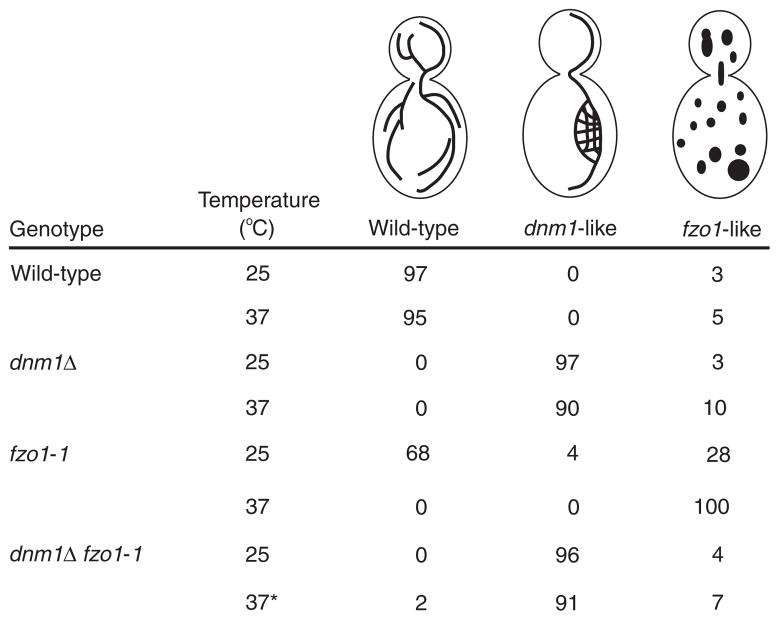
Maintenance of a tubular mitochondrial network is regulated by opposing fission and fusion events17. The fusion reaction requires the transmembrane GTPase Fzo1, which localizes to the outer mitochondrial membrane9. As reported previously, shifting a conditional fzo1-1 mutation to the non-permissive temperature (37 °C) for 10 minutes results in the rapid and complete fragmentation of the mitochondrial network (Figs 5c, d, 6; ref. 9). We postulated that this fragmentation was caused by ongoing mitochondrial fission events regulated by the Dnm1 GTPase. To test this possibility, we analysed mitochondrial morphology at 25 °C and 37 °C in fzo1-1 dnm1Δ double mutant strains. At 25 °C, fzo1-1 dnm1Δ cells contained collapsed mitochondrial membranes and nets (Figs 5e, f, 6). These mitochondrial morphologies were similar to those observed in the single dnm1Δ mutant at both 25 °C and 37 °C (Figs 1d, f, 2, 5f, 6). Shifting fzo1-1 dnm1Δ cells to 37 °C for up to 3 hours had little effect on organelle morphology; wild-type networks were rarely seen (Figs 5i, j, 6), and mitochondrial membranes remained tubular and exhibited dnm1-like morphologies despite the absence of FZO1 function at the higher temperature (Figs 5g, h, 6). Thus, maintenance of a pre-formed net does not appear to require ongoing mitochondrial fusion. We also observed mitochondria with dnm1-like morphologies (95%) in fzo1Δ dnm1Δ strains at 25 °C, although these cells did not contain detectable nets. These results indicate that the dnm1 mitochondrial-morphology defect is epistatic to the fzo1-1 mitochondrial-morphology defect, and that the loss of Dnm1 function in the fzo1-1 dnm1Δ double mutant blocks the mitochondrial fission and fragmentation normally observed in fzo1-1 cells at the non-permissive temperature.
Figure 5. dnm1 mutations block mitochondrial fragmentation in fzo1-1 cells.
Morphology of GFP-labelled mitochondrial membranes (carrying plasmid pVT100UGFP) in b, wild-type (strain JSY3162), d, fzo1-1 (JSY3161), f, dnm1Δ (JSY3164), and h, j, dnm1Δ fzo1-1 (JSY3163) cells at 37 °C. The corresponding differential interference contrast images are shown in a, c, e, g, i. The four strains are sister spores from a single tetrad. Scale bar represents 5 μm.
We showed previously that mitochondrial fragmentation in fzo1 cells results in secondary defects in mitochondrial DNA (mtDNA) maintenance and the loss of mitochondrial genomes9. This mtDNA loss is prevented when a dnm1Δ mutation is introduced into fzo1-1 or fzo1Δ cells. The retention of mtDNA in these strains presumably results from the unusual mitochondrial morphology in dnm1Δ cells. Consistent with this idea, we find that dnm1Δ mutations also prevent or reduce mtDNA loss in several other yeast mitochondrial-morphology mutants (S.W. Gorsich and J.M.S., unpublished observations).
Mitochondrial fusion is not restored in dnm1 fzo1-1 cells
The finding that dnm1 mutations block mitochondrial fragmentation in fzo1-1 cells is consistent with a role for Dnm1 in mitochondrial fission. Alternatively, activation of a FZO1-independent fusion pathway could prevent mitochondrial fragmentation in the fzo1-1 dnm1Δ double mutant. To test this idea, we studied mitochondrial fusion in fzo1-1 dnm1Δ cells during mating. We assessed mitochondrial fusion by labelling mitochondria in cells of opposite mating type with either a mitochondrial-targeted form of the green fluorescent protein (mito-GFP) or MitoTracker CMXRos (Molecular Probes) and examining the distribution of the fluorescent markers in budded zygotes formed at the permissive (25 °C) and non-permissive (37 °C) temperatures17. In wild-type zygotes formed at 25 °C or 37 °C, mitochondrial fluorescent markers co-localized, indicating that the mitochondrial membranes had fused and the mitochondrial contents had mixed (Fig. 7a–d, Table 2). Mitochondrial fusion also occurred efficiently in fzo1-1 × fzo1-1 zygotes formed at 25 °C (Table 2). As shown previously, mitochondrial networks fragmented and failed to fuse in fzo1-1 × fzo1-1 zygotes formed at 37 °C, indicating that fusion requires FZO1 function (Fig. 7e–h, Table 2; ref. 9). Consistent with its proposed role in mitochondrial fission, DNM1 function is not required for mitochondrial fusion, as indicated by the mixing of mitochondrial content observed in dnm1Δ × dnm1Δ zygotes at both temperatures (Fig. 7i–l, Table 2). Mitochondrial fusion observed in dnm1Δ cells is, however, strictly dependent on FZO1 function. Mitochondrial fusion is blocked in zygotes formed from fzo1-1 dnm1Δ haploid cells at 37 °C, but not 25 °C (Fig. 7m–p, Table 2). Taken together, these results show that maintenance of mitochondrial tubules in fzo1-1 dnm1Δ mutants results from a block in mitochondrial fission rather than the activation of an alternate fusion pathway in these cells. Moreover, with respect to mitochondrial fusion during mating, the fzo1 mutation is epistatic to the dnm1 mutation.
Figure 7. Mitochondrial fusion is not restored in dnm1 fzo1-1 cells.
Wild-type (a–d), fzo1-1 (e–h), dnm1Δ (i–l) and dnm1Δ fzo1-1 (m–p) cells of opposite mating type were labelled with either mito-GFP (b, f, j, n) or MitoTracker CMXRos (c, g, k, o) and mated at 37 °C. Images of zygotes were obtained by confocal microscopy and mitochondrial fusion was assessed by examining the overlayed mito-GFP and MitoTracker images (d, h, l, p). The ‘z’ in i designates the zygote visualized in j–l. Scale bars represent 2 μm.
Table 2.
Mitochondrial fusion in budded zygotes
| Strains crossed | Temperature (°C) | Fused mitochondria |
|---|---|---|
| FZO1, DNM1 × FZO1, DNM1 | 25 | 152/152 (100%) |
| 37 | 104/105 (99%) | |
|
| ||
| FZ01, dnm1Δ × FZ01, dnm1Δ | 25 | 114/115 (99%) |
| 37 | 104/105 (99%) | |
|
| ||
| fzo1-1, DNM1 × fzo1-1, DNM1 | 25 | 56/59 (95%) |
| 37 | 3/65 (1.5%) | |
|
| ||
| fzo1-1, dnm1Δ × fzo1-1, dnm1Δ | 25 | 122/128 (95%) |
| 37 | 2/94 (2%) | |
Discussion
The process of mitochondrial fission is likely to require several steps, including the recruitment of Dnm1 (and any accessory proteins) to sites on the sides of mitochondrial tubules, the constriction of mitochondrial tubules at these sites, and the coordinated division of the outer and inner mitochondrial membranes to generate new tubule ends. Using immunogold labelling, we have characterized a pattern of Dnm1 localization that is consistent with each of these steps (Fig. 4). First, we detected Dnm1 clustered on the outer membrane of mitochondrial tubules that are not constricted. We suggest that these Dnm1 clusters represent sites of future constriction and fission. Second, Dnm1 was found concentrated at constriction sites in mitochondrial tubules. Although these constricted regions could represent sites of mitochondrial fission or fusion, our genetic studies do not support a role for the Dnm1 GTPase in fusion9,11. Thus, these constriction sites most likely represent mitochondrial tubules in the process of dividing. Third, Dnm1 was found clustered at the ends or tips of mitochondrial tubules. In some sections (Fig. 4d, e), the labelled mitochondrial tip was located close to another, unlabelled, tip, indicating that these structures may have been derived from newly divided mitochondrial tubules. We never detected gold particles on both tips in cases where two tips were juxtaposed. It is possible that the Dnm1 protein remains associated with only one of the new mitochondrial ends formed by fission. In vivo, these Dnm1 ‘caps’ may quickly disassemble, leaving behind a free mitochondrial end. Alternatively, tips coated with Dnm1 may ultimately form stable interactions with other cellular structures.
Mitochondrial tubules in dnm1 mutant cells form nets of varying sizes. Although these nets could arise in several ways, one explanation is that the ends of mitochondrial tubules have fused with one another or with the sides of adjacent tubules to form an interconnected web. Presumably, these nets also form transiently in wild-type cells but are rarely observed because they are quickly resolved by DNM1-mediated fission events that release tubule ends. According to this scenario, formation of these tubular nets in fzo1-1 dnm1Δ cells requires that the block in mitochondrial fission precede the block in mitochondrial fusion. In addition, ongoing fusion does not appear to be required to maintain mitochondrial nets. We observe that mitochondrial nets in fzo1-1 dnm1Δ cells persist for several hours after a shift to the non-permissive temperature at which mitochondrial fusion is blocked (Figs 5, 6). Finally, mitochondrial nets may not form under conditions in which both mitochondrial fission and fusion are blocked. Although fzo1Δ dnm1Δ cells contain mitochondria with dnm1-like morphologies (95% at 25 °C), mitochondrial nets are not visible in these cells. Although we cannot rule out the possibility that nets are present but cannot be resolved in fzo1Δ dnm1Δ cells, these results indicate that FZO1-dependent fusion may be required to form mitochondrial nets in the absence of fission.
Figure 6. Mitochondrial-membrane morphology in dnm1 and dnm1 fzo1-1 mutant cells.
The asterisk indicates that 5% of dnm1Δ fzo1-1 cells contain visible nets at 37 °C.
An alternative possibility is that mitochondrial nets represent an intermediate stage in the formation of the wild-type network. Mitochondrial membranes that have lost their cytoskeletal attachments form spherical organelles18–21. These spherical compartments could be converted into tubular nets in several steps. First, the organelle must be flattened to bring regions of the inner membrane that face the mitochondrial matrix into close proximity. Second, periplasmic fusion between sections of the inner and outer mitochondrial membranes could open ‘holes’ in the mitochondrial compartment. The resulting structure would resemble the mitochondrial nets observed in dnm1 mutant strains. Subsequent fission events, mediated by the Dnm1 GTPase, and fusion events, mediated by the Fzo1 GTPase, would then be required to remodel the net, establishing and maintaining the branched reticulum characteristic of wild-type cells.
Methods
Yeast methods
Standard methods were used for growth, transformation and genetic manipulation of Saccharomyces cerevisiae. All rich (YPDextrose, YPGlycerol) and synthetic (SDextrose, SGlycerol) media were prepared as described22.
Yeast strains
All yeast strains are isogenic to strain FY10 (ref. 23). The following strains were used in this study: JSY1781, MATa/MATα leu2Δ1/leu2Δ1 HIS3/his3Δ200 ura3-52/ura3-52DNM1-HAc/DNM1-HAc; JSY3094, MATa leu2Δ1 his3Δ200 ura3-52 mdm20Δ::LEU2; JSY3095, MATa leu2Δ1 his3Δ200 ura3-52 mdm20Δ::LEU2 dnm1Δ::HIS3; JSY3096, MATα leu2Δ1 his3Δ200 ura3-52; JSY3097, MATα leu2Δ1 his3Δ200 ura3-52 dnm1Δ::HIS3; JSY3161, MATa his3Δ200 ura3-52 fzo1-1; JSY3162, MATα his3Δ200 ura3-52; JSY3163, MATa his3Δ200 ura3-52 fzo1-1 dnm1Δ::HIS3; and JSY3164 MATα his3Δ200 ura3-52 dnm1Δ::HIS3. JSY3094–JSY3097 and JSY3161–JSY3164 are sister spores from the same tetrad.
Quantification of mitochondrial morphology phenotypes
Mitochondrial morphology was scored in wild-type and mutant cells expressing mitochondrial-targeted forms of GFP from plasmids (pDO12, ref. 11, and pVT100UGFP, provided by B. Westermann and W. Neupert). pVT100UGFP efficiently targets GFP to both rho+ (containing mtDNA) and rho° (lacking mtDNA) mitochondria. Alternatively, 100 μl culture was stained with 100 nM 3,3′-dihexyloxacarbocyanine (DiOC6) (Molecular Probes) as described16. Cells were grown overnight in YPD or synthetic medium at 25 °C, diluted to 0.5 optical-density units per ml in fresh medium and incubated at 25 °C or 37 °C for an extra 3 h before viewing. Numbers in Figs 2, 6 represent the percentage of an asynchronously growing culture with a particular mitochondrial morphology (n=100–300). Comparable results were obtained using both staining methods, indicating that the mitochondrial-morphology phenotypes observed in mutant cells were not due to the presence of the Cox4–GFP fusion protein. Digital microscopic images of cells were acquired as described9,11.
Mitochondrial-fusion assay
To examine mitochondrial fusion during mating, we transformed haploid strains JSY3161–JSY3164 of opposite mating type with the mito-GFP plasmid (pVT100UGFP) or labelled them with MitoTracker CMXRos (Molecular Probes); the strains were mated at 25 °C or 37 °C, and analysed as described17.
Electron microscopy
dnm1 mdm20Δ cells were fixed and embedded as described9. Serial sections (75 nm) were viewed with a Hitachi H-7100 transmission electron microscope. Digital images obtained from 12 consecutive sections were used to produce three-dimensional reconstructions of mitochondrial nets using Voxblast software (VayTek, Fairfield, IA).
Immunoelectron microscopy was performed essentially as described24. Exponentially growing cells (30 °C) were fixed in suspension for 15 min by adding an equal volume of freshly prepared 8% formaldehyde in 1 × PBS, pH 7.4. The cells were pelleted, resuspended in fresh fixative (4% formaldehyde, 1 × PBS, pH 7.4) and incubated for an extra 18–24 h at 4 °C. The cells were washed briefly in PBS and resuspended in 1% low-gelling-temperature agarose. The agarose blocks were trimmed into pieces of 1 mm3, cryoprotected by infiltration with 2.3 M sucrose/20% polyvinyl pyrrolidone (10 K)/PBS (pH 7.4) for 2 h, mounted on cryopins and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on a Leica UCT ultramicrotome equipped with an FC-S cryo-attachment and collected onto formvar/carbon-coated nickel grids. The grids were washed through several drops of 5% fetal calf serum (FCS) plus PBS containing 10 mM glycine (pH 7.4), blocked in 10% FCS for 30 min and incubated overnight in 20 μg ml–1 monoclonal anti-haemagglutinin antibody (Covance Research Products, Richmond, CA). After washing, the grids were incubated for 2 h in 5-nm gold donkey anti-mouse conjugate (Jackson ImmunoResearch). The grids were washed through several drops of PBS followed by several drops of ddH2O. Grids were then embedded in an aqueous solution containing 3.2% polyvinyl alcohol (10 K)/0.2% methyl cellulose (400 centiposes)/0.1% uranyl acetate. The sections were examined and photographed on a Philips 420 transmission electron microscope at 80 kV.
Acknowledgments
We thank D. Otsuga for identifying the net phenotype in the dnm1 mutant. This work was supported by grants from the NIH and the American Cancer Society (to J.M.S.) and a grant from the NSF (to J.N.).
References
- 1.Williamson DH, et al. In: Genetics, Biogenesis and Bioenergetics of Mitochondria. Bandlow W, Schweyen RJ, Thomas DY, Wolf K, Kaudewitz F, editors. Walter de Gruyter; Berlin: 1976. pp. 99–115. [Google Scholar]
- 2.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 3.Tourte M, Mignotte F, Mounolou JC. Organization and replicative activity of the mitochondria of oogenic and previtellogenic oocytes in Xenopus laevis. Dev Growth Differ. 1981;23:9–21. doi: 10.1111/j.1440-169X.1981.00009.x. [DOI] [PubMed] [Google Scholar]
- 4.Fuller MT. In: The Development of Drosophila melanogaster. Bate M, Martinez-Arias A, editors. Cold Spring Harb. Lab. Press; Cold Spring Harbor: 1993. pp. 71–147. [Google Scholar]
- 5.Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- 6.Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Techn. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 7.Johnson LV, Walsh ML, Chen LB. Localization of mitochondria in living cells with rhodamine 123. Proc Natl Acad Sci USA. 1980;77:990–994. doi: 10.1073/pnas.77.2.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- 9.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 11.Otsuga D, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnova E, Shurland DL, Ryazantsev SM, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 14.Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 15.Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 16.Hermann GJ, King EJ, Shaw JM. The yeast gene, MDM20, is necessary for mitochondrial inheritance and organization of the actin cytoskeleton. J Cell Biol. 1997;137:141–153. doi: 10.1083/jcb.137.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nunnari J, et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boldogh I, Vojtov N, Karmons S, Pon LA. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p. J Cell Biol. 1998;141:1371–1381. doi: 10.1083/jcb.141.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess SM, Delannoy M, Jensen RE. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol. 1994;126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherman F, Fink GR, Hicks JB, editors. Methods in Yeast Genetics. Cold Spring Harb. Lab. Press; Cold Spring Harbor: 1986. pp. 1–186. [Google Scholar]
- 23.Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S228C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 24.Rieder SE, Banta LM, Köhrer K, McCaffery JM, Emr SE. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]