Abstract
Background
Tissue banking has become a major initiative at many oncology centers. The influence of warm ex-vivo ischemia times, storage times, and biobanking protocols on RNA integrity and subsequent microarray data is not well documented.
Methods
A prospective institutional review board–approved protocol for the banking of abdominal neoplasms was initiated at Memorial Sloan-Kettering Cancer Center in 2001. Sixty-four representative pancreas cancer specimens snap-frozen at various ex-vivo procurement times (≤10 min, 11–30 min, 31–60 min, >1 h) and banked during three time periods (2001–2004, 2004–2006, 2006–2008) were processed. RNA integrity was determined by microcapillary electrophoresis using the RNA integrity number (RIN) algorithm and by results of laser-capture microdissection (LCM).
Results
Overall, 42% of human pancreas cancer specimens banked under a dedicated protocol yielded RNA with a RIN of ≥7. Limited warm ex-vivo ischemia times did not negatively impact RNA quality (percentage of tissue with total RNA with RIN of ≥7 for ≤10 min, 42%; 11–30 min, 58%; 31–60 min, 33%; >60 min, 42%), and long-term storage of banked pancreas cancer biospecimens did not negatively influence RNA quality (total RNA with RIN of ≥7 banked 2001–2004, 44%; 2004–2006, 38%; 2006–2008, 50%). RNA retrieved from pancreatic cancer samples with RIN of ≥7 subject to LCM yielded RNA suitable for further downstream applications.
Conclusions
Fresh-frozen pancreas tissue banked within a standardized research protocol yields high-quality RNA in approximately 50% of specimens and can be used for enrichment by LCM. Quality of tissues of the biobank were not adversely impacted by limited variations of warm ischemia times or different storage periods. This study shows the challenges and investments required to initiate and maintain high-quality tissue repositories.
Propelled by the emergence of new genomic technologies, particularly tissue-based RNA and DNA microarrays, the understanding and treatment of human cancer is currently undergoing a major transformation. This so-called genomic revolution aims to capture the biology and phenotype of cancer by gene expression profiles and tumor genomic alterations rather than by their shared morphologic criteria.1 These genomic data rely on the availability and quality of tissue repositories.
The lack of reproducibility of gene signatures of human breast cancer tissues raises concern as to variations in quality of tissue used for such studies.2,3 Despite ongoing research efforts by the Office of Biorepositories and Biospecimen Research, the Translational Research Working Group, the Cooperative Human Tissue Network of the National Cancer Institute, and other initiatives, few data exist on the quality of banked human tissue.4–6
Many surgical departments have developed tissue banks, some linked to their clinical database. Recently, M. D. Anderson Cancer Center reported on the development of their integrated biospecimen and multidisciplinary clinical database for pancreas cancer.7 This study documented a methodology for the collection of biospecimens and highlighted the importance of linking these repositories to carefully maintained clinicopathological databases.
The goal of the current study was to assess the quality of pancreas tissue procured in our integrated biospecimen bank. We investigated a panel of 64 fresh-frozen pancreatic adenocarcinoma and 18 intraductal papillary mucinous neoplasms (IPMN) at various time points after surgery for RNA integrity and suitability for laser-capture microdissection (LCM).
MATERIALS AND METHODS
Tissue Banking and Tissue Collection
Patients are provided information regarding tissue banking at their preoperative clinic visit, usually within 2 weeks before operation. Trained research assistants and the responsible surgeon provide detailed explanations of the process of serum and tissue collections, and patients are given the option to consent for general research or research, which entailed molecular and genetic analysis of the procured biomaterial. The Internal Review Board (IRB) of Memorial Sloan-Kettering Cancer Center (MSKCC) and the MSKCC Ethics Committee gave approval for the banking and use of the accrued specimens (MSKCC IRB protocol 00-032).
Patient serum, plasma, and buffy coat were obtained from blood samples procured during preadmission testing. All patients undergoing pancreatic resection for all histological diagnoses are eligible.
Banking Process
All patients consenting to MSKCC IRB protocol 00-032 are identified by the tissue procurement service (TPS) the day before operation. At the time of resection, the specimens are oriented by the attending surgeon and are brought immediately in prelabeled containers to the pathology facility, where time of arrival is recorded and the specimen is immediately processed by pathology attending or assistant personnel. Because of the close proximity of the two facilities, the transport time is generally <2 min.
Banking Process
The specimen is oriented, measured, and described. The external surface is painted with tissue-marking dyes, and the pancreatic duct and common bile duct are cannulated where appropriate. The gland is then opened along these ducts and processed as previously described.8 The cut surfaces are inspected and areas of tumor sought. An area of 0.8 × 0.8 cm suggesting the presence of tumor is excised with a scalpel blade, which is then divided into two similar halves. A similar area from uninvolved pancreatic parenchyma is sampled as normal control. The specimen then enters the routine histopathological processing.
Immediately after removal, the tumor and normal tissue aliquots are coated in OCT compound (Sukura Finetek, Torrance, CA) and snap-frozen in liquid nitrogen. The frozen tissue is stored in a labeled CryoVial and stored at −80°C.
Biobank Collection Times
All biospecimens accrued under IRB protocol 00-032 have their procurement times recorded. Time to procurement is defined by time of arrival of the specimen in TPS to cryopreservation (time of snap freezing). Time of specimen handoff as recorded by the nursing log-in time of specimen receipt to time of arrival in TPS was approximately 2 min. Queries of the pancreas database at MSKCC TPS are performed by bank’s informatics staff. For this study, data were sorted into four groups: procurement times of ≤10 min, 11–30 min, 31–60 min, and >60 min. Data were also grouped according to procurement periods 2001–2004, 2004–2006, and 2006–2008. These data were graphed by Microsoft Excel. Pearson χ2 tests were performed to compare the proportion of pancreas tumor specimens with high-quality RNA (RIN of ≥7) banked within different ex-vivo times and during different procurement periods.
RNA Quality
Sixty-four pancreaticoduodenectomy specimens with ductal adenocarcinoma and 18 pancreaticoduodenectomy specimens with IPMN were randomly selected. For histopathologic assessment, 5-µm sections of frozen tissues were made at −20°C. OCT blocks were quickly refrozen at −80°C. A histopathologist with experience in pancreas pathology (U.B.) reviewed all hematoxylin and eosin stained sections and selected representative tumor areas. Corresponding blocks were rethawed, and tumor areas marked on hematoxylin and eosin–stained samples were macrodissected. Each specimen was homogenized in Tri-Zol Reagent (Invitrogen, Carlsbad, CA), and total RNA was extracted according to manufacturers’ instructions. The isolated RNA was analyzed on the Agilent 2001 Bioanalyzer. To assess RNA integrity, RNA integrity numbers (RINs) are calculated by Agilent software. By means of this tool, sample integrity is no longer determined by the ratio of the ribosomal bands but by the entire electrophoretic trace of the RNA sample (total of 10 features), including the presence or absence of degradation products.
Laser-Capture Microdissection
Pancreatic tumor tissue is often surrounded by stromal and inflammatory tissue that reduces cellular homogeneity by 20% to 80%. To minimize the impact of stromal and inflammatory tissue on analysis of pancreatic cancer, tissue cell enrichment by laser microdissection is necessary. The Arcturus VeritasTX LCM System was used to minimize contamination of tumor tissue with stromal cells, and to achieve pancreatic cancer tissue homogeneity of >80%. The system combines ultraviolet laser cutting and laser capture using an infrared laser source, an advantage especially valuable in tissue with high ribonuclease activity like pancreas. Eight cases of pancreas cancer in which initial quality control has demonstrated RNA of high quality with RINs of ≥7 were subject to LCM. Only specimens with total RNAs and RINs of ≥7 were chosen for LCM. Frozen sections (8 µm) were obtained and immediately fixed in cold 95% ethanol. Sections were briefly (5 to 10 seconds) stained with hematoxylin with Arcturus HistoGene Staining Solution and were dehydrated in 100% ethanol followed by xylene (described in the Arcturus HistoGene LCM Frozen Section Staining Kit protocol). Five thousand to 10,000 cells from pancreatic adenocarcinoma were obtained by LCM with the Veritas Laser microdissection system (Arcturus). Digital images of tissue before and after LCM were captured. Each session of LCM was completed within 20 min. RNA was isolated with the Arcturus PicoPure RNA Isolation Kit following the manufacturer’s instructions. DNAse digestion was performed with the Qiagen RNase-Free DNAse Set (Qiagen, Valencia, CA). RNA quality was assessed on the Agilent 2001 Bioanalyzer.
RESULTS
A total of 561 pancreatic tissue specimens have been banked since the protocol opened. Ductal adenocarcinoma was the most common diagnosis (n = 404), followed by IPMN without invasive cancer (n = 44) and serous cystadenoma (n = 35). Pancreaticoduodenectomy specimens comprised most cases (n = 415), followed by distal pancreatectomy with or without splenectomy (n = 131). Ex-vivo times were divided into time periods of ≤10 min, 11–30 min, 31–60 min, and >60 min, and banking periods were divided by the years 2001–2004, 2004–2006, and 2006–2008.
Impact of Procurement Period on Biobank Collection Times
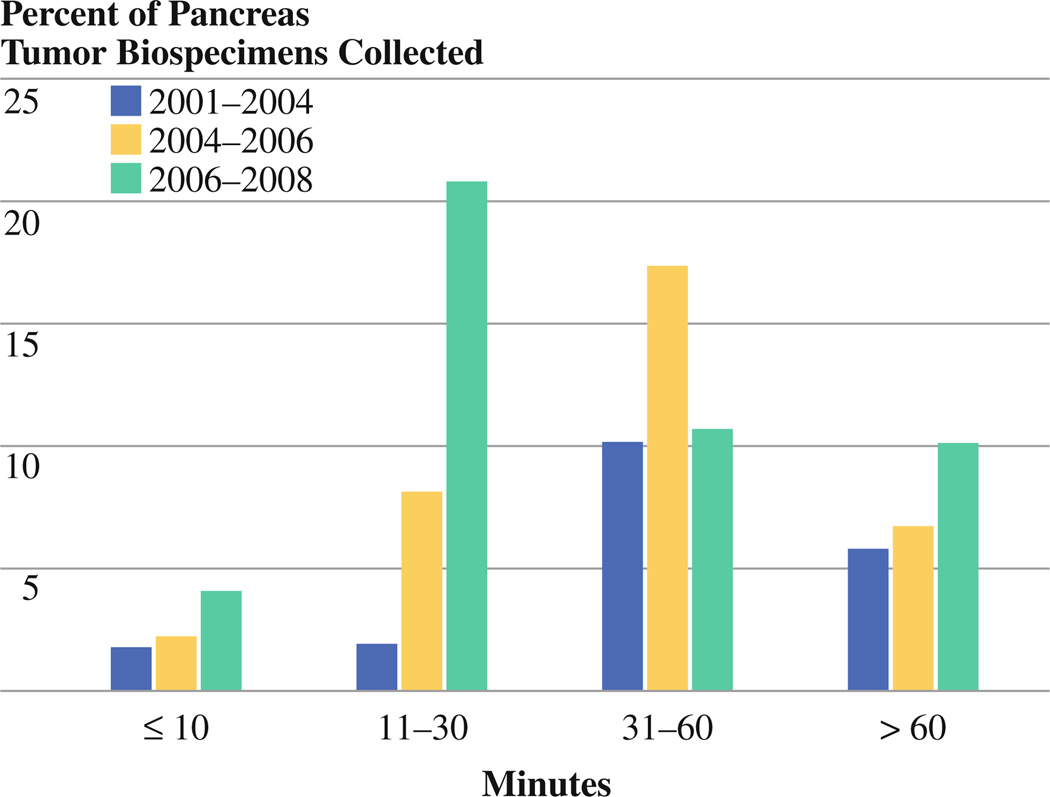
The proportion of pancreas tumor biospecimens collected within each of the four time groups (≤10, 11–30, 31–60, and >60 min) stratified by time period (2001–2004, 2004–2006, and 2006–2008) is shown in Fig. 1. Whereas only 3.7% (21 of 568) of pancreatic biospecimens collected during 2001–2004 were cryopreserved within 30 min, this proportion increased to 10.4% (59 of 568) and 24.8% (142 of 568) during the 2004–2006 and 2006–2008 periods. This increase in biospecimens banked with shorter procurement times was accompanied by a 71% increase in the annual procurement rate (from 37 to 130 specimens per year) during the study time.
FIG. 1.
Relation between ex-vivo procurement times (in minutes, from excision to cryopreservation) and time periods 2006–2008, 2004–2006, and 2001–2004
Relationship Between Ex-Vivo Ischemia Time and RNA Integrity
To assess the degree of degradation as a function of procurement time, electropherograms from isolated total RNAs from all 64 adenocarcinoma and 18 IPMN were obtained. As the standard for RNA integrity, the RIN was calculated for all specimens. Figure 2 shows 20 representative electrophoreses with their corresponding RINs.
FIG. 2.
Integrity of total RNA isolated from banked fresh-frozen pancreatic cancer samples. Representative electropherograms (Agilent 2100 Bioanalyzer) show RNA integrity measured by RNA integrity number (RIN) of 20 representative samples (bottom)
There was no marked decline of RNA quality in samples with prolonged procurement times (Fig. 3). A standard of RIN of ≥7 was set as a cutoff for RNA quality sufficient for RNA microarray analyses and RNA expression arrays. A RIN of ≥7 was present in 42% of analyzed pancreatic cancer samples. Proportion of samples with high-quality RNA within the different time periods did not statistically differ between cases with warm ischemia times of >60 min and shorter procurement times (P = 0.81; Fig. 3). The period of procurement did not negatively impact RNA integrity. No difference in the proportion of cases with RNA suitable for microarray analyses was identified between recently procured cases (2006–2008) and cases procured 5 years before (P = 0.83; Fig. 4). The proportion of IPMN specimens with a RIN of ≥7 was lower than that for pancreatic adenocarcinoma (6 of 18). No impact of warm-ischemia time (<1 h) or length of storage on RNA quality of IPMN tissues was observed (data not shown).
FIG. 3.
Percentage of samples suitable for gene expression arrays (RNA integrity number [RIN] of ≥7) at different ex-vivo procurement times (time from surgical removal to cryopreservation) for a ≤10 min, b 11–30 min, c 31–60 min, and d >1 h. For each group, total RNA from 12 fresh-frozen tissue blocks (total of 48) was extracted and RIN determined after electropherogram and analysis on Agilent 2100 Bioanalyzer
FIG. 4.
Impact of procurement period on RNA degradation. a 2001– 2004. b 2004–2006. c 2006–2008. Percentage of cases with RNA integrity numbers (RINs) of ≥7 for each group is shown
Impact of LCM on RNA Quality of Procured Pancreas Tissue
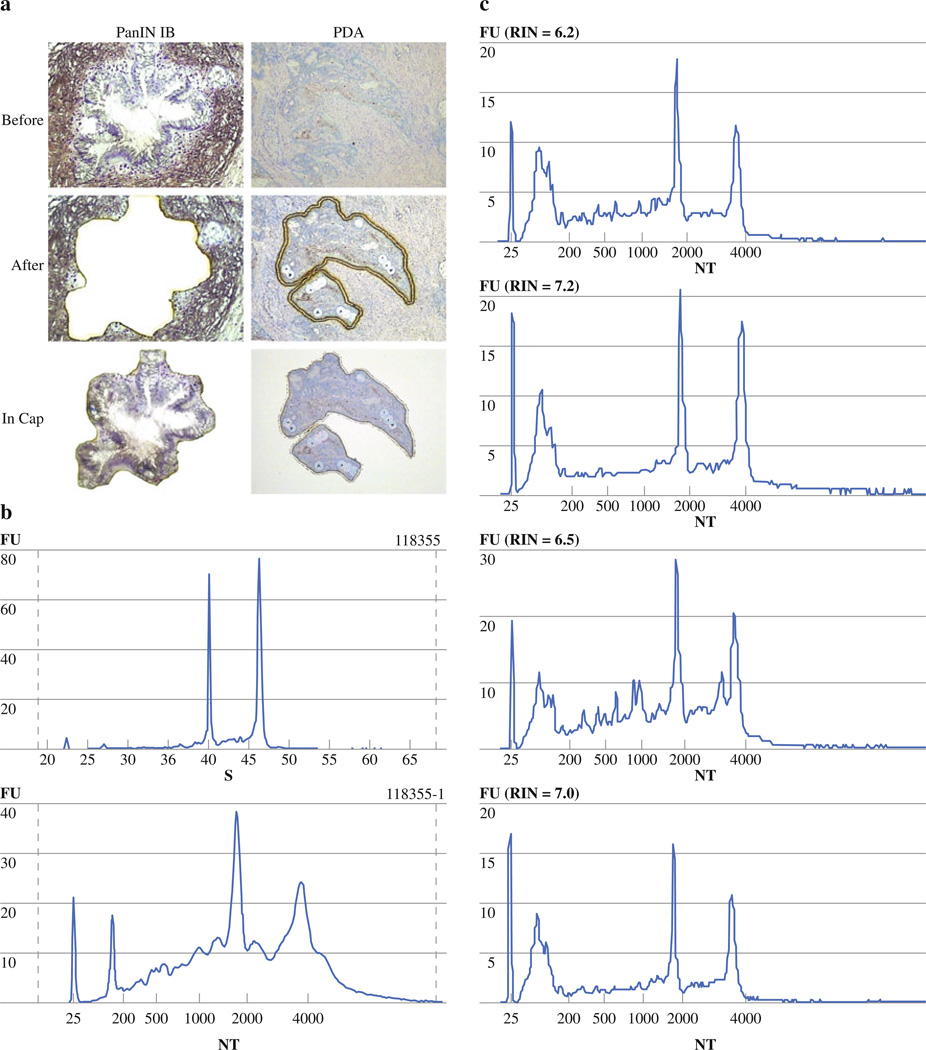
Figure 5a shows successful LCM of low-grade PanIN lesion (PanIN-1B) and invasive cancer. Cellular homogeneity was >80%. Figure 5b shows the electropherograms of four representative samples of RNA extracted from LCM invasive pancreatic cancer samples. RNA isolated from 5,000 to 10,000 microdissected pancreatic cancer cells yielded RINs between 6 and 7. On an average, slide preparation, staining, and laser capture were completed within 20 to 30 min. RINs dropped by one to two points as a result of the process of microdissection but remained >6 (Fig. 5b). A representative example of pre- and post-LCM electropherograms with the corresponding RINs is shown in Fig. 5c.
FIG. 5.
Efficiency of laser-capture microdissection (LCM) using procured fresh-frozen pancreatic tissue. a Representative samples of LCM of invasive, ductal adenocarcinoma of the pancreas and PanIN IB lesion using dual Arcturus system. b Electropherograms of total RNA extracted from laser-capture microdissected pancreas tissue (5,000–10,000 cells) of four representative sample including their RNA integrity numbers (RINs) are shown. c Pre- and post-LCM electropherograms of representative pancreas cancer specimen shows RINs of 9 and 7.2
DISCUSSION
The identification of molecular cancer subtypes and the development of prognostic and predictive gene signatures through microarray gene expression profiling has become a powerful tool to capture the biological heterogeneity of cancer.9,10 Microarray gene expression data in the form of gene signatures are particularly needed in pancreas cancer, where improved predictive and prognostic tools compared to the current staging system would allow better risk stratification and allocation of limited treatment options.
The quality of gene expression data from microarray analysis is inextricably linked to (1) the quality of biospecimens and (2) the degree of degradation of RNA extracted from these specimens.4,11–13 For operatively derived specimens, certain variables of specimen quality include in-vivo ischemia time, degree of surgical manipulation, or anesthesia, which are fixed during the process of specimen retrieval. Few studies have emerged in the literature that examined the effects of preanalytic biospecimen collection variables on analysis of RNA quality and gene expression analysis.14–20 Previous gene expression studies in both invasive adenocarcinoma and precursor lesions (PanIN and IPMN) did not indicate how to control for these variables or provide detailed data on RNA integrity and used biospecimens.21–24
We hypothesized that tissues procured under a prospective and IRB-approved research protocol at a tertiary referral center for pancreatic diseases would represent a valid source of good-quality biospecimens and RNA. We tested this assumption by evaluating 64 randomly selected, fresh-frozen human pancreatic adenocarcinoma and 18 IPMN tissue samples from pancreaticoduodenectomy specimens procured after various ex-vivo ischemic times and during different time periods. Quality of banked tissue was assessed by the absence of RNA degradation as well as the ability to retrieve nondegraded RNA from specimens subject to LCM. RNA integrity for each tissue was assessed by determination of the RIN, which integrates 10 features from RNA electropherograms and which has been shown to be superior to previous standards of RNA quality like 18S to 28S rRNA ratio in the past.11–13,25,26
Our results indicate that limited ex-vivo ischemia times of fresh-frozen pancreas cancer tissue procured within a dedicated TPS have no marked adverse impact on RNA quality, and that overall, slightly less than half of banked specimens under the specified protocol, regardless of the procurement period, will yield high-quality RNA. These results are consistent with previously published studies on this subject. In the largest analysis to date on molecular quality of human tissues, an experience from the Cooperative Human Tissue Network, Jewell et al. examined RNA and DNA quality of various epithelial cancer specimens and found no marked decline in RNA quality within the first 5 hours after surgical excision.14 Overall, 60% of stored human tissue had good RNA quality as judged by routine gel electrophoresis and Northern blot testing. In breast cancer, Ohashi et al. found no loss of RNA integrity in normal breast tissue for up to 3 hours after surgical removal, a finding reproduced by the study of Barnes et al., which showed no RNA degradation as assessed by Northern blot testing for up to 24 hours after specimen removal.18,19 Similar findings exist from the analysis of procured lung tissue, tonsil, and colon tissue.14,27 On the other hand, recently presented research on both the influence of in-vivo and warm ex-vivo ischemia time in prostate cancer specimens on gene expression made different observations.15–17,20 Two recently conducted studies demonstrated that marked gene transcript level alterations occur early during prostatectomy.17,20 These changes were most marked in known hypoxia- and stress-response genes, as demonstrated by expression changes in JUNB, EGR1, and IER2, but comprised <5% of the overall examined genes.
The current study highlights the heavy investments of time and resources necessary to yield suitable RNA from pancreas tissues in approximately 50% of cases. Protocol development, consent in clinic, quick transport to TPS, personnel for pathological dissection, clerical database support, and monthly doctor review at pancreas staging conferences are only some examples of the staffing investments necessary to maintain such a service.
One important limitation to this study is that we did not subject the isolated RNA to gene expression microarray analysis but used the calculated RINs as surrogate markers for high-quality RNA. Although the exact cutoff for the optimal RIN for microarray analysis has been shown to depend on the chosen individual platform as well as the investigators specifications for false inclusion and false exclusion, the RIN has consistently been shown to detect reliable RNA and avoid misclassification as reported for the 28S/18S ratio or manual methods.11–13,25,26 Strand et al., using a cutoff of a RIN value of 6, showed that microarray profiles from breast tumor samples change considerably when comparing samples with RIN values of <6 to samples with RIN values of ≥6.12 Our cutoff of RIN of ≥7 is slightly lower than the one of 8 recommended by Fleige and Pfaffl, which describe samples with RIN of ≥8 as perfect for downstream application, taking the very high ribonuclease contents in human pancreatic tissue into account.28 Although the RIN required for biomarker development via reverse transcriptase–polymerase chain reaction might be lower than that for full-scale genomic microarray or sequencing analyses, this value is not known for many of the novel emerging genomic techniques such as serial analysis of gene expression, tag sequencing, or micro-RNA analysis. Because intact biomolecules will ensure maximum benefit from these novel applications, biobanking protocols should strive for the best possible conversation of these molecules.
As a second quality control measurement, we chose LCM as a further test for tissue integrity. Most experiments that aim to develop biomarkers or gene signatures specific for pancreatic adenocarcinoma will require on the one hand sufficiently homogenous cell populations representing the histopathological entity of interest void of the large number of contaminating inflammatory and stromal cells. On the other hand, LCM will aid in the elucidation of gene expression perturbations of the epithelium and surrounding stroma, and biomarkers derived from genetic alterations in the tumor microenvironment, either induced by tumor progression or as a result of endogenous genetic polymorphisms of the host, might be applied to clinical decision making in the near future as they have recently been shown to harbor predictive value in cancer.29
Several strengths of the study should be noted: our sample size of 64 fresh-frozen adenocarcinomas and 18 IPMN specimens represents one of the largest tissue quality control series. Second, our study cohort was standardized and homogeneous because it was limited to two histopathological diagnoses and one operative procedure. Third, although the procurement process was conducted within a research protocol and did not change during the study period, we examined the influence of different procurement periods on RNA quality. Barnes et al. had observed in their study a difference in RNA quality obtained from tissues procured in the pre-2000 era, an observation we did not confirm.19 The authors attributed this finding to the introduction of stringent standard operating procedures into biobanking after 2000. However, despite such rigorous procurement protocols, including the one presented in this study, up to 50% of banked specimens might not yield high-quality RNA. We attribute this dropout rate to factors that are beyond the control of surgeons, pathologists, TPS personnel, and institutional investments like surgical trauma, in-vivo hypoxia, or other types of cellular stress. The cause for the observed decreased RNA quality extracted from IPMN specimens remains to be determined: although previous studies suggest an association between increased mucin tissue content and inferior RNA quality, increased degradation might be solely due to the substantially smaller amounts of RNA yielded from these cystic pancreatic neoplasms.30
This study evaluated the quality of banked tissue from our prospective pancreas tissue bank. Fresh-frozen pancreas tissue procured within a standardized research protocol yielded high-quality RNA in approximately 50% of the cases, and the samples can be used with good results for further cellular applications like LCM. Quality of tissues of the biobank as assessed by RNA integrity was not adversely impacted by limited variations of warm ischemia times or different time periods of tissue procurement. However, the presented methods and data should be interpreted as an attempt of further standardization of tissue sampling and biobanking, and it must be cautioned that quality control of RNA integrity at the tissue level does not substitute for internal controls in the actual microarray experiments. High-quality gene expression data may be derived from high-quality and accurate tissue repositories. The surgeon’s role in developing these repositories is critical; however, the financial and time requirements necessary should not be underestimated.
REFERENCES
- 1.Garman KS, Nevins JR, Potti A. Genomic strategies for personalized cancer therapy. Hum Mol Genet. 2007;16(Spec No. 2):R226–R232. doi: 10.1093/hmg/ddm184. [DOI] [PubMed] [Google Scholar]
- 2.Acharya CR, Hsu DS, Anders CK, et al. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. JAMA. 2008;299:1574–1587. doi: 10.1001/jama.299.13.1574. [DOI] [PubMed] [Google Scholar]
- 3.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 4.Compton C. Getting to personalized cancer medicine: taking out the garbage. Cancer. 2007;110:1641–1643. doi: 10.1002/cncr.22966. [DOI] [PubMed] [Google Scholar]
- 5.National Cancer Institute. NCI best practices for biospecimen resources. 2007 http://www.biospecimens.cancer.gov/practices/.
- 6.Srivastava S, Gray JW, Reid BJ, et al. Translational Research Working Group. Translational Research Working Group developmental pathway for biospecimen-based assessment modalities. Clin Cancer Res. 2008;14:5672–5677. doi: 10.1158/1078-0432.CCR-08-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang RF, Wang H, Lara A, et al. Development of an integrated biospecimen bank and multidisciplinary clinical database for pancreatic cancer. Ann Surg Oncol. 2008;15:1356–1366. doi: 10.1245/s10434-008-9833-1. [DOI] [PubMed] [Google Scholar]
- 8.Albores-Saavedra J, Heffess C, Hruban RH, Klimstra D, Longnecker D. Recommendations for the reporting of pancreatic specimens containing malignant tumors. The Association of Directors of Anatomic and Surgical Pathology. Am J Clin Pathol. 1999;111:304–307. doi: 10.1093/ajcp/111.3.304. [DOI] [PubMed] [Google Scholar]
- 9.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 10.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 11.Copois V, Bibeau F, Bascoul-Mollevi C, et al. Impact of RNA degradation on gene expression profiles: assessment of different methods to reliably determine RNA quality. J Biotechnol. 2007;127:549–559. doi: 10.1016/j.jbiotec.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 12.Strand C, Enell J, Hedenfalk I, Fernö M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis—a comparison of three evaluation methods using microcapillary electrophoresis traces. BMC Mol Biol. 2007;8:38. doi: 10.1186/1471-2199-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weis S, Llenos IC, Dulay JR, et al. Quality control for microarray analysis of human brain samples: The impact of postmortem factors, RNA characteristics, and histopathology. J Neurosci Methods. 2007;165:198–209. doi: 10.1016/j.jneumeth.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Jewell SD, Srinivasan M, McCart LM, et al. Analysis of the molecular quality of human tissues: an experience from the Cooperative Human Tissue Network. Am J Clin Pathol. 2002;118:733–741. doi: 10.1309/VPQL-RT21-X7YH-XDXK. [DOI] [PubMed] [Google Scholar]
- 15.Dash A, Maine IP, Varambally S, et al. Changes in differential gene expression because of warm ischemia time of radical prostatectomy specimens. Am J Pathol. 2002;161:1743–1748. doi: 10.1016/S0002-9440(10)64451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spruessel A, Steimann G, Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004;36:1030–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 17.Lin DW, Coleman IM, Hawley S, et al. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006;24:3763–3770. doi: 10.1200/JCO.2005.05.1458. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi Y, Creek KE, Pirisi L, Kalus R, Young SR. RNA degradation in human breast tissue after surgical removal: a timecourse study. Exp Mol Pathol. 2004;77:98–103. doi: 10.1016/j.yexmp.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Barnes RO, Parisien M, Murphy LC, Watson PH. Influence of evolution in tumor biobanking on the interpretation of translational research. Cancer Epidemiol Biomarkers Prev. 2008;17:3344–3350. doi: 10.1158/1055-9965.EPI-08-0622. [DOI] [PubMed] [Google Scholar]
- 20.Schlomm T, Näkel E, Lübke A, et al. Marked gene transcript level alterations occur early during radical prostatectomy. Eur Urol. 2008;53:333–344. doi: 10.1016/j.eururo.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 21.Iacobuzio-Donahue CA, Maitra A, Shen-Ong GL, et al. Discovery of novel tumor markers of pancreatic cancer using global gene expression technology. Am J Pathol. 2002;160:1239–1249. doi: 10.1016/S0002-9440(10)62551-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato N, Fukushima N, Maitra A, et al. Gene expression profiling identifies genes associated with invasive intraductal papillary mucinous neoplasms of the pancreas. Am J Pathol. 2004;164:903–914. doi: 10.1016/S0002-9440(10)63178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchholz M, Braun M, Heidenblut A, et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24:6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 24.Prasad NB, Biankin AV, Fukushima N, et al. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–1626. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 25.Imbeaud S, Graudens E, Boulanger V, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:56. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroeder A, Mueller O, Stocker S, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micke P, Ohshima M, Tahmasebpoor S, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86:202–211. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 28.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–139. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Blansfield JA, Caragacianu D, Alexander HR, 3rd, et al. Combining agents that target the tumor microenvironment improves the efficacy of anticancer therapy. Clin Cancer Res. 2008;14:270–280. doi: 10.1158/1078-0432.CCR-07-1562. [DOI] [PubMed] [Google Scholar]
- 30.Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J. 2005;19:1356–1358. doi: 10.1096/fj.04-3552fje. [DOI] [PubMed] [Google Scholar]