Abstract
Every cancer is different and cancer cells differ from normal cells, in particular, through genetic alterations. HLA molecules on the cell surface enable T lymphocytes to recognize cellular alterations as antigens, including mutations, increase in gene product copy numbers or expression of genes usually not used in the adult organism. The search for cancer-associated antigens shared by many patients with a particular cancer has yielded a number of hits used in clinical vaccination trials with indication of survival benefit. Targeting cancer-specific antigens, which are exclusively expressed on cancer cells and not on normal cells, holds the promise for much better results and perhaps even a cure. Such antigens, however, may specifically appear in very few patients or may be mutated appearing just in one patient. Therefore, to target these in a molecularly defined way, the approach has to be individualized.
Keywords: antigens, cancer immunotherapy, genome sequencing, HLA ligandome, immunogenicity, immunomonitoring, mutations, T cells, vaccination
The concept of cancer immunotherapy looks back on a long history with phases of frequent failures. Paul Ehrlich hypothesized that the immune system constantly eliminates cancer cells, and that cancer incidence would be much higher in the absence of such immunosurveillance. It took 100 years until this was proven to be correct. Although Klein et al., in 1960, definitely demonstrated the existence of tumor-associated antigens that can lead to rejection of cancer by the mouse that had developed in this very mouse [1], the observation that T-cell-deficient nude mice did not have more frequent cancer [2] almost killed the belief in cancer immunosurveillance. Subsequently, a large body of evidence based on animals deficient for immune genes and showing increased cancer incidence, eventually led to the acceptance of Ehrlich’s hypothesis [3]. Clinical oncologists as well as main stream cancer biologists still did not consider cancer immunity as something of importance – the ‘bible’ of cancer biology in 2000 did not even list immune evasion as a cancer hallmark [4] eventually changing this in 2011 by including ‘avoiding immune destruction’ as an emerging one [5].
Recent developments
The field of cancer immunotherapy changed in the recent years and now is in a phase of exponential growth with regard to its reputation. An early clinical successes was the curative T-cell-mediated therapy of leukemia by donor lymphocyte transfusion [6]. Another success is the rise of passive immunotherapy with monoclonal antibodies [7]. A more recent development is the approval of two active immunotherapies, sipuleucel-T [8] and, in particular, ipilimumab. Finally, the very recently reported clinical successes of nivolumab and other PD-1/PD-L1 antibodies [10] delivering objective response rates of up to 50% in advanced melanoma patients when used in combination with ipilumimab [11] have convinced most clinical oncologists that immunotherapy will play a vital role in cancer therapy in the very near future. In addition, the use of adoptive T-cell transfer, in particular of chimeric antigen receptor-modified T cells has shown impressive clinical benefit [12,13].
Whereas sipuleucel-T contains a single antigen of the tumor-associated kind, applied as a vaccine in a complicated way and with only limited clinical success, antibodies blocking CTLA-4 and PD-1/PD-L1 represent a new principle of cancer immunotherapy: modulation of checkpoints controlling immune regulation. By blocking the inhibitory molecule CTLA-4 or PD-1 on T cells, immune responses otherwise underlying suppression or termination are activated and enhanced. In the Phase III study for ipilimumab preceding approval, a fraction of advanced melanoma patients not only showed prolonged overall survival as compared to the control group but also reached a long-lasting stability long after the end of drug application that is not seen with conventional cytotoxic or cytostatic drugs or even with small molecule inhibitors. This long-lasting beneficial effect is ascribed to the induction of specific immunity against cancer-specific antigens, although immunomonitoring for the reaction against defined antigens had not been performed. It is to be assumed that cancer-specific antigens, including non-synonymous mutations, are the target of some of the T cells induced or relieved from suppression by ipilimumab. In addition to the beneficial immune responses against the cancer cells, however, responses against self-antigens have been induced, as to be judged from the range of adverse events. These were mainly immune-related gastroenterological or dermatological events and 2% of the patients died of immune-related complications. Thus, the blockade of this immune regulatory checkpoint presumably induced or activated immune responses to cancer-specific antigens as well as normal self antigens expressed in normal tissues far away from the cancer sites. Although the recent successes represent a hallmark in the history of cancer therapy, there is still a lot of room for improvement. Long-lasting survival benefit is observed only in approximately 20% of ipilimumab-treated melanoma patients and the translation of the remarkable objective response rates achieved with PD-1/PD-L1 antibodies into a significant survival benefit still needs to be confirmed. Remarkably, preliminary biomarker data suggests that response to PD1/PD-L1 antibodies is strongly associated with prior infiltration of T cells in the tumor implying that such therapies would require a preexisting endogenous or vaccine-induced antigen-specific immune response in the tumor [14].Adoptive transfer of chimeric antigen receptor-modified T cells specific for the B-cell-specific antigen CD19, shared by malignant B-cell leukemia cells, resulted in complete remission in patients. Since such cells are so highly efficient, the selection of antigens has to be performed very carefully since expression on normal cells essential for survival has to be avoided. Thus, the choice of new antigens is rather limited.
Antigen-specific vaccination
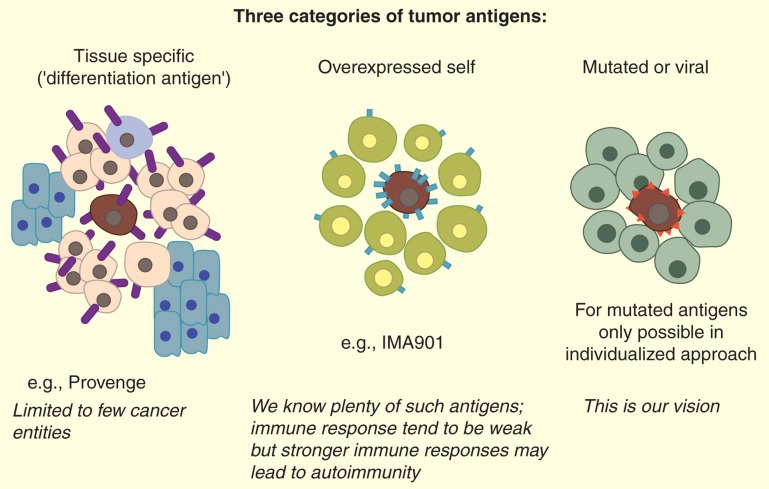
The alterations distinguishing cancer cells from their normal counterparts are reflected in differences in expressed gene products, manifested in higher copy numbers of a normal cellular protein, or in expression of a protein not usually expressed in the tissue of cancer origin (e.g., cancer testis antigens), or neoantigens caused by mutations [15–21] or viral infection [22]. For simplicity, we distinguish three types of cancer antigens that are considered for therapeutic vaccination: i) neoantigens, including mutations and viral antigens, ii) self proteins that are either overexpressed or usually not expressed in most of the adult body and iii) tissue-specific gene products in case the cancer affects a tissue or cell type not essential for the life of the patient (e.g., B cells, melanocytes, or prostate) (Figure 1). Cancer testis antigens [23,24] are a especially versatile and useful subcategory of type 2: they are self antigens and as such prone to mediate negative T-cell selection in the thymus, but they are not expressed on normal adult tissues. Thus, cancer testis antigens should not cause antigen target-related toxicities even if high-affinity T cells can be achieved.
Figure 1.
A simplified classification of the numerous types of tumor antigens.
All three types of antigens can be used for vaccination in various forms, for example, as protein [8], virus constructs [25], DNA [26], RNA [27], long peptides [22] or peptides representing exactly the natural HLA ligands [28] on tumor cells [29,30]. Although each of the antigen formats have specific merits, the use of short synthetic peptides, typically 8–12 amino acids long for HLA class-I binding peptides and 15–20 amino acids long for HLA class II-binding peptides, identical to the natural substances on the tumor cell, have the advantage of relatively easy production in clinical grade. Additionally, they offer the possibility of easy immunomonitoring using exactly the ingredients of the vaccine [29]. These aspects favor synthetic peptides identical to HLA ligands for use in personalized approaches.
Personalized approach – mutated antigens
Ideally, one would like to have the efficient immune responses against cancer-specific antigens that presumably were the cause for the observed long-term survival in the ipilimumab Phase III studies in the absence of the self-directed responses that likely have led to the adverse events. New immune system checkpoint modulators are in development [31,32], such as antibodies against PD-1 or PD-L1, and is believed to balance the immune responses more to tumor-related antigens on account of the predominant expression of PD-L1 in the tumor microenvironment. The ultimate limitation of immune responses against tumor-specific antigens, however, requires the immunization against these antigens directly. This is the goal of the attempt at using a comprehensive analysis of the HLA ligandome (also termed immunopeptidome) of cancer cells to pursue tumor antigen discovery for a personalized vaccine approach [33].
This approach (Figure 2), also known as the Tübingen approach or XPRESIDENT™ [33–35], always requires a piece of tumor tissue from the very patient for whom the vaccine is to be prepared, essentially as we had anticipated in a 2002 review [35]. The tumor sample ideally should be surgically removed, judged by a pathologist for tumor cell content, and should be ideally accompanied by a sample of normal tissue from the organ of tumor origin. This is feasible for many entities, for example, for renal or hepatocellular carcinoma, but not for others, for example, ovarian carcinoma. For these cases, other tissues or cells of the patient, for example, peripheral blood cells, can serve as a source of reference material at least for excluding abundant normal self-peptides from consideration. Other methods such as those described in [36] are able to find interesting T-cell epitopes but do not directly give information on HLA ligands which are differentially expressed between tumor and normal cells.
Figure 2.
XPRESIDENT platform, also known as the Tübingen approach, for identification, selection and validation of tumor-associated peptides.
LC-MS/MS: Liquid chromatography coupled to tandem mass spectrometry; NOMAP: Non-tumor associated peptide;TUMAP: Tumor associated peptide.
Then, HLA molecules are precipitated from the tissues samples, and HLA-bound peptides are eluted by established [37] and now optimized [38] methods. We expect that at least 10,000 different peptides are present on the HLA molecules of a cell, be it cancer or normal [39,40]. Present technology probably allows to separate and identify about 30% of these; further improvement of HPLC and mass spectrometry technology is expected to increase this rate [38,41,42].
Since reliable mass spectrometric identification of peptides requires knowledge of the underlying genomic sequences present in the tissue under analysis, exome and transcriptome sequencing is performed in addition. The masses of mutated sequences predicted to be presented by the patient’s HLA molecules, typically 9 amino acids long [43,101] are calculated and searched for in the mass spectrometry data. If the mass of a predicted mutated peptide is found, the fragmentation pattern is being determined. If it fits to the predicted sequence, a synthetic peptide of this sequence, labeled with an isotope variant amino acid, for example, 15N-Valin, is spiked in for calibration and definitive confirmation of the mutated peptide as an HLA ligand. A faster alternative to this procedure would be an improved mass spectrometry system allowing unequivocal de novo sequence determination.
Once a mutated HLA ligand (present in tumor cells but not in any other cell) has been found, this can be used for inclusion in a cancer vaccine in this particular patient. As can be appreciated from this outline, the complexity of the finding procedure requires some time – in our experience presently a few months that may be condensed to a few weeks in the future.
Recently, the therapeutic potential of mutated antigens has been started to be validated preclinically using next-generation sequencing of mouse tumors followed by successfully inducing anticancer immunity in these mice using mutated peptide vaccines [44] and clinically by the observation that a complete and durable regression in an advanced melanoma patient treated with adoptively transferred tumor-infiltrating lymphocytes appeared to be predominantly mediated by specific immune responses to a mutated neoantigen [45].
Personalized approach – non-mutated antigens
The identification of non-mutated HLA ligands overexpressed on tumor cells, as compared to normal cells of the same patient available for analysis, is much faster. Such peptides should be also considered for inclusion in a personalized vaccine, since for most tumor-associated HLA ligands, we find a vast heterogeneity of expression in tumors from different patients and for some individuals very strongly overpresented or even presumably specifically presented tumor self-antigens can be found. Here, however, a difficulty is the analysis of their expression in other tissues of the same patient that in many cases are not available for analysis. This problem can be addressed by using standard gene, protein or even better HLA ligand expression data from normal tissues of unrelated individuals (as gene expression does not absolutely correlate with HLA ligandome expression [46]), and on information about the cancer relatedness of the gene of origin of the peptide. Overexpressed, tumor-associated HLA ligands selected to be shared by a high proportion of patients have been successfully used in clinical Phase I and II trials in HLA-A*02 patients with renal cell carcinoma (multipeptide vaccine IMA901) [29] and colorectal carcinoma (multipeptide vaccine IMA910). While such predefined multipeptide vaccines have the advantage to be applied ‘off-the-shelf’, in addition to antigens shared abundantly by a patient population, individually and highly overexpressed tumor antigens do exist. Thus, the use of such individually presented peptides ideally identified as natural HLA ligands on the tumor of the patient to be vaccinated are considered an additional therapeutic advantage.
Based on our experience in the few experimental attempts so far with patient-individualized cancer vaccination, we foresee the following three-step standard procedure for the near future.
Three-step strategy for individualized immunotherapy
First step – as fast as possible (e.g., right after surgery): HLA allele typing and vaccination with off-the-shelf, that is, tumor-associated peptides predefined as abundantly present on the majority of tumors of a given tumor entity.
Second step – after individual HLA ligand analysis: vaccination with suitable individually overexpressed peptides stemming from known tumor-associated gene products. As in the first step, these peptides can be predefined (i.e., manufactured ready for clinical application in a peptide warehouse) but due to their lower abundance in all patients of a given tumor entity, they would be individually composed to unique drug products from the warehouse peptides. The advantage of this warehouse approach is that this can be done within relatively little time but still individualized.
Third step – after completed genome and transcriptome sequencing and mass spectrometric identification of mutated HLA ligand candidates: vaccination with mutated peptides in addition to continuing vaccination with second step peptides.
Problems to be solved
Briefly, these are the challenges: (1) identification of antigens, in particular mutations; (2) induction of productive immune responses in clinical trials; (3) monitoring of the immune responses and their correlation with clinical outcome; (4) potentially further cycles of points 1 to 3 to improve the processes based on the findings. The following items aim at these problems.
Technical problems in the identification of mutated peptides
As far as the identification of individualized cancer-specific antigens, that is, mutations, is concerned, there are still a number of technical problems: i) The reliability of genome and transcriptome sequencing results. In order to minimize false positive mutation detection, technical as well as biological replicates appear mandatory. ii) The heterogeneity of tumor cells contained in a surgical tumor sample. This can be partially judged by the number of reads for particular mutations; to be efficient, an individualized vaccine should aim at combining several antigens to increase the chance to hit every tumor cell with at least one T-cell specificity.
Better adjuvants, better immunomodulation
So far, therapeutic cancer vaccines do not generally induce strong immune responses. However, as the breath of immune responses directly associates with the clinical benefit, in particular overall survival, as shown by the IMA901 trials in renal cell cancer patients [23], it can be assumed that increasing the frequency as well as the quality of vaccine-induced T cells would also lead to broader and thus more efficacious immune response. To induce better immune responses, the route of administration (intradermal, subcutaneous and intramuscular), the choice of adjuvants (e.g., montanide), local immunomodulators (e.g., GM-CSF, defined TLR ligands) and the preceding (e.g., cyclophosphamide) or accompanying (e.g., cytokines such as type I interferon, IL-2, IL-12 or IL-15) systemic immunomodulators have to be empirically evaluated, ideally in multi-arm or multi-cohort Phase I/II trials.
Determination of immunogenicity of candidate peptides
In our experience, only a fraction of (non-mutated) peptides identified as HLA ligands are immunogenic in vivo. However, testing immunogenicity in vitro [42], using peripheral blood T cells from healthy individuals, proved to be of highly predictive value for actual immunogenicity in vivo [Walter et al., Manuscript in Preparation]. Thus, for composition of an individualized multipeptide cocktail, one should aim to include an in vitro immunogenicity test beforehand. Whether this is needed also for mutated peptides is not known yet.
Regulatory issues
Regarding the personalized approach, the regulatory issues need to be solved together with the responsible authorities (such as EMA, US FDA and Paul Ehrlich Institute). To prepare this, members of the Regulatory Research Group (RRG) of the Association of Cancer Immunotherapy (CIMT; [102]) held a meeting recently at the EMA on the issue of such patient-individualized vaccines termed actively personalized vaccines (APVACs) by the CIMT RRG. It appeared that the existing regulations are in principle compatible with the needs of this approach but specific points occurring with APVACs only and not with off-the-shelf therapeutics still need to be addressed and are currently being worked out by CIMT RRG in collaboration with industry and academic partners pursuing such pioneering APVAC projects.
Expert commentary
Taking into consideration our present knowledge on cancer development on the one hand and on adaptive immunity and its interdependence on innate immunity on the other, we come to the conclusion that the interaction between a spontaneously developing human cancer and the surrounding host environment must be a formidably complex problem of systems biology, with a large number of variables evolving over the years or even decades of tumor development. A major component of the host environment is the host immune system, as is well documented in mouse experiments [47,48]. For human cancer, this has been most convincingly documented by the prognostic value of the quantity and quality of tumor infiltrating immune cells in colorectal carcinoma and other cancers [49–51].
Every single human cancer patient harbors a distinct microcosm of malignant cells. Within this microcosm, considerable heterogeneity is found that by itself already points to selection events, in the sense that those cancer cell clones managing best to deal with the environment (i.e., the host) will thrive best. Thus, the cancer in a patient must consist of many different clones. It is assumed that these are all derived from a founder population that had acquired enough genetic alterations to escape the controls of the normal tissue community, in particular those alterations known as ‘hallmarks of cancer’ [5], and that these clonal populations are genomically instable. During growth of the cancer, or more precisely, during its evolution in the host, permanent Darwinian selection must take place, whereby the cancer cell community manipulates its environment to its own benefit, so that a particular tumor microenvironment surrounds or intercalates with the cancer cells. In addition to the selection pressure by host factors, the cancer clones in an actual cancer patient, in a world region with multiple available therapeutic interventions, are also exposed to the selective pressure inflicted by attempted cancer therapies. The spectrum of gene mutations is, therefore, unique for every individual cancer patient and differs from the set of mutations in tumors of other patients with probably little overlap. The genes coding for tumor drivers, or for protection from cell death, or for surveillance of genome integrity, are frequently found to be affected by mutations or other genetic alterations; however, the exact site of the alteration or mutation is widely different from patient to patient. These considerations have led to the concept of individualized, mutation-targeted vaccination against cancer [33,35,44,52].
Five-year view
Multipeptide vaccines targeting overexpressed HLA ligands but not mutated peptides will be continued and improved, based on biomarker analysis of previous trials and by combining with better immune modulation through immunomodulators or wise combination with checkpoint inhibitors and standard therapy that has strong synergistic potential with immunotherapy.
There will be two opposing but eventually mutually complementary trends in multipeptide vaccination: On the one hand, complex multipeptide cocktails containing potentially 20 or more peptides covering shared tumor-associated, non-mutated peptides for a given tumor entity fitting to several frequent HLA allotypes addressing the vast majority of the world’s population will be developed. On the other hand, there will be the consequent development of the individualized approach, targeting HLA ligands identified on tumor samples of the give patient to be treated, including mutated peptides. The complementary nature of the two approaches is in the potential of sequential application, as outlined above in the section on the three-step strategy for individualized immunotherapy: The off-the-shelf peptides could be administered quickly as ‘one-fits-all’ cancer antigen cocktail, and the individualized antigen cocktails would be given later, after detailed molecular analysis of the patient’s tumor.
For those patients in advanced disease state, where the mounting of an effective immune responses may take too much time, adoptive T-cell transfer carrying engineered T-cell receptors may be used as life-saving interventions. This approach currently depends on antigens shared by many patients, mainly due to the long time it requires to identify and select an appropriate T-cell receptor. However, for adoptive cellular therapy an individualized approach is also conceivable. Similar to a peptide warehouse for vaccination, a warehouse of T-cell receptors could be built readily available for transfection into the autologous patient T cells.
Financial & competing interests disclosure
HG Rammensee is co-founder of Immatics Biotechnologies GmbH, CureVac GmbH and Synimmune GmbH, all active in the field of immunotherapy of cancer. H Singh-Jasuja is co-founder and employee of Immatics Biotechnologies GmbH. The support of DFG (German Research Council) (SFB685) and DKTK (DKFZ German Cancer Consortium) is acknowledged. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Key issues
HLA ligands (peptides) expressed on tumor cells are targets for T-cell-mediated immunotherapy.
We distinguish the following three categories of tumor peptides to be considered for cancer immunotherapy.
Overexpressed peptides shared by as many patients of a given HLA type and a given cancer entity, to be used in multipeptide cocktails (e.g., IMA 901) for all patients with that HLA type and that cancer entity.
Overexpressed peptides identified by mass spectrometry in the tumor sample of an individual patient to be used for an individualized vaccination for this patient only. It will also frequently incorporate off-the-shelf peptides.
True tumor-specific mutated peptides identified by genome and transcriptome sequencing, mass spectrometry of the HLA ligandome and verification with synthetic peptide, to be used in individualized vaccination.
The logistic and regulatory challenges are multiple: The individualized peptide vaccination approach (APVAC) requires drug substance production (peptide synthesis) and drug formulation (mixing the peptide cocktails) according to the current regulatory boundaries, that is, under GMP conditions. Approval of such clinical trials and finally marketing approval will be a novel approach for the regulatory authorities (EMA and US FDA). However, first interactions appear that regulatory agencies are open and interested to encourage such activities in light of the therapeutic potential.
Combination therapies are likely to be advantageous, for example, peptide vaccination with immune modulators or checkpoint modulators or standard therapy with immunogenic potential, for example, radiotherapy or selected targeted small molecule or chemotherapies.
References
Papers of special note have been highlighted as:
of interest
•• of considerable interest
- 1.Klein G, Sjogren HO, Klein E, Hellstrom KE. Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host . Cancer Res. 1960;20:1561–1572. [PubMed] [Google Scholar]
- 2.Old LJ, Boyse EA. Current enigmas in cancer research . Harvey Lect. 1973;67:273–315. [PubMed] [Google Scholar]
- 3.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape . Nat. Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer . Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation . Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Van Rhee F, Kolb HJ. Donor leukocyte transfusions for leukemic relapse . Curr. Opin. Hematol. 1995;2(6):423–430. doi: 10.1097/00062752-199502060-00005. [DOI] [PubMed] [Google Scholar]
- 7.Rodt H, Kolb HJ, Netzel B et al. GVHD suppression by incubation of bone marrow grafts with anti-T-cell globulin: effect in the canine model and application to clinical bone marrow transplantation . Transplant. Proc. 1979;11(1):962–966. [PubMed] [Google Scholar]
- 8.Kantoff PW, Higano CS, Shore ND et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer . N. Engl. J. Med. 2010;363(5):411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]; • This study describes the first FDA approved therapeutic tumor vaccination.
- 9.Wolchok JD, Neyns B, Linette G et al. Ipilimumab monotherapy in patients with pretreated advanced melanoma: a randomised, double-blind, multicentre, phase 2, dose-ranging study . Lancet Oncol. 2010;11(2):155–164. doi: 10.1016/S1470-2045(09)70334-1. [DOI] [PubMed] [Google Scholar]
- 10.Hamid O, Robert C, Daud A et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma . N. Engl. J. Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolchok JD, Kluger H, Callahan MK et al. Nivolumab plus ipilimumab in advanced melanoma . N. Engl. J. Med. 2013;369(2):122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This report impressively shows the power of the immune system to destroy cancer tissues.
- 12.Brentjens RJ, Davila ML, Riviere I et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia . Sci. Transl. Med. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grupp SA, Kalos M, Barrett D et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia . N. Engl. J. Med. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powderly JD, Koeppen H, Hodi FS et al. Biomarkers and associations with the clinical activity of PD-L1 blockade in a MPDL3280A study . J. Clin. Oncol. 2013;31(Suppl., Abstract 3001) [Google Scholar]
- 15.Agrawal N, Jiao Y, Bettegowda C et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma . Cancer Discov. 2012;2(10):899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanda M, Matthaei H, Wu J et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia . Gastroenterology. 2012;142(4):730–733. e739. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodis E, Watson IR, Kryukov GV et al. A landscape of driver mutations in melanoma . Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, Jiao Y, Dal Molin M et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways . Proc. Natl Acad. Sci. USA. 2011;108(52):21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsons DW, Li M, Zhang X et al. The genetic landscape of the childhood cancer medulloblastoma . Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood LD, Parsons DW, Jones S et al. The genomic landscapes of human breast and colorectal cancers . Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 21.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control . Nat. Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 22.Kenter GG, Welters MJ, Valentijn AR et al. Vaccination against HPV-16 oncoproteins for vulvar intraepithelial neoplasia . N. Engl. J. Med. 2009;361(19):1838–1847. doi: 10.1056/NEJMoa0810097. [DOI] [PubMed] [Google Scholar]
- 23.Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary . Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- 24.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy . Immunol. Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 25.Kantoff PW, Schuetz TJ, Blumenstein BA et al. Overall survival analysis of a phase II randomized controlled trial of a Poxviral-based PSA-targeted immunotherapy in metastatic castration-resistant prostate cancer . J. Clin. Oncol. 2010;28(7):1099–1105. doi: 10.1200/JCO.2009.25.0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirayama M, Nishikawa H, Nagata Y et al. Overcoming regulatory T-cell suppression by a lyophilized preparation of Streptococcus pyogenes . Eur. J. Immunol. 2013;43(4):989–1000. doi: 10.1002/eji.201242800. [DOI] [PubMed] [Google Scholar]
- 27.Rittig SM, Haentschel M, Weimer KJ et al. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients . Mol. Ther. 2011;19(5):990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules . Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 29.Walter S, Weinschenk T, Stenzl A et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival . Nat. Med. 2012 doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]; •• This study shows immune responses against vaccine peptides to be associated with clinical benefit and is the first controlled study in humans showing the effect of low-dose cyclophosphamide on regulatory T cells.
- 30.Feyerabend S, Stevanovic S, Gouttefangeas C et al. Novel multi-peptide vaccination in Hla-A2+ hormone sensitive patients with biochemical relapse of prostate cancer . Prostate. 2009;69(9):917–927. doi: 10.1002/pros.20941. [DOI] [PubMed] [Google Scholar]
- 31.Oosterwijk E, Boerman OC, Oyen WJ, Old LJ, Mulders PF. Antibody therapy in renal cell carcinoma . World J. Urol. 2008;26(2):141–146. doi: 10.1007/s00345-008-0236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howland SW, Tsuji T, Gnjatic S, Ritter G, Old LJ, Wittrup KD. Inducing efficient cross-priming using antigen-coated yeast particles . J. Immunother. 2008;31(7):607–619. doi: 10.1097/CJI.0b013e318181c87f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinschenk T, Gouttefangeas C, Schirle M et al. Integrated functional genomics approach for the design of patient-individual antitumor vaccines . Cancer Res. 2002;62(20):5818–5827. [PubMed] [Google Scholar]
- 34.Singh-Jasuja H, Emmerich NP, Rammensee HG. The Tubingen approach: identification, selection, and validation of tumor-associated HLA peptides for cancer therapy . Cancer Immunol. Immunother. 2004;53(3):187–195. doi: 10.1007/s00262-003-0480-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rammensee HG, Weinschenk T, Gouttefangeas C, Stevanovic S. Towards patient-specific tumor antigen selection for vaccination . Immunol. Rev. 2002;188:164–176. doi: 10.1034/j.1600-065x.2002.18815.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang RF, Rosenberg SA. Human tumor antigens for cancer vaccine development . Immunol. Rev. 1999;170:85–100. doi: 10.1111/j.1600-065x.1999.tb01331.x. [DOI] [PubMed] [Google Scholar]
- 37.Rammensee HG, Falk K, Rotzschke O. Peptides naturally presented by MHC class I molecules . Annu. Rev. Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 38.Kowalewski DJ, Stevanovic S. Biochemical large-scale identification of MHC class I ligands . Methods Mol. Biol. 2013;960:145–157. doi: 10.1007/978-1-62703-218-6_12. [DOI] [PubMed] [Google Scholar]
- 39.Hillen N, Stevanovic S. Contribution of mass spectrometry-based proteomics to immunology . Expert Rev. Proteomics. 2006;3(6):653–664. doi: 10.1586/14789450.3.6.653. [DOI] [PubMed] [Google Scholar]
- 40.Stevanovic S, Schild H. Quantitative aspects of T cell activation--peptide generation and editing by MHC class I molecules . Semin. Immunol. 1999;11(6):375–384. doi: 10.1006/smim.1999.0195. [DOI] [PubMed] [Google Scholar]
- 41.Nishikawa H, Kato T, Tawara I et al. Accelerated chemically induced tumor development mediated by CD4+CD25+ regulatory T cells in wild-type hosts . Proc. Natl Acad. Sci. USA. 2005;102(26):9253–9257. doi: 10.1073/pnas.0503852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutoit V, Herold-Mende C, Hilf N et al. Exploiting the glioblastoma peptidome to discover novel tumour-associated antigens for immunotherapy . Brain. 2012;135:1042–1054. doi: 10.1093/brain/aws042. Pt 4. [DOI] [PubMed] [Google Scholar]
- 43.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs . Immunogenetics. 1999;50(3)(4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 44.Castle JC, Kreiter S, Diekmann J et al. Exploiting the mutanome for tumor vaccination . Cancer Res. 2012;72(5):1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 45.Lu YC, Yao X, Li YF et al. Mutated PPP1R3B is recognized by T Cells used to treat a melanoma patient who experienced a durable complete tumor regression . J. Immunol. 2013;190(12):6034–6042. doi: 10.4049/jimmunol.1202830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinzierl AO, Lemmel C, Schoor O et al. Distorted relation between mRNA copy number and corresponding major histocompatibility complex ligand density on the cell surface . Mol. Cell Proteomics. 2007;6(1):102–113. doi: 10.1074/mcp.M600310-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Matsushita H, Vesely MD, Koboldt DC et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting . Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion . Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 49.Fridman WH, Dieu-Nosjean MC, Pages F et al. The immune microenvironment of human tumors: general significance and clinical impact . Cancer Microenviron. 2012;6(2):117–122. doi: 10.1007/s12307-012-0124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study illustrates the power of the patients' immune response to fight their cancer.
- 50.Galon J, Franck P, Marincola FM et al. Cancer classification using the Immunoscore: a worldwide task force . J Transl. Med. 2012;10(1):205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• This study uses quality analysis of immune responses within the tumor for prognosis.
- 51.Charoentong P, Angelova M, Efremova M et al. Bioinformatics for cancer immunology and immunotherapy . Cancer Immunol. Immunother. 2012;61(11):1885–1903. doi: 10.1007/s00262-012-1354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rammensee HG. Some considerations on the use of peptides and mRNA for therapeutic vaccination against cancer . Immunol. Cell Biol. 2006;84(3):290–294. doi: 10.1111/j.1440-1711.2006.01442.x. [DOI] [PubMed] [Google Scholar]
Websites
- 101.SYFPEITHI - a database of MHC ligands and peptide motifs (Ver. 1.0) www.syfpeithi.de. doi: 10.1007/s002510050595. [DOI] [PubMed]
- 102.CIMT. www.cimt.eu