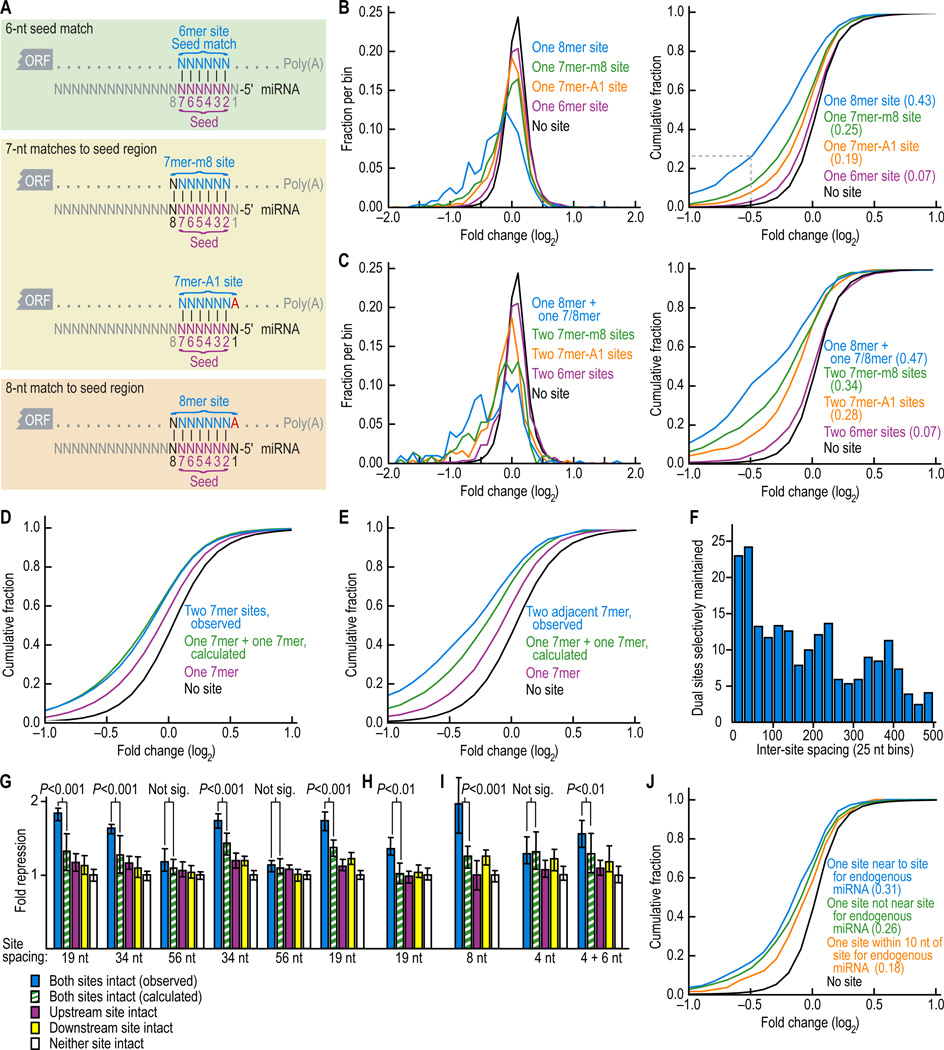
Figure 1. Downregulation of messages with 6-8mer sites.
(A) Canonical miRNA complementary sites.
(B) Effectiveness of single canonical sites. Changes in abundance of mRNAs following miRNA transfection were monitored with microarrays. Distributions of changes (0.1 unit bins) for messages containing the indicated single sites in their UTRs are shown (left), together with the cumulative distributions (right). The dashed line in the cumulative distributions indicates that 27% of mRNAs with UTRs containing a single 8mer were down-regulated at least 29% (2−0.5 = 0.71). Results of eleven experiments, each performed in duplicate and each transfecting a duplex for a different miRNA (Table S2), were consolidated. Results shown were an amalgam of the data from all 11 miRNAs; the relative strengths of the different sites were consistent when examining each transfection individually. For the cumulative plots, the minimal fraction of downregulated genes in that distribution is reported (parentheses), based on comparison with the no site distribution. Repression from UTRs containing an 8mer site was significantly more than that from UTRs with a 7mer-m1 site (P < 10−20, 1-sided K-S test); similar comparisons between UTRs containing a 7mer-m8 site versus a 7mer-A1 site, a 7mer-A1 versus a 6mer, and, a 6mer versus no site were also significant (P < 10−6, P < 10−20 and P < 10−31, respectively).
(C) Increased effectiveness of dual sites. Changes in mRNA abundance following miRNA transfection, represented as in B, except mRNAs with 3'UTRs containing the indicated pairs of sites were monitored. Repression from UTRs containing both an 8mer and either a 7mer or 8mer site was significantly more than that from UTRs with two 7mer-m8 sites (P < 10−3, 1-sided K-S test); similar comparisons between UTRs containing two 7mer-m8 sites versus two 7mer-A1 sites, two 7mer-A1 versus two 6mer, and, two 6mer versus no site were also significant (P = 0.034, P < 10−11 and P < 10−6 respectively).
(D) Independence of most dual sites. Cumulative distributions of changes in mRNA levels following miRNA transfection for messages containing the indicated combinations of miRNA binding sites. Simulated values for 3'UTRs containing two 7mer sites (green) were calculated by combining the effect of two single 7mers; actual values for 3'UTRs containing two 7mers are in blue and those with length-matched UTRs containing single 7mers are in purple; otherwise as shown for Figure 1B.
(E) Synergism between closely spaced sites. Cumulative distributions of changes in mRNA levels as for (D), except the plot for two observed sites (blue) only considered 3'UTRs containing two closely spaced 7mers (within 100 nt of each other). Repression from UTRs containing two adjacent sites was significantly increased compared to simulated UTRs containing one plus one site (P = 0.040, 1000 resampling iterations), whereas repression from UTRs containing two sites irrespective of distance did not significantly differ from simulated UTRs containing one plus one site (P = 0.81).
(F) Selective maintenance of dual sites spaced at different intervals. Human 3'UTRs with exactly two 7mer sites to the same miRNA were binned based on inter-site distance (counting the number of nucleotides between the 3' nt of the first site and the 5' nt of the second site). The number of conserved dual sites exceeding the background (as estimated from the average of control cohorts) was plotted after performing for each bin site-conservation analysis analogous to that in Lewis et al (2005), using the miRNA families conserved broadly among vertebrates (Table S1).
(G) MicroRNA-mediated repression of luciferase reporter genes fused to 3'UTR fragments containing two miR-124 target sites with different spacing intervals. After normalizing to the transfection control, luciferase activity from HeLa cells cotransfected with each reporter construct and its cognate miRNA (miR-124) was normalized to that from cotransfection of each reporter with its non-cognate miRNA (miR-1). Plotted are the normalized values, with error bars representing the third largest and third smallest values among 12 replicates. P values (Wilcoxon rank-sum test) indicate whether repression from a reporter containing both sites (blue) was significantly greater than expected from multiplicative effects (green). For modeling independent activity of sites, repression expected from a reporter with two sites was the product of repression observed from otherwise identical reporters containing single intact sites (purple and yellow; pairing off the repression values in the order that they were generated). Reporters of the rightmost quintet were identical to those of the leftmost quintet, except the point substitutions disrupting target sites differed.
(H) Repression observed for the reporter constructs as in the leftmost quintet of (G) but modified such that both miR-124 sites were substituted with miR-1 sites. miR-196 served as the noncognate miRNA.
(I) Repression of reporter constructs containing 3'UTR fragments with naturally closely spaced miR-1 and miR-133 sites, compared to that of mutant derivatives of these fragments. A mixture of miR-1 and miR-133 was co-transfected as the cognate miRNA, and miR-196 served as the noncognate control.
(J) Cooperativity between sites to transfected and endogenous miRNAs in HeLa. Endogenous sites considered were those for let-7 RNA, miR-16, miR-21, miR-23, miR-24, miR-27, and miR-30 (Tom Tuschl, personal communication). 7mer-m8 sites at a cooperative distance (>7 nt and <40 nt) from an endogenous miRNA 7-8mer site were significantly more downregulated than sites that were either too close to an endogenous miRNA (≤7 nt, including overlapping sites; P = 0.0054, 1-sided K-S test), or not close to an endogenous site (≥40 nt, or no endogenous site, P = 0.036, 1-sided K-S test).