Abstract
APOBEC3 proteins are DNA cytosine deaminases that restrict the replication of human immunodeficiency virus deficient in the counterdefense protein Vif. Here, we address the capacity of APOBEC3F to restrict via deaminase-dependent and -independent mechanisms by monitoring spreading infections in diverse T cell lines. Our data indicate that only a deaminase-proficient protein is capable of long-term restriction of Vif-deficient HIV in T cells, analogous to prior reports for APOBEC3G. This indicates that the principal mechanism of APOBEC3F restriction is deaminase-dependent.
Keywords: APOBEC3F, APOBEC3G, deaminase, HIV restriction, Vif
Introduction
APOBEC3 proteins restrict human immunodeficiency virus type 1 (HIV) in the absence of the viral accessory protein Vif [reviewed in (Albin, 2010; Malim and Bieniasz, 2012; Refsland and Harris, 2013)]. It is generally agreed that enzymatic function is an important component of the restriction mechanism, which results in viral hypermutation and decreased cDNA accumulation [e.g. (Harris et al., 2003; Mangeat et al., 2003; Yu et al., 2004; Zhang et al., 2003) and recently reviewed in (Albin and Harris, 2013)]. In addition, many have proposed diverse deaminase-independent mechanisms of restriction [e.g. (Bishop et al., 2006; Bishop et al., 2008; Guo et al., 2006; Holmes et al., 2007; Iwatani et al., 2007; Li et al., 2007; Luo et al., 2007; Mbisa et al., 2007; Mbisa et al., 2010; Newman et al., 2005; Shindo et al., 2003; Yang et al., 2007)]. It is possible, however, that some of these deaminase-independent mechanisms may represent transient overexpression artifacts since stable expression of deaminase-deficient APOBEC3G (A3G) fails to inhibit viral replication (Browne et al., 2009; Miyagi et al., 2007; Schumacher et al., 2008). Although APOBEC3F (A3F) was initially thought to have more prominent deaminase-independent effects (Holmes et al., 2007), stable expression of A3F zinc coordination mutants in HeLa cells also fails to restrict Vif-deficient HIV in a single cycle of replication (Miyagi et al., 2010).
We describe herein a series of studies designed to extend to A3F these prior analyses of APOBEC3 deaminase activity in HIV restriction using a spreading infection model that might better represent conditions of natural infection. Like our predecessors, we find using novel cell lines stably expressing A3G catalytic mutants that such mutants are incapable of efficiently restricting Vif-deficient HIV. Similarly, we find that A3F catalytic mutants exert little to no restrictive activity under similar conditions of stable T cell expression. We subsequently confirm the efficacy of both endogenously expressed and stably transfected wildtype A3F for the restriction of HIV using Vif mutants selectively susceptible to A3F but not A3G. We therefore conclude that deaminase activity is required for efficient restriction of Vif-deficient HIV by A3F.
Materials and Methods
Plasmid generation
Wildtype A3F and A3G are identical to GenBank NM_145298 and NM_021822, respectively, as previously described (Albin et al., 2010a; Albin et al., 2010b; Haché et al., 2008). Variants A3F E251Q and A3G E259Q were generated by QuikChange site-directed mutagenesis (Stratagene) and confirmed by DNA sequencing. V5-tagged derivatives as shown in Fig. 2 were derived by similar methods as previously described (Albin et al., 2013; Albin et al., 2010a; Albin et al., 2010b).
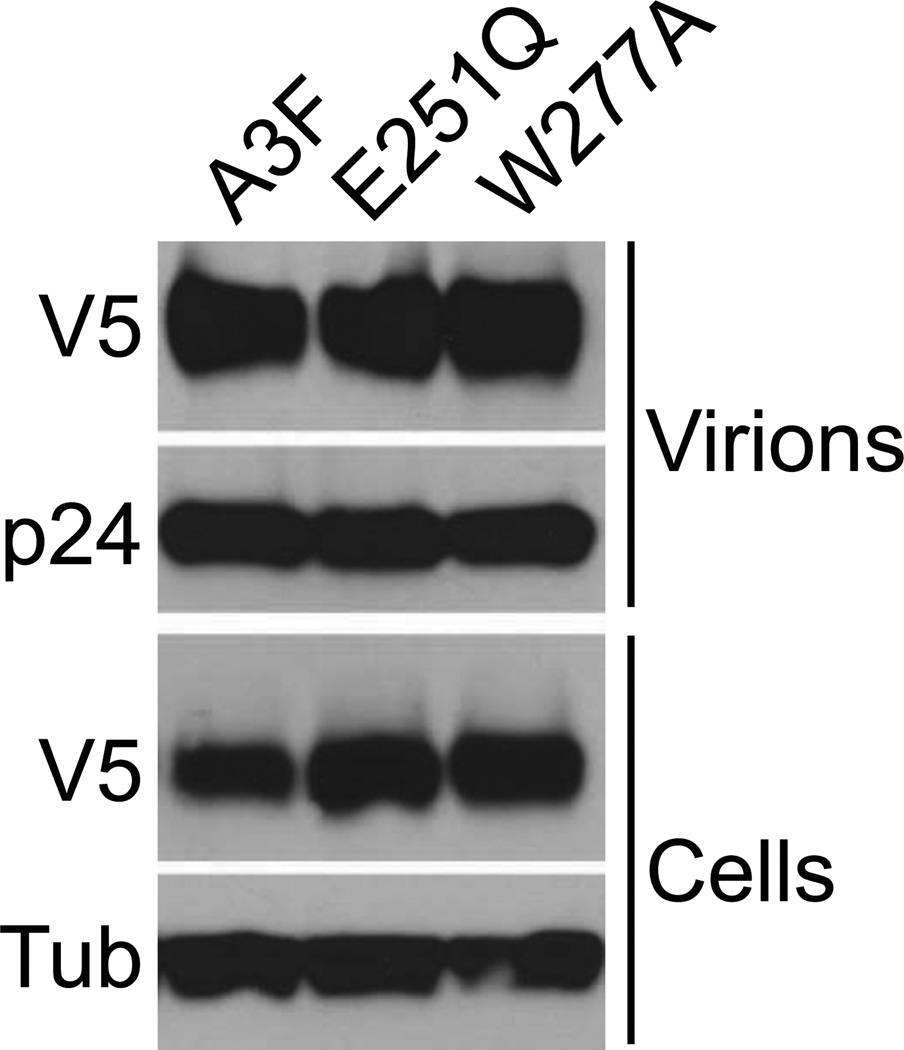
Fig. 2. Encapsidation of A3F catalytic mutants E251Q and W277A.
Plasmids expressing A3F-V5 or either of two catalytic mutant derivatives were cotransfected with Vif-deficient HIVIIIB. Virus-containing supernatants and producer cells were harvested approximately two days post-transfection, and expression and encapsidation were analyzed by western blot.
HIVIIIB and a derivative containing tandem stop codons at positions 26 and 27 of vif are identical to GenBank EU541617 with the exception of an A200C mutation that corrects an aberrant upstream open reading frame that would otherwise inhibit viral gene expression, as previously described (Albin et al., 2013; Albin et al., 2010a; Albin et al., 2010b; Haché et al., 2009; Haché et al., 2008). Vif mutants were generated by site-directed mutagenesis in a shuttle vector containing the vif-vpr segment of HIVIIIB, which was subsequently subcloned back into a full-length proviral background using the SwaI and SalI sites, as previously described (Albin et al., 2010a).
Cell lines
CEM-GFP and SupT11-derived T cell lines were maintained in RPMI media containing 10% fetal bovine serum, penicillin/streptomycin and β-mercaptoethanol. 293T cells were cultured in DMEM containing 10% fetal bovine serum and penicillin/streptomycin.
The generation of SupT11-derived cell lines expressing single wildtype or variant APOBEC3 proteins has been described previously (Albin et al., 2010a; Albin et al., 2010b). SupT11 is a clonal derivative of SupT1 isolated by limiting dilution and found to have undetectable expression A3F and A3G and little if any expression of other APOBEC3 proteins (Refsland et al., 2010). Briefly, SupT11 cells were electroporated with 20 μg linearized plasmid, plated at several dilutions in 96-well plates and selected in G418-containing RPMI. Single-cell clones were then expanded and analyzed by western blot for expression of A3F or A3G, as indicated below and shown in Fig. 1.
Fig. 1. APOBEC3 expression levels in the cell lines used in this study.
(A) A3F levels found in SupT11 cells stably transfected with untagged A3F (code letter B followed by a number to indicate a distinct, clonal line), A3F E251Q (code letter D) or a vector control (code letter A) in comparison with naturally nonpermissive lines CEM2n and H9. (B) A3G levels found in SupT11 cells stably transfected with untagged A3G (code letter C), A3G E259Q (code letter E) or a vector control (code letter A) in comparison with naturally nonpermissive lines CEM2n and H9.
Encapsidation
A3F-V5 or catalytic mutant derivatives E251Q or W277A were cotransfected with Vif-deficient HIVIIIB. Two days post-transfection, transfected cells and virus-containing supernatants were harvested. Viruses were pelleted through 20% sucrose cushions and lysed in a 5× sample buffer consisting of 62.5 mM Tris pH 6.8, 20% glycerol, 2% sodium dodecyl sulfate and 5% β-mercaptoethanol. A3F-V5 levels expressed or encapsidated were then analyzed by western blot methods similar to those described below for the analysis of APOBEC3 expression levels in T cell lines, probing for V5 (Invitrogen), p24 (antibodies purified from hybridoma cell line 183-H12-5C from the AIDS Research and Reference Reagent Program) or tubulin (Covance).
Western blots
Expression of APOBEC3 proteins in SupT11-derived cell lines was assessed by lysing individual lines in a buffer consisting of 25 mM HEPES pH 7.4, 150 mM NaCl, 1 mM MgCl2, 10% glycerol and 1% Triton X-100 with 50 µM MG132 and complete protease inhibitor (Roche). Lysates were then mixed with the 5× sample buffer described above to a final concentration of 2×. This mix was boiled for 10 minutes and loaded onto 10% sodium dodecyl sulfate polyacrylamide gels for fractionation by electrophoresis prior to transfer to polyvinylidene fluoride membranes.
Membranes were blocked with 4% milk in 0.1% Tween diluted in phosphate-buffered saline. These were then probed either with rabbit anti-A3F (Dr. Michael Malim, #1474 via the AIDS Research and Reference Reagent Program), rabbit anti-A3G (Dr. Jaisri Lingappa, #10201 via the AIDS Research and Reference Reagent Program) or mouse anti-tubulin (Covance) followed by horseradish peroxidase (HRP) conjugated secondary antibodies as appropriate. HRP activity was then detected by treatment with HyGLO chemiluminescent HRP detection reagent (Denville) and exposure to film. Between distinct primary antibodies, membranes were stripped with a buffer containing 62.5 mM Tris pH 6.8, 2% sodium dodecyl sulfate and 100 mM β-mercaptoethanol at 50 °C and washed in phosphate-buffered saline with 0.1% Tween.
Spreading infections
Viruses were produced by transfecting 293T cells in 10 cm dishes with 5–10 µg proviral plasmid DNA. Virus-containing supernatants were then harvested, passed through 0.45 µm filters and frozen. An aliquot of each virus preparation was thawed and titrated on CEM-GFP cells in 96-well plates, and the volume corresponding to a given percentage of cells infected was calculated to normalize starting quantities.
Volumes of wildtype or Vif-deficient HIVIIIB corresponding to MOI 0.01 or 0.05 were used to initiate spreading infections on four SupT11-derived cell lines each expressing A3F, A3F E251Q, A3G or A3G E259Q as well as SupT11-derived lines stably transfected with an empty vector control and naturally nonpermissive CEM2n cells. Infections utilizing vif mutants as described in the main text were initiated in similar fashion. Following infection of 50,000 target cells with normalized virus quantities in a total volume of 1 mL in 24-well plates, viral spread was monitored periodically by using 150 µL of supernatant to infect 25,000 CEM-GFP cells in 96-well plates in a total volume of 250 µL. Three days later, infected CEM-GFP reporter cells were fixed in 4% paraformaldehyde and analyzed by flow cytometry using a FACSCalibur instrument (Becton-Dickinson) with subsequent data analysis in FlowJo software (Tree Star). Infected 24-well plate cultures were periodically split and fed to prevent cell overgrowth.
Results
Stable T cell expression of A3F catalytic mutants fails to restrict Vif-deficient HIV
To determine whether deaminase-deficient A3F is capable of restricting HIV in T cells, we stably transfected a permissive T cell line, SupT11, with wildtype A3F, A3G or catalytic mutant derivatives and isolated clones by limiting dilution as described previously [e.g. (Albin et al., 2010a; Albin et al., 2010b; Hultquist et al., 2011)]. Expression levels of untagged A3F and A3G in all resultant lines were comparable between wildtype and mutant sets (Fig. 1). We specifically used mutants of the catalytic glutamates in A3F (E251Q) and A3G (E259Q) since these show little or no discernible encapsidation defect despite loss of deaminase activity [e.g. (Holmes et al., 2007; Schumacher et al., 2008) and Fig. 2, which shows encapsidation in virions produced by 293T cotransfection with A3F-V5 and Vif-deficient HIV expression plasmids]. Although Fig. 2 does not directly demonstrate equal encapsidation efficiency of A3F and its catalytic mutants in SupT11 cells, it does demonstrate equal inherent encapsidation potential of the proteins in question, which may not be assumed for all catalytic mutants [e.g. (Holmes, et al., 2007)]. One may thus infer that A3F E251Q is unlikely to be inherently disadvantaged in its ability to encapsidate.
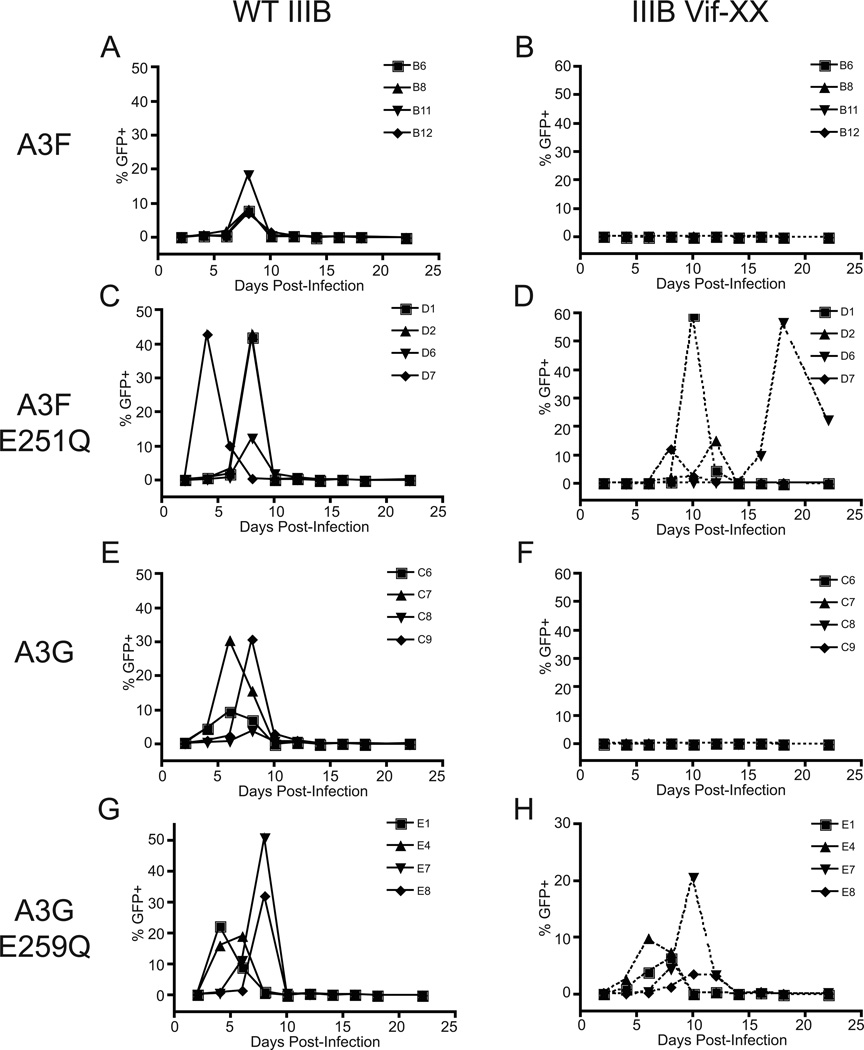
Infections of all of these lines were initiated in parallel at multiplicities of infection (MOIs) of 0.01 and 0.05 with either wildtype or Vif-deficient HIVIIIB containing tandem stop codons at positions 26 and 27 of vif (Albin et al., 2010a; Albin et al., 2010b; Haché et al., 2008). We have previously shown that similar conditions result in restriction of Vif-deficient HIVIIIB by wildtype A3F for at least three months or until the acquisition of adaptive viral mutations (Albin et al., 2010a). Consistent with these data, stable expression of either wildtype A3F or A3G in SupT11 restricts the spread of Vif-deficient HIVIIIB for the duration of the experiment (Figs. 3B and 3F). In contrast, Vif-deficient viruses peak within approximately two weeks on cell lines expressing catalytic mutants of each APOBEC3 protein with perhaps a modest delay among A3F E251Q lines in comparison with A3G E259Q (Figs. 3D and 3H). Thus, the catalytic glutamate and most likely deaminase activity itself is required for efficient restriction of HIV by stably expressed A3F or A3G in T cell lines.
Fig. 3. Deaminase activity is required for restriction of Vif-deficient HIV by both A3F and A3G.
Representative spreading infection curves infected at MOI 0.05 from one of three independent experiments demonstrating the inability of catalytic mutants of A3F or A3G to efficiently restrict Vif-deficient HIV. (A–B) Wildtype or Vif-deficient HIVIIIB on four independent cell lines expressing wildtype A3F. (C–D) On four independent cell lines expressing A3F E251Q. (E–F) On four independent cell lines expressing wildtype A3G. (G–H) On four independent cell lines expressing A3G E259Q. Cell lines in all panels are named in as Fig. 1. Variation in the time to peak and in the height of peaks reflects the fact that each cell line used is an independent subclone. Note also that the peak of infectious virion production is typically followed shortly thereafter by a rapid drop in detected infectious virus. This reflects viral cytopathic effects in permissive cultures where HIV successfully spreads, which results in the death of the permissive producer cell cultures shortly after peak infectious virion production. In the absence of surviving producer cells, no infectious virion production is detected.
Selectively susceptible vif alleles confirm the restrictive efficacy of endogenous A3F
Although A3F restricts Vif-deficient HIV efficiently at the levels of expression demonstrated in our stably transfected cell lines, another recent report found that the levels of A3F in ex vivo peripheral blood mononuclear cells derived from several donors are inadequate for restriction (Mulder et al., 2010). To assess the effects of A3F in primary cells, the authors of this study utilized a W11R Vif mutant that is capable of neutralizing A3G but not A3F. For our part, we had previously abandoned similar lines of inquiry on finding that mutants of the putative A3F-interacting region, 14DRMR17 (D14A), but not of the putative A3G-interacting region, 40YRHHY44 (Y40A), faithfully reproduce their transient overexpression phenotypes in stably transfected T cells [(Russell and Pathak, 2007) and (Albin and Harris, unpublished data)].
To determine whether the levels of A3F or A3G in naturally nonpermissive cells are adequate to restrict, we carried out additional experiments characterizing the spreading infection phenotypes of the Vif W11R mutant along with three mutants of the relevant putative A3F- or A3G-interacting regions that we had identified incidentally in unrelated long-term passage experiments: R15G, H43N and H43N/E117K (Albin and Harris, unpublished data). Mutation of each of these residues has been previously shown to ablate Vif anti-A3F (residues 11 and 15) or anti-A3G (residue 43) activity in transient overexpression systems (He et al., 2008; Mulder et al., 2010; Nagao et al., 2010; Pery et al., 2009; Russell and Pathak, 2007; Simon et al., 2005; Tian et al., 2006; Yamashita et al., 2008; Yamashita et al., 2010; Zhang et al., 2008).
Consistent with transient overexpression data, each of the mutants predicted to be susceptible to A3F, W11R or R15G, was restricted in SupT11-A3F lines and in naturally nonpermissive CEM2n cells that express endogenous A3F at levels slightly lower than those found in primary CD4+ T cells (Refsland et al., 2012) (Fig. 4A and 4C). The same mutants were not restricted in a SupT11-A3G line (Fig. 4B). In comparison, mutants H43N and H43N in combination with E117K were capable of spreading efficiently in the presence of both A3F and A3G and showed a modest delay in CEM2n cells, somewhat more pronounced for H43N versus the H43N/E117K mutant (Fig. 4). These results are consistent with prior long-term passages of 40YRHHY44 mutants in nonpermissive CEM cells, wherein the authors observed no reversion of the mutated residues, implying lack of selective pressure and, by extension, largely intact anti-A3G function (Russell et al., 2009). Combined with the prior observation that mutation of the putative A3F-interacting region 14DRMR17 can result in the neutralization of Vif-resistant A3G D128K (Schröfelbauer et al., 2006), these data suggest that the losses of function associated with each region may represent something more complex than simple loss of direct interaction with Vif.
Fig. 4. Mutation of the putative A3F-interacting region of Vif but not the putative A3G-interacting region of Vif yields viruses selectively susceptible to the predicted APOBEC3 protein.
(A) Growth of wildtype, Vif-deficient and the indicated Vif mutant derivatives of HIVIIIB in the presence of A3F. (B) Growth of wildtype, Vif-deficient and the indicated Vif mutants in the presence of A3G. (C) Growth of wildtype, Vif-deficient and the indicated Vif mutants in naturally nonpermissive CEM2n cells. Results are representative of 2–3 independent experiments. The symbols associated with viruses showing no growth are boxed to facilitate comparison.
Discussion
We have described here T cell lines stably expressing A3F E251Q that, in contrast to similar lines expressing wildtype A3F, fail to restrict Vif-deficient HIV. These results mirror those previously observed with A3G (Browne et al., 2009; Miyagi et al., 2007; Schumacher et al., 2008), and together, we propose that both A3F and A3G require catalytic activity for the efficient restriction of Vif-deficient HIV. Although unlikely based on many experiments in our laboratory and others over the past decade, we cannot exclude the hypothetical possibility that the catalytic glutamate residue of these proteins may have an unknown function in restriction beyond its described role catalyzing DNA cytosine deamination. It is also formally possible that the encapsidation efficiency of A3F and its E251Q derivative may differ in T cells. The results shown in Fig. 2 with particles produced on 293T cells, however, suggest that there is no inherent difference in the encapsidation potential of these proteins, as previously shown for A3G and A3G E259Q [e.g. Schumacher et al., 2008).
It also remains possible that expression of A3F and A3G in vivo might hypothetically reach levels at which any of the deaminase-independent mechanisms of restriction previously described in transient overexpression studies might be active. This could be particularly relevant for A3F, which may have greater deaminase-independent effects than A3G both in transient overexpression experiments and in primary CD4+ T cells (Gillick et al., 2013; Holmes et al., 2007). It is telling, however, that even under conditions of likely overexpression in SupT11 cells, A3F E251Q and A3G E259Q are unable to restrict Vif-deficient HIV (Fig. 3). In the absence of data concerning the exact induction conditions under which deaminase-independent restriction might occur, we must for the time being conclude that the predominant mechanism of restriction by A3F and A3G is deaminase-dependent.
Recent papers have also called into question the ability of even wildtype A3F to restrict Vif-deficient HIV when stably or endogenously expressed (Miyagi et al., 2010; Mulder et al., 2010). To confirm the results seen in our analyses of wildtype versus catalytic mutant stable expression in SupT11, we also used Vif mutants selectively susceptible to either A3F or A3G (Mulder et al., 2010; Russell and Pathak, 2007; Simon et al., 2005). As shown in Fig. 4, both the W11R and the R15G mutants in or around the 14DRMR17 region thought to be important for A3F interaction fail to spread both in SupT11 lines stably transfected with A3F and in naturally nonpermissive CEM2n cells, which have been previously shown to express endogenous A3F at levels even lower than those in primary CD4+ cells (Refsland et al., 2012), implying the efficacy of A3F in restriction. It is likely that A3D also contributes to restriction in CEM2n (and in the peripheral mononuclear blood cells utilized in other studies) and thereby may lead to overestimates of the efficacy of A3F (Hultquist et al., 2011; Refsland et al., 2012). However, given the ability of multiple lines stably transfected with A3F to restrict over many studies (Albin et al., 2010a; Albin et al., 2010b; Haché et al., 2008; Han et al., 2008; Hultquist et al., 2011) and the apparently weaker restrictive abilities of A3D (Hultquist et al., 2011), we think this unlikely.
It has been previously proposed that the inability of peripheral mononuclear blood cells to restrict HIV was a result of low A3F expression levels in the samples used (Mulder et al., 2010). This is possible, albeit at odds with the restriction of vif mutants W11R and R15G by CEM2n cells in Fig. 4, but it is notable that a more recent analysis of the relative restrictive capacities of A3D, A3F and A3G failed to demonstrate long-term restriction by even A3G in primary CD4+ T cells (Chaipan et al., 2013). Like Mulder et al., these authors observed robust restriction of a Vif mutant susceptible to all APOBEC3 proteins but not of a Vif mutant susceptible to one or two APOBEC3 proteins. Rather than signaling a hierarchy of relative importance as implied by the modest differential delays observed by Chaipan et al., it may therefore be more accurate to say that restriction in primary cells reflects the cumulative effects of multiple APOBEC3 proteins. Alternatively, there may be something fundamentally different about the mechanism of restriction and/or the requirement for Vif in primary cells that accounts for these results.
A curious observation made in the course of these studies is the inability of A3G to robustly restrict T cell spread by Vif-proficient viruses with a mutation in the 40YRHHY44 region thought to be important for binding to A3G (Russell and Pathak, 2007). Although not widely noted, such results have been implied twice before (Chaipan et al., 2013; Russell et al., 2009). The Russell et al. manuscript is particularly notable between the two of these because it describes long-term passages of 40YRHHY44 mutant viruses in naturally nonpermissive CEM cells. Despite ample opportunity for the virus to revert these presumably deleterious mutations, the 40YRHHY44 mutations remained intact throughout passage (Russell et al., 2009), implying a lack of selective pressure in this spreading infection system despite what would be predicted based on single-cycle replication experiments. Combined with the ability of mutants in the putative A3F-binding 14DRMR17 region to reverse the Vif-resistant phenotype of A3G D128K (Schröfelbauer et al., 2006), it seems unlikely that 14DRMR17 and 40YRHHY44 are simply regions Vif that directly interact with A3F or A3G, respectively.
Conclusions
In conclusion, we have shown that the restriction of HIV by A3F requires intact deaminase activity. We have also confirmed prior reports demonstrating the importance of deaminase activity for HIV restriction by A3G. These results suggest that the predominant driving force for the maintenance of Vif function by HIV centers on the deaminase-dependent restriction mechanisms previously described. We further note that not all vif mutations thought to yield selective susceptibility to one APOBEC3 protein or another have notable spreading infection phenotypes. This may complicate the attempt to use such mutants in parsing the in vivo contributions of various APOBEC3 proteins and further suggests that the growing literature on loss of function vif mutations will need to be carefully interpreted as regards defining the regions of Vif that directly interact with A3F and A3G [reviewed in (Albin, 2010; Smith et al., 2009)]. Insofar as one can trust the phenotypes of these loss of function mutants, however, they appear to support a model wherein A3F is at least as relevant as A3G to the overall nonpermissive phenotype in nonpermissive T cell lines. Knowledge of the in vivo relevance of distinct APOBEC3 proteins, by contrast, is not as well defined.
Highlights.
Catalytic activity of A3F is required for efficient restriction of Vif-deficient HIV
Stably expressed A3F restricts HIV Vif mutants selectively susceptible to A3F
Putative A3F/G-interaction Vif mutants show divergent spreading infection phenotypes
Acknowledgements
We thank Judd Hultquist, Brett Anderson, and Leah Evans for thoughtful comments. This research was funded by the National Institute of Allergy and Infectious Diseases (R01 AI064046 to RSH) and by the National Institute of General Medical Sciences (P01 GM091743 to RSH). JSA was supported in part by the National Institute on Drug Abuse (F30 DA026310) and by the University of Minnesota Medical Scientist Training Program (T32 GM008244).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Reviews in Molecular Medicine. 2010;12:1–26. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Anderson JS, Johnson JR, Harjes E, Matsuo H, Krogan NJ, Harris RS. Dispersed sites of HIV Vif-dependent polyubiquitination in the DNA deaminase APOBEC3F. J Mol Biol. 2013;425:1172–1182. doi: 10.1016/j.jmb.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Haché G, Hultquist JF, Brown WL, Harris RS. Long-term Restriction by APOBEC3F Selects Human Immunodeficiency Virus Type 1 Variants with Restored Vif Function. Journal of Virology. 2010a;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Harris RS. APOBECs and their role in proviral DNA synthesis. In: Le Grice SF, Goette M, editors. Human Immunodeficiency Virus Reverse Transcriptase: A Bench-to-Bedside Success. New York, NY: Springer; 2013. pp. 253–280. [Google Scholar]
- Albin JS, LaRue RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. The Journal of Biological Chemistry. 2010b;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. Journal of Virology. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C, Smith JL, Hu WS, Pathak VK. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. Journal of Virology. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillick K, Pollpeter D, Phalora P, Kim EY, Wolinsky SM, Malim MH. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. Journal of Virology. 2013;87:1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. Journal of Virology. 2006;80:11710–11722. doi: 10.1128/JVI.01038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haché G, Abbink TE, Berkhout B, Harris RS. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. Journal of Virology. 2009;83:5956–5960. doi: 10.1128/JVI.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haché G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wang X, Dang Y, Zheng YH. APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog. 2008;4:e1000095. doi: 10.1371/journal.ppat.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. The Journal of Biological Chemistry. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Hultquist JF, Lengyel JA, Refsland EW, Larue RS, Lackey L, Brown WL, Harris RS. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. Journal of Virology. 2011;85(21):11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Research. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. The Journal of Biological Chemistry. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. Journal of Virology. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. Journal of Virology. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. Journal of Virology. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Brown CR, Opi S, Khan M, Goila-Gaur R, Kao S, Walker RC, Jr, Hirsch V, Strebel K. Stably expressed APOBEC3F has negligible antiviral activity. Journal of Virology. 2010;84:11067–11075. doi: 10.1128/JVI.01249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. Journal of Virology. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder LC, Ooms M, Majdak S, Smedresman J, Linscheid C, Harari A, Kunz A, Simon V. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. Journal of Virology. 2010;84:9613–9617. doi: 10.1128/JVI.02630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao T, Yamashita T, Miyake A, Uchiyama T, Nomaguchi M, Adachi A. Different interaction between HIV-1 Vif and its cellular target proteins APOBEC3G/APOBEC3F. J Med Invest. 2010;57:89–94. doi: 10.2152/jmi.57.89. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Pery E, Rajendran KS, Brazier AJ, Gabuzda D. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. Journal of Virology. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Harris RS. The APOBEC3 Family of Retroelement Restriction Factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Research. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. Journal of Virology. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer B, Senger T, Manning G, Landau NR. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. Journal of Virology. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Haché G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. Journal of Virology. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. The Journal of Biological Chemistry. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Bu W, Burdick RC, Pathak VK. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol Sci. 2009;30:638–646. doi: 10.1016/j.tips.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. Journal of Virology. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Kamada K, Hatcho K, Adachi A, Nomaguchi M. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes and Infection / Institut Pasteur. 2008;10:1142–1149. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Nomaguchi M, Miyake A, Uchiyama T, Adachi A. Status of APOBEC3G/F in cells and progeny virions modulated by Vif determines HIV-1 infectivity. Microbes and Infection / Institut Pasteur. 2010;12:166–171. doi: 10.1016/j.micinf.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo F, Cen S, Kleiman L. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007;365:92–100. doi: 10.1016/j.virol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chen G, Niewiadomska AM, Xu R, Yu XF. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS ONE. 2008;3:e3963. doi: 10.1371/journal.pone.0003963. [DOI] [PMC free article] [PubMed] [Google Scholar]