Abstract
Background
The prognosis of MBM is variable with prolonged survival in a subset. It is unclear whether MBMs differ from extracranial metastases (EcM) and primary melanomas (PrM).
Methods
To study the biology of MBM we performed histopathologic analysis of tumor blocks from patients' craniotomy samples and whole genome expression profiling (WGEP) with confirmatory immunohistochemistry (IHC).
Results
Mononuclear infiltrate and low intratumoral hemorrhage were associated with prolonged overall survival (OS). Pathway analysis of WGEP data from 29 such craniotomy tumor blocks demonstrated that several immune-related BioCarta gene sets were associated with prolonged OS. WGEP analysis of MBM in comparison with same-patient EcM and PrM showed that MBM and EcM were similar—but both differ significantly from PrM. IHC analysis revealed that peritumoral CD3+ and CD8+ cells were associated with prolonged OS.
Conclusions
MBMs are more similar to EcM compared to PrM. Immune infiltrate is a favorable prognostic factor for MBM.
Keywords: Melanoma, Brain Metastases, Craniotomy Immune Infiltrate, Gene Expression Profiling
Introduction
Metastases to the brain are the most frequent intracranial tumors in adults.(1) Melanoma is the most frequent solid cancer to metastasize to the brain.(2) B-Raf inhibitors and ipilimumab are active against MBM.(3, 4) Mutations in B-Raf and N-Ras proteins are associated with higher incidence of MBM.(5) No association between B-RafV600E mutations in MBM and OS was found.(6) MBM exhibit higher levels of phosphoinositide 3-kinase pathway activation compared to EcM.(7, 8)
The brain microenvironment differs from that of extracranial sites. It lacks lymphatics and contains glial cells that may influence tumor growth.(9) Blood vessels in brain tumors differ from normal brain vessels.(10) Immune cells have also been described in a handful of metastatic solid tumors to the brain, although their prognostic significance is unknown.(11)
To gain insight into the biology of MBM we performed histopathologic analysis followed by WGEP and confirmatory IHC and in situ hybridization (ISH) analysis of craniotomy tumor specimens. Our study proposes new prognostic factors for MBM, and may explain clinical efficacy of immunotherapies in patients with MBM.
Patients and Methods
Patients and Tumors
Under IRB-approved protocols, de-identified cases from patients who underwent craniotomy for MBM at the University of Pittsburgh Medical Center (UPMC) were obtained by performing a CoPATH pathology search followed by retrieval of formalin-fixed paraffin-embedded (FFPE) tumor blocks from craniotomy, EcM, and PrM specimens.
Histopathologic Assessment of MBMs
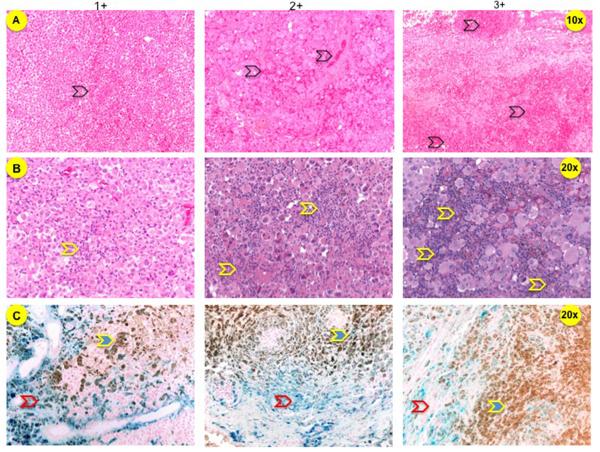
Hematoxylin and eosin (H&E)-stained sections were reviewed by a neuropathologist (RH) who was blinded to patient history. Immune infiltrate, hemorrhage, gliosis, pigmentation, and necrosis, were semi-quantitatively scored on a 0–3+ scale with 0–1+ being absent/low and 2–3+ being high. Tumor hemorrhage was defined by the presence of fresh hemorrhage, hematoidin-laden macrophages, organized blood clot, and ruptured vessels or hemorrhage adjacent to necrotic areas. Hemorrhage was quantitated as low with 0–2 foci of hematoidin, fresh blood, or organized clot; high with blood occupying >1/3 of the specimen (Figure 1A). Immune infiltrate was quantitated by the presence of mononuclear cells around blood vessels and/or within the tumor parenchyma. Low infiltrate was defined as 0–2 perivascular infiltrates and high infiltrate was defined as >2 perivascular and/or any infiltrates within the tumor parenchyma (Figure 1B). Melanin was assessed using a combination of H&E and Gomori's modified iron stain (Figure 1C). Gliosis was defined as the presence of reactive astrocytes only near the tumor. Necrosis was estimated as a percentage of necrotic tumor.
Figure 1. Tumor sections of MBMs.
A. H&E-stained sections that were scored high and low hemorrhage (black open arrows). B. H&E-stained sections that show high and low immune infiltrate (yellow open arrows). C. Sections were stained with potassium ferrocyanide and counterstained with nuclear fast red (Kernechtrot), if adjacent H&E-stained sections showed pigmentation. Hemosiderin is stained blue (red open arrows) and melanin remains brown (yellow arrows).
WGEP Analysis
Using a blade under the guidance of H&E-staining of every 10th adjacent 5-micron section, tissues corresponding to non-necrotic tumor devoid of immune infiltrate, hemorrhage, and glial cells were microdissected. Deparaffinization of tissue pellets, RNA extraction, purification, and incubation with the Human Ref8 v3 BeadChips (Illumina Inc., San Diego, CA) followed by scanning on the Illumina BeadStation GX were performed in the UPCI Cancer Biomarkers Illumina Platform Facility.(12)
Sources of Antibodies
The following primary antibodies were used: CD3 (DAKO, Carpinteria, CA), CD4 (Vector Laboratories, Burlingame, CA), CD8 (DAKO), CD14 (Vector), CD19 (Leica Microsystems, Buffalo Grove, IL), Forkhead box P3 (FoxP3, eBiosciences, San Diego, CA), CD247 (Sigma-Aldrich, St. Louis, MO), transforming growth factor beta (TGFβ, Abcam, Cambridge, MA), and glial fibrillary acidic protein (GFAP, DAKO). The following antibodies were generated in Dr. Ferrone's lab: HLA class I (HC-10/HC-A2 clones), HLA class II (LGII-612.14), and tapasin (TO-3).
IHC, ISH and Scoring Definitions
For IHC, FFPE tumor sections were probed with antibodies and stained with Vulcan Fast Red (Biocare Medical, Concord, CA). CD3, CD4, CD8, CD14, CD19, and CD247-positive mononuclear cells were scored separately (RH and SM) for peritumoral and intratumoral compartments, as previously described (Figure 2).(13) Expression of TGFβ, HLA class I/II, and tapasin by melanoma cells were assessed using the H-score whose median was used as the cutoff value between high versus low expression. ISHs were performed (BFJ and TAR) using human-specific sequences for detection of chemokine (CXCL13, CCL19, CCL21) mRNAs. Autoradiographic exposure times of 35S-labeled riboprobes were 7–10 days.(14)
Figure 2. IHC staining of brain tumor sections.
Tumor sections were stained with Vulcan Fast Red and counterstained with hematoxylin. A. CD3 stain. Representative pictures (20× magnification) show low and high degrees of intratumoral (upper row) or peritumoral-perivascular infiltrate (lower row). B. GFAP stain. GFAP+ cells (blue arrows) are not only present in tumor periphery (left image) but also within the tumor (middle and right image).
Statistical Analysis
Cox proportional-hazards modeling was used to identify clinicopathologic variables associated with OS defined as time-to-death (TTD) from first craniotomy, by fitting a univariate regression model for each variable. Kaplan–Meier (KM) survival analysis using the log-rank significance test was also performed for dichotomous variables. Patients who were still alive between the first craniotomy and March 12, 2012 were censored at the date of last follow-up. Curves were constructed using IBM SPSS Statistics release 19.0.0 software (IBM Company, Armonk, NY). Variables that were significantly associated with OS in univariate analysis were subjected to multivariate Cox regression analysis.
Bioinformatics Analysis
Bioinformatics analysis of WGEP data was performed using the Biometric Research Branch-Array (BRB) Tools software (http://linus.nci.nih.gov/BRB-ArrayTools.html). Data were quantile-normalized using the lumi R package. Probe sets were excluded from further analysis if <20% of gene expression data values had ≥1·5-fold change in either direction of probe set's median value and the percentage of data missing exceeded 50%. To assess whether specific cellular pathways were associated with OS, we performed survival pathway analysis on the MBM dataset using the Survival Gene Set analysis (GSA) tool. The Efron-Tibshirani maxmean test was applied to identify gene sets at a p=0·05 significance level.
Hierarchical clustering analysis was used to assess the degree of association between PrM, EcM, and MBM using either all genes that passed the filtering criteria or highly discriminant genes. Highly discriminating genes were selected as differentially expressed between two same-patient different sites (MBM-EcM, MBM-PrM, and EcM-PrM) using paired t-test. Analysis of variance (ANOVA) using all genes that passed the filtering criteria was used to assess whether MBM are significantly different from EcM and/or PrM. Enrichment analysis of differentially expressed probe sets to determine their biological annotation to specific cellular pathways was performed for the same patient EcM-MBM tumor samples using DAVID database (http://david/abcc.ncifcrf.gov). Enrichment p-value was set to 0.05 after Benjamini-Hochberg multiple comparison correction testing.
Results
Clinical Characteristics of Patients with MBM
115 patients underwent craniotomy at UPMC between November 1995 and July 2011 (Table 1). 27% and 30% of patients originally presented either with unknown primary(15) or thin (AJCC stage I) melanoma, respectively. No patients had received vemurafenib or radiation therapy prior to craniotomy and only 2 out of 9 patients with B-RafV600 mutation received vemurafenib following craniotomy.
Table 1.
Clinicopathologic characteristics of patients who underwent craniotomy.
| Age (Median, range) | 51 (14–83) |
| Sex | |
| Male | 74 |
| Female | 41 |
| Melanoma Origin | |
| Cutaneous | 79 |
| Non-cutaneous | 2 |
| Unknown primary (All Criteria) | 22 |
| Unknown primary (Partial Criteria) | 8 |
| Unknown | 4 |
| B-Raf Mutation Status | |
| Known | 19 |
| B-RafV600E | 6 |
| B-RafV600K | 3 |
| B-Raf wild type | 10 |
| Unknown | 96 |
| Stage (AJCC 2009) | |
| I | 34 |
| II | 23 |
| III | 11 |
| IV | 30 |
| Unknown | 17 |
Patients were considered to have `partial' criteria for unknown primary(15) if they had no knowledge of excision of a pigmented lesion.
High Immune Infiltrate and Low Hemorrhage in MBM are Associated with Prolonged OS
Due to tumor block availability (n=106), absence of viable tumor cells (n=3), or cytology specimens (n=2) our final analysis was based on 101 of cases. As shown in Table 2 immune infiltrate (p=0.006), hemorrhage (p=0.04), recursive partitioning analysis (RPA) class (p<0.0001), ECOG performance status (p=0.024), and local therapy after craniotomy (p=0.019) were significantly associated with OS. No significant correlation between B-Raf mutation status and immune infiltrate or hemorrhage was noted (Pearson's chi-square test p=0.57 and p=0.49, respectively). Figure 3 shows the Kaplan-Meier OS curves partitioned by immune infiltrate and hemorrhage. A significant interaction between the `immune infiltrate' and `hemorrhage' variables was noted (Cox regression test, p=0.0031). In particular, patients with high immune infiltrate and low/absent hemorrhage had prolonged OS compared to all remaining patients (543 vs. 164 days, log-rank p<0.001). Cox multiple regression analysis showed that immune infiltrate (p=0.008), ECOG performance status (p=0.043), RPA (p<0.0001), and local therapy (p=0.0005) remained significant predictors of OS.
Table 2.
Clinicopathologic factors in relation to OS in patients with MBMs.
| Variable | No of Pts | Median Survival (days) | Log-Rank P | KM Estimator HR (95% CI) |
|---|---|---|---|---|
|
| ||||
| Age at Craniotomy | ||||
| <65 | 80 | 189 | 0.33 | 1.30 (0.75–2.15) |
| >65 | 21 | 137 | ||
|
| ||||
| Sex | ||||
| Male | 63 | 154 | 0.10 | 1.44 (0.94–2.26) |
| Female | 38 | 228 | ||
|
| ||||
| ECOG Performance Status | ||||
| 0 | 12 | 481 | ||
| 1 | 61 | 191 | 0.024 | 1.71 (1.05–2.70) |
| 2 | 24 | 142 | ||
| 3 | 4 | 8 | ||
|
| ||||
| Extracranial Disease | ||||
| Absent | 48 | 143 | 0.36 | 0.82 (0.54–1.26) |
| Present | 51 | 191 | ||
| Unknown | 2 | 143 | ||
|
| ||||
| RPA | ||||
| 1 | 26 | 475 | ||
| >1 | 74 | 151 | <0.0001 | 3.4 (1.97–6.23) |
|
| ||||
| Systemic Therapy Prior to Craniotomy | ||||
| Immunotherapy | 31 | 244 | 0.81 (0.50–1.29) | |
| Chemotherapy | 8 | 185 | 0.66 | 1.26 (0.55–2.52) |
| Sequential or Combination Therapy | 4 | 335 | 0.78 (0.23–1.90) | |
| No systemic therapy | 58 | 154 | ||
|
| ||||
| Number of brain lesions | ||||
| 1 | 49 | 181 | 0.61 | 0.88 (0.52–1.44) |
| >1 | 29 | 212 | ||
|
| ||||
| Systemic Therapy After Craniotomy | ||||
| Immunotherapy | 7 | 212 | 0.43 (0.13–1.06) | |
| Chemotherapy | 33 | 222 | 0.13 | 0.95 (0.60–1.48) |
| Sequential or Combination Therapy | 4 | 1011 | 0.35 (0.09–0.98) | |
| No systemic therapy | 57 | 133 | ||
|
| ||||
| Local Therapy After Craniotomy | 0.019 | |||
| Stereotactic radiosurgery only | 50 | 248 | 0.34 (0.20–0.58) | |
| Whole brain irradiation only | 7 | 253 | 0.37 (0.14–0.88) | |
| Repeat Craniotomy | 4 | 170 | 0.76 (0.22–2.00) | |
| Combination local therapy | 17 | 232 | 0.41 (0.21–0.78) | |
| No local therapy | 23 | 57 | ||
|
| ||||
| Immune Infiltrate | ||||
| Low | 56 | 160 | ||
| High | 44 | 259 | 0.006 | 0.54 (0.35–0.84) |
| Non assessable | 1 | |||
|
| ||||
| Hemorrhage | ||||
| Low | 42 | 234 | ||
| High | 58 | 160 | 0.04 | 1.58 (1.02–2.48) |
| Non assessable | 1 | |||
|
| ||||
| Necrosis | ||||
| Low | 56 | 156 | 0.56 | 0.88 (0.57–1.34) |
| High | 44 | 202 | ||
| Non assessable | 1 | |||
|
| ||||
| Melanin | ||||
| Low | 73 | 181 | ||
| High | 23 | 191 | 0.28 | 0.75 (0.44–1.24) |
| Non assessable | 5 | |||
|
| ||||
| Gliosis | ||||
| Low | 9 | 120 | 0.25 | 0.64 (0.32–1.48) |
| High | 45 | 164 | ||
| Non assessable | 47 | |||
Presented p-values are from univariate analysis.
Figure 3. Kaplan-Meier curves of prognostic histopathologic factors in MBM.
A. Immune infiltrate. B. Intratumoral hemorrhage. C. Immune infiltrate and intratumoral hemorrhage.
WGEP of MBM Identifies Pathways Associated with Outcome
To gain insight regarding cellular processes associated with survival in MBM, we performed WGEP of RNA obtained from FFPE brain sections. Only 29 patients' tumor blocks were suitable for microdissection. Survival analysis using the log-rank test of the entire 101-patient cohort compared with the 29-patient subset revealed no significant difference in time to death from craniotomy [p=0.17, median survival is 177 versus 246 days, hazard ratio (HR) 0.72, 95% confidence intervals (95% CI) 0.49–1.13]. 15,067 probe sets (out of a total of 24,526) that passed the filtering criteria were used to perform Cox proportional hazards model to identify gene sets associated with survival. Table 3 shows 49 (out of a total of 284) prognostic BioCarta pathways. The leading gene sets that were associated with prolonged OS are immune-related, with the T-cell receptor (TCR) function pathway being the most significant category. In contrast, pathways associated with shortened OS involved genes associated with hypoxia, the lissencephaly gene (LIS1) in neuronal migration and development, and oxidative stress, among others.
Table 3.
Biocarta pathways that were differentially expressed in MBMs.
| Pathways Associated with Good Prognosis | GSA test p-value | |
|---|---|---|
| 1 | CD3 Complex | < 0.005 |
| 2 | T helper (Th) Surface Molecules | < 0.005 |
| 3 | HIV-Induced T-Cell Apoptosis | < 0.005 |
| 4 | B-Cell Surface Molecules | < 0.005 |
| 5 | Th1/Th2 Differentiation | < 0.005 |
| 6 | Role of Tob in T-Cell Activation | < 0.005 |
| 7 | Activation of Csk Inhibits Signaling through the TCR | 0.005 |
| 8 | Lck and Fyn Kinases Initiate TCR Activation | 0.005 |
| 9 | Cells/Molecules Involved in Local Acute Inflammatory Response | 0.005 |
| 10 | Dendritic Cells Regulate Th1/Th2 Development | 0.005 |
| Pathways Associated with Adverse Prognosis | ||
| 1 | West Nile Virus | < 0.005 |
| 2 | Y Branching of Actin Filaments | 0.005 |
| 3 | Hypoxia-Inducible Factor | 0.005 |
| 4 | Regulation of ck1/cdk5 by type 1 Glutamate Receptors | 0.005 |
| 5 | fMLP-Induced Chemokine Expression in HMC-1 cells | 0.005 |
| 6 | Free Radical-Induced Apoptosis | 0.01 |
| 7 | Repression of Pain Sensation by DREAM | 0.015 |
| 8 | Transcription Regulation by Methyltransferase of CARM1 | 0.015 |
| 9 | Cadmium Induces DNA Synthesis and Proliferation in Macrophages | 0.02 |
| 10 | Lissencephaly Gene in Neuronal Migration and Development | 0.025 |
Validation of WGEP and Histopathologic Data
Since one of the gene set categories associated with prolonged OS was the TCR pathway, we performed IHC analysis of MBM for immune cell subsets. High peritumoral CD3+ and CD8+ mononuclear cells were significantly associated with prolonged OS (Table 4). To assess whether melanoma-infiltrating immune cells were functional within MBMs, we stained sections with CD247, the ζ chain of the TCR that is essential for amplification of TCR signaling and frequently lost in cancer.(16) Although CD247+ mononuclear cells were detectable, high numbers of CD247+ cells were infrequently observed. In addition, neither peritumoral CD4+ and CD14+ mononuclear cells nor the presence of any intratumoral mononuclear cell population tested were associated with prolonged OS (Table 4).
Table 4.
Prognostic significance of IHC and ISH variables assessed in MBM tissues.
| Variable | Median Survival (days) | Proportional Hazard p | KM Estimator HR (95%CI) | Variable | Median Survival (days) | Proportional Hazard p | KM Estimator HR (95%CI) |
|---|---|---|---|---|---|---|---|
|
| |||||||
| CD3+ (n=40) | CD8+ (n=37) | ||||||
| Peritumoral | Peritumoral | ||||||
| High (n= 7) | 688 | 0.006 | 0.24 (0.07–0.63) | High (n=10) | 540 | 0.042 | 0.43 (0.18–0.95) |
| Low (n=33) | 156 | Low (n=27) | 164 | ||||
| Intratumoral | Intratumoral | 370 | 0.12 | 0.53 (0.22–1.15) | |||
| High (n=10) | High (n=12) | 171 | |||||
| Low (n=29) | 191 | 0.19 | 0.57 (0.22–1.27) | Low (n=24) | |||
| N/A (n= 1) | 156 | N/A (n= 1) | |||||
|
| |||||||
| CD4+ (n=36) | CD14+ (n=38) | ||||||
| Peritumoral | Peritumoral | ||||||
| High (n= 6) | 370 | 0.24 | 0.57 (0.19–1.36) | High (n=11) | 281 | 0.51 | 0.76 (0.32–1.67) |
| Low (n=29) | 164 | Low (n=20) | 160 | ||||
| N/A (n= 1) | N/A (n= 7) | ||||||
| Intratumoral | 51 | 0.13 | 2.5 (0.59–7.30) | Intratumoral | |||
| High (n= 3) | 191 | High (n=13) | 156 | 0.54 | 1.25 (0.58–2.53) | ||
| Low (n=32) | Low (n=25) | 250 | |||||
| N/A (n= 1) | |||||||
|
| |||||||
| CD19+ (n=16) | FoxP3+ (n=38) | ||||||
| High (n= 0) | N/A | N/A | N/A | High (n=19) | 246 | 0.87 | 0.94 (0.46–1.93) |
| Low (n=16) | 293 | Low (n=19) | 178 | ||||
|
| |||||||
| CD247+ (n=32) | HLA class I (n=20) | ||||||
| Peritumoral | |||||||
| High (n= 5) | 156 | 0.72 | 0.82 (0.24–2.18) | High (n=10) | 219 | 0.90 | 0.94 (0.37–2.35) |
| Low (n=27) | 246 | Low (n=10) | 236 | ||||
| Intratumoral | |||||||
| High (n= 4) | 536 | 0.84 | 0.88 (0.20–2.61) | ||||
| Low (n=28) | 222 | ||||||
|
| |||||||
| HLA class II (n=18) | Tapasin (n=19) | ||||||
| High (n=9) | 148 | 0.60 | 1.29 (0.48–3.41) | High (n=10) | 281 | 0.41 | 0.67 (0.24–1.75) |
| Low (n=9) | 246 | Low (n= 9) | 212 | ||||
|
| |||||||
| TGFβ (n=21) | CCL19 (n=31) | ||||||
| High (n=11) | 246 | 0.67 | 0.72 (0.26–1.89) | Present (n= 9) | 273 | 0.68 | 0.84 (0.35–1.87) |
| Low (n=10) | 370 | Absent (n=22) | 184 | ||||
|
| |||||||
| CXCL13 (n=30) | CCL21 (n=27) | ||||||
| Present (n=17) | 293 | 0.79 | 0.9 (0.41–2.07) | Present (n= 3) | 536 | 0.24 | 0.43 (0.07–1.48) |
| Absent (n=13) | 143 | Absent (n=24) | 260 | ||||
Presented p-values are from univariate analysis. All but chemokines (CCL19, CCL21, and CXCL13) were assessed using IHC. Regarding CD19+ immune cells, values could not be estimated because none of the tissues contained high numbers (N/A).
We then asked whether the higher immune cell infiltrate is secondary to either higher expression of molecules involved in antigen presentation machinery by melanoma cells (e.g. HLA class I, class II, tapasin) or to lower expression of the immunosuppressive cytokine TGFβ by melanoma cells or to the expression of the chemokines CCL19, CCL21, and CXCL13 by mononuclear cells (17) or to lower abundance of naturally-occurring T regulatory cells (e.g. FoxP3+). The expression-abundance of none of these molecules-immune cell subsets was associated with prolonged OS (Table 4).
MBM are Biologically Closer to EcM than PrM
To assess whether MBM differ from other EcM or PrM in their WGEP, we performed WGEP of 72 samples. Same-patient MBM-EcM, MBM-PrM, and EcM-PrM pairs were available from 26, 12, and 12 patients, respectively. ANOVA using all probe sets showed that MBM and EcM cluster significantly differently from PrM (p<0.0001). Shown in Figure 4 is a dendrogram of the 72 samples that was constructed using all 15,067 probe sets. To measure the degree of similarity between MBM, EcM, PrM we used the Euclidean distance metric and average linkage and calculated the distances between same-patient MBM-EcM, MBM-PrM, and EcM-PrM. The distance between MBM-EcM was significantly smaller, compared to distances calculated between the other two groups (MBM-PrM and EcM-PrM) when clustering was performed on all 72 samples using all 15,067 probe sets (mean: 124 versus 169 and 162, respectively; ANOVA p=0.09, t-test to compare the distance between MBM-EcM and MBM-PrM as well as between MBM-EcM and EcM-PrM have a p-value of 0.04 for both).
Figure 4. Assessment of the degree of similarity between MBM, EcM, and PrM.

Dendrogram plot shows the gene expression relatedness of the 72 samples based on the 15,067 probe sets that passed the filtering criteria. Each sample is labeled by its site (B, brain, red; E, extracranial, blue; P, primary, black) and patient number. Brackets below patient samples link same-patient MBM and EcM (black closely related; grey distantly related; red, closely related distant metastases, EcM or MBM, with PrM). Numbers shown above nodes (red dots) are Euclidean distances calculated between two samples (e.g. B1-E1, B15-E15, B31-E31, E9-P9) based on the Euclidean distance scale shown to the left. Euclidean distance is one of the metrics used to quantify the difference between two points in space, which in our study are each individual tumor tissue samples. The difference between each tumor tissue samples is based on expression data from gene that are common to the sites. The larger the Euclidean distance, the larger the separation between tumor pairs.
One of the probe sets that was significantly upregulated in MBM, as opposed to EcM and PrM, was GFAP, a protein expressed by reactive astrocytes. IHC analysis confirmed that GFAP was detectable only in astrocytes of 21 MBM and absent in all six stained EcM. GFAP+ cells were not only surrounding melanoma cells, but were also detectable in clusters or islands within the tumor (Figure 2B). DAVID analysis of the differentially expressed genes between same-patient EcM-MBM tumor samples failed to reveal cellular processes that were significantly different between EcM and MBM.
Discussion
Patients with metastatic melanoma live longer, in the current era of effective new therapies.(18, 19) Clinical trials using these agents in patients with MBM have shown efficacy(3, 4) although the mechanism for their effects in MBM is unclear. Our immunohistochemical and GSA of MBM show that high degree of immune infiltrate and low intratumoral hemorrhage in MBM tumor tissues are favorable prognostic factors for patients who undergo craniotomy for MBM. Equally important, WGEP of MBM demonstrates similarity to EcM and both expression profiles differ from PrM. The role of stromal components, such as the immune infiltrate or reactive glial (GFAP+) cells interdigitating with melanoma, along with the similarity between EcM and MBM in gene expression suggests that signals from the stroma, and to a lesser extent the biology of melanoma cells, play an important role in determining prognosis. The clinical-prognostic significance of the tumor microenvironment mediating cross-talk with cancer cells has been shown in several other solid tumors.(13)
The prognostic significance of the high CD3+/CD8+ immune infiltrate in MBM is supported by histopathologic, IHC/ISH, and WGEP analyses. A previous study in patients with glioblastoma multiforme showed that glia-associated CD8+ infiltrates correlate with long-term survival.(20) However, peritumoral, but not intratumoral, T-cells were associated with prolonged OS, as previously described.(21) The lack of high numbers of intratumoral infiltrates as well as active (e.g. CD247+) mononuclear cells may account for the short OS seen in patients with MBM. However, the presence of immune cells provides an opportunity for immunotherapeutic approaches. The lack of association between other mononuclear populations, or expression of various components of the antigen expression machinery, immunosuppressive cytokines (e.g. TGFβ), and chemokines by melanoma cells with OS suggests that other factors may be involved.
This study provides new insights regarding the biology of MBM in relation to EcM and PrM. Although several genes overexpressed in PrM include among others, keratins, which may imply contamination, several keratins are indeed expressed by melanoma cells.(22) In addition, a significant number of genes that were previously found differentially expressed between PrM and/or human epidermal melanocytes versus metastatic melanomas were also found in our 514 gene list (e.g. KRT6B/14/16/17, LOR, KLK7, GJB6, PITX1, CST, DSC).(23) In contrast, we were surprised to find that only 23 out of the 514-gene list were differentially expressed between EcM and MBM. The gene expression similarity between MBM and EcM, which remained significant using all 15,067 probe sets for the clustering analysis, is in line with a recent study regarding the concordance of B-Raf and N-Ras mutations in melanoma metastases(24, 25) as well as the concordance of gene copy number variations between same-patient EcM and MBM.(25) This similarity is clinically relevant, because it explains results from ongoing clinical trials using B-Raf inhibitors in patients with MBM, which show that the response rate of MBMs is not significantly different from that of EcM.(3) The biological similarity and interconnectivity between EcM and MBM may explain the fact that the outcome of patients with MBM is not solely dependent on the effective control of MBM, but also relates to control of EcM since only 50% of patients with MBM die from events directly associated with MBM.
Our study was unable to investigate whether melanoma cells within the brain acquire neuronal cell characteristics, as was previously described.(26) However proteins, such as the LIS1, which is associated with neuronal migration and its corresponding BioCarta pathway was associated with shortened OS in patients with MBM in our study (Table 3) was expressed by melanoma cells in MBMs as well as melanoma cell lines (data not shown). Despite the similarities between EcM and MBM, our WGEP analysis identified a number of genes that were differentially expressed between MBM and EcM, such as GFAP (Figure 4A). Since GFAP is solely expressed by glial cells, our frequent observation of glial cells interspersed within, but not solely surrounding melanoma lesions, should imply an intimate relation with melanoma cells that may have clinical implications, such as, for example, the glial-mediated protection of melanoma cells from chemotherapies(27) or the glial-mediated upregulation of the PI3K pathway in neighboring melanoma cells that have metastasized to the brain.(7, 8) This latter finding may imply that targeted therapies against the PI3K pathway may be relevant in patients with MBM, especially those who escape B-Raf inhibition.(8) Therefore, the role of the glial microenvironment needs to be further characterized as it may provide clues regarding inferior responses of traditional cytotoxics or failures to the brain from treatment with B-Raf inhibitors.(8)
In summary, we present a translational analysis of a patient cohort with MBM, EcM, and PrM tumor biopsies that illuminates the biological factors that are important in established MBM. Although the tumor tissues that were selected and analyzed for histopathology, immunohistochemistry and WGEP were derived from patients who underwent craniotomy within a single large institution, the findings and conclusions have important clinical implications. First, patients with MBM have variable prognosis and high levels of immune infiltrate and low levels of hemorrhage identify a patient subgroup with improved prognosis. Second, the pursuit of the expanding number of systemic immunotherapies in MBM may be indirectly supported by these findings.
Supplementary Material
Acknowledgements
We would like to thank the University of Pittsburgh Health Sciences Tissue Bank (M Bisceglia), Ms. Mary Horak (Melanoma Center), and outside pathologists, especially Drs. Rabkin (Rabkin Dermatopathology) and Silverman (Dermpath-AmeriPath Diagnostics) for providing tumor blocks for this study.
Financial Support: Samuel and Emma Winters Foundation for Cancer Research (SM), the UPCI Core Grant P30CA047904 (MR), P01 NS040923-09 (RH), P01 CA132714 (TR), P50 CA121973 (JK), R01 CA110249 (SF).
Footnotes
The authors declare no financial disclosures.
References
- 1.Preusser M, Capper D, Ilhan-Mutlu A, Berghoff AS, Birner P, Bartsch R, et al. Brain metastases: pathobiology and emerging targeted therapies. Acta Neuropathol. 2012;123:205–22. doi: 10.1007/s00401-011-0933-9. [DOI] [PubMed] [Google Scholar]
- 2.Davies MA, Liu P, McIntyre S, Kim KB, Papadopoulos N, Hwu WJ, et al. Prognostic factors for survival in melanoma patients with brain metastases. Cancer. 2011;117:1687–96. doi: 10.1002/cncr.25634. [DOI] [PubMed] [Google Scholar]
- 3.Long GV, Trefzer U, Davies MA, Kefford RF, Ascierto PA, Chapman PB, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain (BREAK-MB): a multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13:1087–95. doi: 10.1016/S1470-2045(12)70431-X. [DOI] [PubMed] [Google Scholar]
- 4.Margolin K, Ernstoff MS, Hamid O, Lawrence D, McDermott D, Puzanov I, et al. Ipilimumab in patients with melanoma and brain metastases: an open-label, phase 2 trial. Lancet Oncol. 2012;13:459–65. doi: 10.1016/S1470-2045(12)70090-6. [DOI] [PubMed] [Google Scholar]
- 5.Jakob JA, Bassett RL, Jr., Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–23. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capper D, Berghoff AS, Magerle M, Ilhan A, Wohrer A, Hackl M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123:223–33. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 7.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009;15:7538–46. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niessner H, Forschner A, Klumpp B, Honegger JB, Witte M, Bornemann A, et al. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. doi: 10.1002/cam4.50. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, Kim JS, Park ES, Lee JS, Lin Q, Langley RR, et al. Astrocytes upregulate survival genes in tumor cells and induce protection from chemotherapy. Neoplasia. 2011;13:286–98. doi: 10.1593/neo.11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salgado KB, Toscani NV, Silva LL, Hilbig A, Barbosa-Coutinho LM. Immunoexpression of endoglin in brain metastasis secondary to malignant melanoma: evaluation of angiogenesis and comparison with brain metastasis secondary to breast and lung carcinomas. Clin Exp Metastasis. 2007;24:403–10. doi: 10.1007/s10585-007-9077-7. [DOI] [PubMed] [Google Scholar]
- 11.Berghoff AS, Lassmann H, Preusser M, Hoftberger R. Characterization of the inflammatory response to solid cancer metastases in the human brain. Clin Exp Metastasis. 2012 doi: 10.1007/s10585-012-9510-4. Jul 1 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.April CS, Fan JB. Gene Expression Profiling in Formalin-Fixed, Paraffin-Embedded Tissues Using the Whole-Genome DASL Assay. Methods Mol Biol. 2011;784:77–98. doi: 10.1007/978-1-61779-289-2_6. [DOI] [PubMed] [Google Scholar]
- 13.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol. 2011;29:1539–46. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhart TA, Rogan MJ, Huddleston D, Rausch DM, Eiden LE, Haase AT. Simian immunodeficiency virus burden in tissues and cellular compartments during clinical latency and AIDS. J Infect Dis. 1997;176:1198–208. doi: 10.1086/514113. [DOI] [PubMed] [Google Scholar]
- 15.Das Gupta T, Bowden L, Berg J. Malignant melanoma of unknown primary origin. Surg Gynecol Obstet. 1963;117:341. [PubMed] [Google Scholar]
- 16.Yu B, Zhang W. Down-regulation of CD3zeta is a breast cancer biomarker associated with immune suppression. Cell Biol Int. 2011;35:165–9. doi: 10.1042/CBI20100346. [DOI] [PubMed] [Google Scholar]
- 17.Izraely S, Klein A, Sagi-Assif O, Meshel T, Tsarfaty G, Hoon DS, et al. Chemokine-chemokine receptor axes in melanoma brain metastasis. Immunol Lett. 2010;130:107–14. doi: 10.1016/j.imlet.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–5. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72:1070–80. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katagata Y, Kondo S. Keratin expression and its significance in five cultured melanoma cell lines derived from primary, recurrent and metastasized melanomas. FEBS letters. 1997;407:25–31. doi: 10.1016/s0014-5793(97)00290-1. [DOI] [PubMed] [Google Scholar]
- 23.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombino M, Capone M, Lissia A, Cossu A, Rubino C, De Giorgi V, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30:2522–9. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 25.Chen G, Kim S-B, Lazar AJ, Wubbenhorst B, Ledoux AA, Stemke-Hale K, et al. Characterization of point mutations and copy number changes in melanoma patients with matched brain and extracranial metastases. Society for Melanoma Research Annual Meeting; Hollywood, CA. 2012. [Google Scholar]
- 26.Park ES, Kim SJ, Kim SW, Yoon SL, Leem SH, Kim SB, et al. Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc Natl Acad Sci USA. 2011;108:17456–61. doi: 10.1073/pnas.1114210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Q, Balasubramanian K, Fan D, Kim SJ, Guo L, Wang H, et al. Reactive astrocytes protect melanoma cells from chemotherapy by sequestering intracellular calcium through gap junction communication channels. Neoplasia. 2010;12:748–54. doi: 10.1593/neo.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.