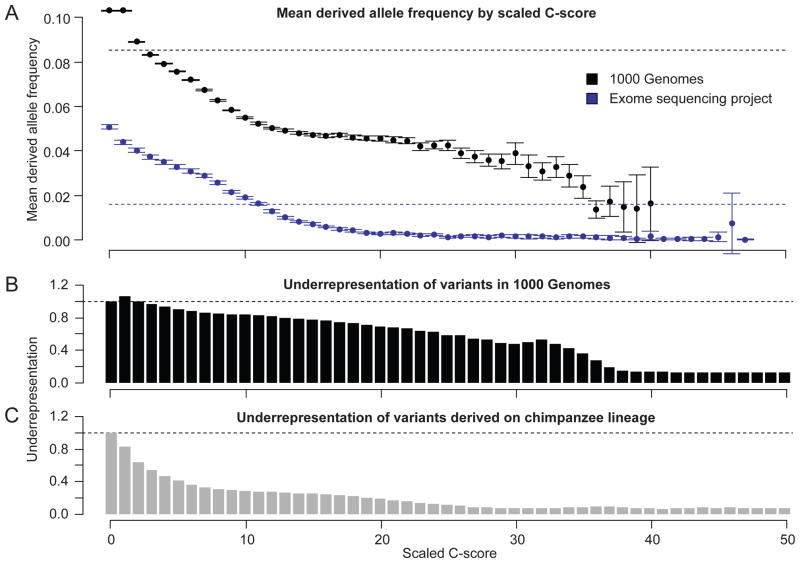
Figure 2.
Relationship between scaled C-scores and: the average derived allele frequency (DAF) of variants identified in the 1000 Genomes Project14 or ESP24 (upper panel); the under-representation of polymorphic sites in 1000 Genomes (middle panel); and chimpanzee lineage derived variants (lower panel). The dashed lines in the upper plot indicate the mean DAF and confidence intervals indicate 1.96x standard errors of the mean (SEM) DAF in each bin. Under-representation is defined as the proportion of 1000 Genomes (middle panel) or chimpanzee-derived (lower panel) variants in a specific scaled C-score bin divided by the frequency with which that scaled C-score is observed for all possible mutations of the human reference assembly (10C-score/−10). The stronger under-representation of chimpanzee-derived variants relative to 1000 Genomes variants is expected given that the former are mostly fixed or high-frequency variants (and have survived many generations of purifying selection) while the latter are mostly low-frequency variants. Depletion values in both panels for C-score bins other than 0 are significantly different from expectation (binomial proportion test, all p-values <10−11).