Abstract
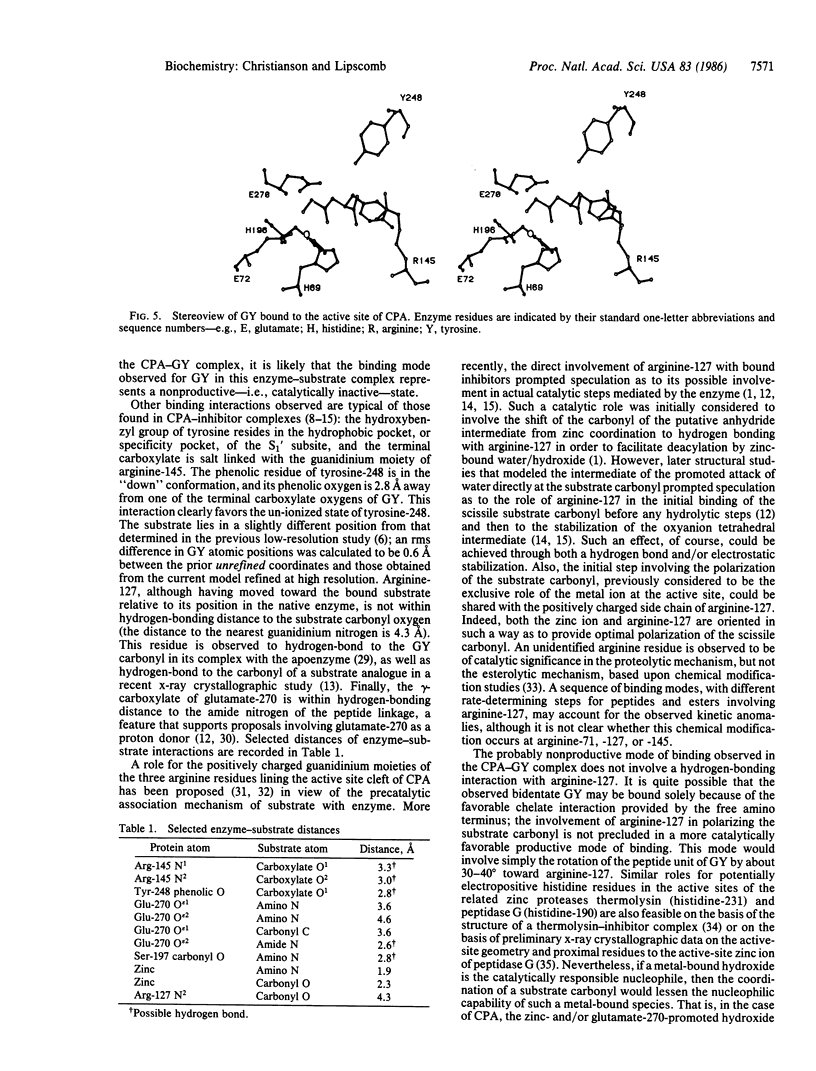
A high-resolution x-ray crystallographic investigation of the complex between carboxypeptidase A (CPA; peptidyl-L-amino-acid hydrolase, EC 3.4.17.1) and the slowly hydrolyzed substrate glycyl-L-tyrosine was done at -9 degrees C. Although this enzyme-substrate complex has been the subject of earlier crystallographic investigation, a higher resolution electron-density map of the complex with greater occupancy of the substrate was desired. All crystal chemistry (i.e., crystal soaking and x-ray data collection) was performed on a diffractometer-mounted flow cell, in which the crystal was immobilized. The x-ray data to 1.6-A resolution have yielded a well-resolved structure in which the zinc ion of the active site is five-coordinate: three enzyme residues (glutamate-72, histidine-69, and histidine-196) and the carbonyl oxygen and amino terminus of glycyl-L-tyrosine complete the coordination polyhedron of the metal. These results confirm that this substrate may be bound in a nonproductive manner, because the hydrolytically important zinc-bound water has been displaced and excluded from the active site. It is likely that all dipeptide substrates of carboxypeptidase A that carry an unprotected amino terminus are poor substrates because of such favorable bidentate coordination to the metal ion of the active site.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Christianson D. W., Lipscomb W. N. Binding of a possible transition state analogue to the active site of carboxypeptidase A. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6840–6844. doi: 10.1073/pnas.82.20.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Dive G., Joris B., Frère J. M., Ghuysen J. M. Structure of a Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase at 2.5 A resolution. Nature. 1982 Sep 30;299(5882):469–470. doi: 10.1038/299469a0. [DOI] [PubMed] [Google Scholar]
- Eklund H., Samma J. P., Wallén L., Brändén C. I., Akeson A., Jones T. A. Structure of a triclinic ternary complex of horse liver alcohol dehydrogenase at 2.9 A resolution. J Mol Biol. 1981 Mar 15;146(4):561–587. doi: 10.1016/0022-2836(81)90047-4. [DOI] [PubMed] [Google Scholar]
- Fink A. L., Petsko G. A. X-ray cryoenzymology. Adv Enzymol Relat Areas Mol Biol. 1981;52:177–246. doi: 10.1002/9780470122976.ch3. [DOI] [PubMed] [Google Scholar]
- Kuo L. C., Makinen M. W. Hydrolysis of esters by carboxypeptidase A requires a penta-coordinate metal ion. J Biol Chem. 1982 Jan 10;257(1):24–27. [PubMed] [Google Scholar]
- Lipscomb W. N., Hartsuck J. A., Reeke G. N., Jr, Quiocho F. A., Bethge P. H., Ludwig M. L., Steitz T. A., Muirhead H., Coppola J. C. The structure of carboxypeptidase A. VII. The 2.0-angstrom resolution studies of the enzyme and of its complex with glycyltyrosine, and mechanistic deductions. Brookhaven Symp Biol. 1968 Jun;21(1):24–90. [PubMed] [Google Scholar]
- Lipscomb W. N. Structure and catalysis of enzymes. Annu Rev Biochem. 1983;52:17–34. doi: 10.1146/annurev.bi.52.070183.000313. [DOI] [PubMed] [Google Scholar]
- Monzingo A. F., Matthews B. W. Binding of N-carboxymethyl dipeptide inhibitors to thermolysin determined by X-ray crystallography: a novel class of transition-state analogues for zinc peptidases. Biochemistry. 1984 Nov 20;23(24):5724–5729. doi: 10.1021/bi00319a010. [DOI] [PubMed] [Google Scholar]
- QUIOCHO F. A., RICHARDS F. M. INTERMOLECULAR CROSS LINKING OF A PROTEIN IN THE CRYSTALLINE STATE: CARBOXYPEPTIDASE-A. Proc Natl Acad Sci U S A. 1964 Sep;52:833–839. doi: 10.1073/pnas.52.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., McMurray C. H., Lipscomb W. N. Similarities between the conformation of arsanilazotyrosine 248 of carboxypeptidase A in the crystalline state and in solution. Proc Natl Acad Sci U S A. 1972 Oct;69(10):2850–2854. doi: 10.1073/pnas.69.10.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., Honzatko R. B., Lipscomb W. N. Structure of an actively exchanging complex between carboxypeptidase A and a substrate analogue. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3288–3291. doi: 10.1073/pnas.77.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., Lewis M., Honzatko R. B., Lipscomb W. N., Hardman K. D. Zinc environment and cis peptide bonds in carboxypeptidase A at 1.75-A resolution. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3408–3412. doi: 10.1073/pnas.78.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., Lewis M., Lipscomb W. N. Refined crystal structure of carboxypeptidase A at 1.54 A resolution. J Mol Biol. 1983 Aug 5;168(2):367–387. doi: 10.1016/s0022-2836(83)80024-2. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lipscomb W. N. Binding of ligands to the active site of carboxypeptidase A. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5455–5459. doi: 10.1073/pnas.78.9.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., Lipscomb W. N. Crystallographic studies on apocarboxypeptidase A and the complex with glycyl-L-tyrosine. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7151–7154. doi: 10.1073/pnas.80.23.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., Lipscomb W. N. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 A resolution. J Mol Biol. 1982 Sep 25;160(3):475–498. doi: 10.1016/0022-2836(82)90309-6. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lipscomb W. N. Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 A resolution. J Mol Biol. 1982 Sep 25;160(3):475–498. doi: 10.1016/0022-2836(82)90309-6. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lipscomb W. N. Structure of the potato inhibitor complex of carboxypeptidase A at 2.5-A resolution. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4633–4637. doi: 10.1073/pnas.77.8.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. F. Functional arginyl residues in carboxypeptidase A. Modification with butanedione. Biochemistry. 1973 Sep 25;12(20):3915–3923. doi: 10.1021/bi00744a020. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Galdes A. The metallobiochemistry of zinc enzymes. Adv Enzymol Relat Areas Mol Biol. 1984;56:283–430. doi: 10.1002/9780470123027.ch5. [DOI] [PubMed] [Google Scholar]
- Wyckoff H. W., Doscher M., Tsernoglou D., Inagami T., Johnson L. N., Hardman K. D., Allewell N. M., Kelly D. M., Richards F. M. Design of a diffractometer and flow cell system for X-ray analysis of crystalline proteins with applications to the crystal chemistry of ribonuclease-S. J Mol Biol. 1967 Aug 14;27(3):563–578. doi: 10.1016/0022-2836(67)90059-9. [DOI] [PubMed] [Google Scholar]


