Abstract
Advances in next-generation technologies have rapidly improved sequencing fidelity and significantly decreased sequencing error rates. However, with billions of nucleotides in a human genome, even low experimental error rates yield many errors in variant calls. Erroneous variants can mimic true somatic and rare variants, thus requiring costly confirmatory experiments to minimize the number of false positives. Here we discuss sources of experimental error in next-generation sequencing and how replicates can be used to abate them.
Introduction
The emergence of next-generation sequencing (NGS) has revolutionized genetics and provided valuable resources for other scientific disciplines. As NGS becomes more widely accessible, its use has extended beyond basic research and into broader clinical contexts. Hence, it is increasingly more important to account for the error that arises in the sequencing process. Error can stem from the bioinformatic analysis1, and also from experimental steps2,3, the latter of which can often be mitigated through the use of replicate experiments.
The use of replicates permeates almost all scientific disciplines. Yet in NGS, many researchers use increased sequencing read depth and bioinformatic filters to address error in lieu of biological replication. This practice is understandable given that replicates can increase study costs substantially. However, sequencing costs have fallen dramatically4, and now is the time to reevaluate the value of replication in sequencing studies.
Here we discuss sources of error in sequencing and the nascent use of replication in published high-throughput sequencing efforts. In addition, we demonstrate how biological replicates can be employed to reduce sequencing error. In particular, replicates can be used to assess the specificity and sensitivity of sequence variant calling methods in a manner that is independent of the algorithms and chemistry used to call variants, thereby guiding the appropriate selection of quality score thresholds.
Experimental Error in NGS
Technological advances and the digital nature of DNA are helping to achieve highly accurate genome sequences. However, sequencing methods are imperfect. NGS applications, such as whole genome sequencing, targeted capture, RNA-Seq and ChIP-Seq, are prone to errors that result in miscalled bases, thus causing short read misalignment and mistakes in genome assembly. Reported sequencing base call accuracy claims for leading high-throughput sequencing technologies vary wildly, ranging from one error in one thousand nucleotides (99.9%)5 to one error in ten million nucleotides (99.9999%)6. Even for methods with the lowest reported rates, the absolute numbers of miscalled genomic variants remain unwieldy, with possibly thousands of false positive variants in a fully sequenced human genome. Furthermore, false positive error masquerades as rare and somatic variants, thereby obfuscating true variants of clinical interest. Known sources for experimental error can be grouped by where they occur in the sequencing workflow (Figure 1a; Box 1), i.e., during sample preparation, library preparation, or sequencing/imaging.
Figure 1. Sources of unexpected and erroneous variation and established post-processing tools used to cope with unexpected variants.

Sequencing experiments involve many steps from sample acquisition to final data analysis, and a major challenge in the process stems from the emergence of unexpected variants a. These can include legitimate somatic mosaicism and rare oncogenic variants. Additionally, many erroneous sequence variants arise during experimental steps (e.g., via sample degradation, PCR amplification, base-calling error). b. Several analytical tools and post-processing mechanisms are often employed for separating true variation from false sequence variants. These include indicators of data quality (e.g., base call and mapping quality scores) and filters that are informed by those indicators. Additional tertiary analyses can also highlight systematic biases through clustering methods and possible false positive variants by accounting for Mendelian inheritance patterns58. Throughout the sequencing and post-processing pipeline, the use of replicated sequencing experiments can help mitigate the impact erroneous variants from the experimental steps and inform post-processing filters. Thus, greater accuracy of germline variant detection can be attained and improved sensitivity can be achieved for true somatic variation.
Box 1. Experimental sources of error abound in sequencing.
The significance and relative impact of each error source on downstream applications depend on many factors, such as sample acquisition, reagents, tissue type, protocol, instrumentation, conditions, analytical application, and the ultimate goal of the study. Sequencing errors can stem from any time point throughout the experimental workflow, including initial sequence preparation, library preparation, and sequencing. Some examples include the following.
Sample preparation
Library preparation
User error (e.g., carry-over of DNA from one sample to the next, contamination from previous reactions)91
PCR amplification errors9
Primer biases (e.g., binding bias, methylation bias, mis-priming, non-specific binding, primer-dimer, hair-pins, interfering pairs, melting temperature too high/low)92,93
3’-end capture bias (poly-A enrichment protocols in RNA-Seq)94
Private mutations (e.g., repeat regions, mispriming over private variation)95
Machine failure (e.g., incorrect PCR cycling temperatures)15
Barcode/adapter errors (e.g., adapter contamination, lack of barcode diversity, incompatible barcodes, over-loading)16,96
Sequencing and imaging
User error (e.g., cluster crosstalk caused by flow cell overloading)97
Dephasing (e.g., incomplete extension, addition of multiple nucleotides instead of single nucleotide)3
Dead fluorophores, damaged nucleotides, and overlapping signals20
Sequence context (e.g., GC-richness, homologous and low-complexity regions, homopolymers)19,98,99
Machine failure (e.g., laser, hard-drive, software, fluidics)
Strand biases98
Sample Preparation
Sequencing error and bias can arise from sample degradation and contamination during sample isolation and preservation. For example, during sample preservation, formalin fixation causes degradation and nucleotide changes7,8. Also, inadequate amounts of high-quality genomic material can increase amplification errors and decrease sequencing read depth9. Finally, contamination poses a challenge when non-tumor cells mask oncogenic somatic variants10, or when exogenous DNA interferes with calls of homo- or heterozygosity11.
Library Preparation
Error also arises during sequencing library preparation, leading to uneven coverage, sequence changes, and interruption of sequence tags. DNA fragmentation can produce length biases, subsequently causing preferential amplification12. Library amplification is subject to unmeasured primer biases, such as primer bias in multiple displacement amplification (MDA)13, mispriming in PCR target enrichment14, and incorporation of sequence errors during clonal amplification and PCR cycling15. When barcodes, adapters, and other pre-defined sequence tags are added to the fragments being sequenced, disruption and inadequate tag design can result in cross-contamination of datasets, read-loss, and decreased read quality2,16. Chimeric reads also can arise in long-insert paired-end libraries17, potentially confounding variant calls and assembly efforts.
Sequencing and Imaging
Current NGS platforms3 have platform-specific sequencing and imaging error types18. For example, substitution error can arise in platforms like Illumina and SOLiD® when incorrect bases are introduced during clonal amplification of templates. Furthermore, Illumina has shown a sequence-specific error profile19 that possibly arises from single-strand DNA folding or sequence-specific alterations in enzyme preference. Pacific Bioscience’s SMRT platform yields long single-molecule reads that are subject to false indels from non-fluorescing nucleotides20,21. Pyrosequencing (e.g., Roche/454 platforms) and semiconductor sequencing (e.g., Ion Torrent) have difficulty counting homopolymer stretches, resulting in carry-forward, insertion and deletion errors22.
Experimental error poses challenges in applications for which accuracy is critical, such as detection of somatic mosaicism23,24 and other clinical applications. Error is often addressed by increasing sequencing read depth, but can also be mitigated by supplementing with careful barcoding strategies25, replicates, orthogonal sequencing technologies26 and knowledge of variant priors27. Together, these approaches can help overcome variations in experimental conditions, stochastic fluctuations, and systematic biases.
Replicates and Experimental Error
Many applications, such as the pursuit of rare causal variants, clinical applications, and somatic variant detection, require high fidelity in sequencing, necessitating confirmatory experiments, such as Sanger sequencing. The standard validation methods used for confirmation tend to be costly and labor-intensive, thus necessitating lower-cost alternatives. An approach that holds promise uses the tried-and-true scientific method of replication to mitigate user error, stochastic differences, and other sources of experimental error. Different types of replication are described below, including sequencing read depth, technical, biological and cross-platform replication.
Sequencing Read Depth
The most straightforward approach to improve sensitivity and accuracy in sequence variant calls is to increase sequencing read depth28,29. By increasing the number of short reads, one can improve variant calling on easily sequenced regions. Consequently, one can reduce the number of missed true variants (false negatives) and sometimes the number of true non-variants that are incorrectly detected as variants (false positives). However, merely increasing sequencing read depth cannot ameliorate issues arising from the wide-spread batch effect phenomenon30 and many other error types introduced in the experimental process. Thus, increased fold coverage is not necessarily an adequate proxy for biological replication and is limited in its ability to mitigate error.
Technical Replicates
The frequency of certain error types can be reduced through technical replication. We define technical replication as the repeat analysis of the exact same sample. For example, technical replicates were used with monozygotic twins, and the data exhibited higher intra-individual correlations than inter-individual correlations31. In another example6, many technical replicate pools were sequenced, each containing dilute DNA. Pools containing haplotypes with incongruent base calls that were suspicious for amplification errors were discarded, and the sequence quality was significantly improved.
Biological replicates
We define biological replication as the preparation and analysis of multiple biological samples under the same conditions from the same host. Biological replicates in genome sequencing can be employed to assess the efficacy of various bioinformatic filters32. Additional benefits gained over technical replicates include the identification of rare somatic mosaicism and differences in transcript abundance. Somatic mosaicism arises from mutations occurring from mutagens and other causes24. Replicates indirectly help uncover somatic mutations in complex and heterogeneous tumors when used to achieve the “normal” baseline sequence in tumor/normal pairs.
Cross-platform replicates
Each sequencing platform introduces unique biases and error types. Thus, integrating sequencing data from different technologies can further mitigate error. For example, sequencing both blood and saliva on two different platforms (Illumina, Complete Genomics), resulted in 88.1% concordance of SNVs across replicates33. Validation rates for variants called on both platforms were higher than variants that were not. In another study, sequencing on three platforms (Illumina, 454, and SOLiD®) showed 64.7% concordance5. This disparity could result from multiple experimental error sources, as well as differences in downstream bioinformatic processing. Cross-platform replicates greatly reduce the number of false positive variants, but the different biases from each sequencing platform may cause many true variants to be overlooked when comparing cross-platform replicates.
Reducing error and replicates
As sequencing further permeates science and medicine, replicates will be invaluable to researchers and clinicians alike. Current efforts in sequencing error mitigation rely mainly on filtering strategies, including filtering for sequencing read depth, base call quality, short read alignment quality, variant call quality, known variants, strand bias, allelic imbalance and sequence context10,21,25,27,34–37. All these post-processing techniques help reduce uncertainty in the final genotyping variant call (Figure 1b).
Bioinformatic filtering techniques can be optimized using technical, biological, and cross-platform replicates to improve specificity and sensitivity32. For example, optimal quality score thresholds for each filter may be selected using replicate genome sequences. An individual human genotype has roughly 3 million variants37; however variant callers can predict >20 million variants of differing quality per genome, mainly from mismapped short reads38, mosaicism, and sequencing error. Consequently, thresholds are chosen to limit the variants called in the individual’s genotype. Ideally, these thresholds are chosen with experimental confirmation39, but this can be costly. We assert that replicates can abet bioinformatic filtering and reduce the number of variants requiring validation, thereby improving the quality of the sequence being mapped or assembled.
To illustrate, we use biological replicates to conduct a simple analysis for assessing the reliability of single nucleotide substitution calls (Figure 2). For genotyping, the number of replicates should be chosen to attain adequate statistical power at the loci in question. However, here we seek a set of likely false positives stemming from experimental error, which thus requires only three replicates for a voting majority. For the replicates, we obtained sequence data from three distinct tissue samples of participant PGP1 in the Personal Genome Project40 (see Supplementary Notes).
Figure 2. Platform-independent method for choosing quality score thresholds using replicate sequencing data.
Variants are called for all replicates and then classified as concordant if the variant calls agree among the replicates or discordant if they differ. Variants are then rank-ordered by the desired metric (e.g., quality scores), and plotted similar to receiver-operator characteristic (ROC) curves. That is, the cumulative distributions of concordant and discordant variants are plotted left to right as the stringency of the confidence score of interest decreases.
Loci were identified in which one or more replicates contain a single nucleotide variant (SNV). In brief, SNV loci are deemed “concordant” when all replicate variant calls agree41, and SNV loci are called “discordant” when other replicates differ from a target replicate. Thus, concordant loci represent true positive variants, and discordant loci signal false positive variants. See the Supplementary Notes for precise definitions of concordance and discordance, for details on choosing a target replicate, and for implementation details.
Once discordant (false positive) and concordant (true positive) variants have been separated from each other, metrics of variant call confidence (e.g., quality scores or read depth) are used to rank-order the target variants. Using the rank-ordered sets, one can plot the accumulation rate for concordant and discordant variants with decreasing score stringency, in a representation similar to a receiver-operator characteristic (ROC) curve. Thus, variant call quality score thresholds can be chosen to maximize the proportion of all concordant variants seen at or below a particular threshold relative to the fraction of all discordant variants. This analysis (Figure 3) suggests that, while adequate read depth across the genome is essential28,29, read depth is not the best measure of reliability of a specific variant call at a particular locus. Indeed, read depth at a particular locus is an inferior filter when compared with error-model-based quality scores. We found that this holds true for quality scores computed by software packages that process genomic35 and expression27,36 data. Even after removing regions with abnormally high read depths (enriched for misalignment errors in low-complexity sequence38), quality scores considered here still outperform read depth as a filter for sequencing error.
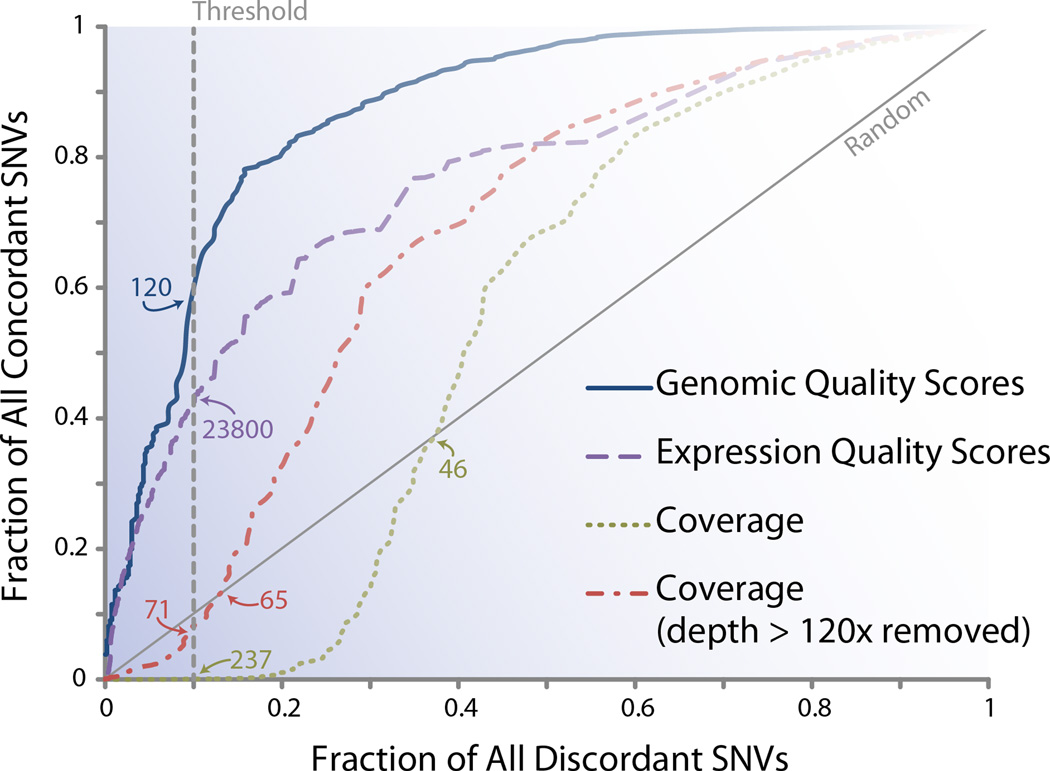
Figure 3. Plotting replicate scores to assess filter efficiency.
The efficiency of different variant call filter metrics can be evaluated by plotting replicate-based SNV concordance and discordance in a manner similar to a ROC curve. As one travels from left to right on the plot, the rank-ordered quality score is reduced in stringency and the fractions of retained concordant and discordant variants increase. Thus, this curve quantifies the proportion of good data (concordant SNVs) retained and bad data (discordant SNVs) discarded as a consequence of variable quality score cut-offs. For the genomes used in our analysis, this graph indicates that filtering variants solely based on locus read depth is inferior to filtering by genomic35 and expression27,36 quality scores35. Furthermore, filtering by expression data quality scores is also inferior to filtering by genomic quality scores (genomic quality scores from Complete Genomics Inc.), but nevertheless both are better than filtering loci by read depth. The read depth curve that excludes outliers (read depth higher than the 99.5th-percentile) outperforms the all-inclusive read depth curve. As an example of how to understand the value of a threshold, note that choosing a threshold score of 120 as a measure for highest quality for the genomic data will include the same fraction of total predicted errors as choosing a threshold quality score of 23800 for the expression data. Meanwhile, when a similar threshold is chosen for read depth, the efficiency at retaining true variants is worse than random.
In addition to comparing disparate error model quality scores, this approach can be used to evaluate the effect of manipulating quality score thresholds for a specific data set of interest. For example, sensitivity of a particular threshold can be evaluated by considering the false negative rate, as estimated by the number of concordant variants that are lost as a result of applying the threshold.
Post-processing Error in NGS
Even with replicates, some types of error cannot be addressed without further technological advances and improvements in bioinformatic processing. For example, insertions and deletions42 as well as paralogs and other repetitive sequence43 often confound NGS short read alignment44,45, resulting in mismapped reads and ultimately, variant call errors. Other sources of error can arise from limitations in software and configuration during secondary analysis, including read clipping and filtering46, allelic bias47, and variant call confidence models48. These cannot be addressed with replicates alone.
Erroneous variant calls also arise from incomplete reference data. This error type arises when reads are mapped to unfinished reference genomes/transcriptomes and drafts containing misassembled regions49. These errors will steadily decrease in frequency as reference genome assemblies and annotations such as GRChr3750 and RefSeq51 are completed and corrected with each new build release.
Lastly, strides in haplotype phasing hold promise not only for reducing amplification errors6, but also for reducing the causal variation search space. For example, only through accurate haplotype phasing can we begin to discern the difference between two dysfunctional gene copies (i.e., a double mutant) and a single normal copy52. This difference can have important implications with regard to phenotype and clinical applications of sequencing. Unfortunately, current mainstream NGS methods do not consistently discern between these two cases. Thus, ad hoc experimental6,53,54 and computational procedures55,56 are required to distinguish the haplotypes of diploid cells.
Concluding Remarks
In these past decades, amazing scientific and technologic advances have provided molecular-level resolution for the inner workings of life. NGS technologies are providing insights into genetic disease associations57–63, differences in human gut microbiota64, amino acid essentiality in proteins65, experimental evolution66–68, biotherapeutic development69–73, protein-DNA interactions74, epigenetics75, cancer genomics39,76 and clinical diagnosis77. Efforts to find biologically and clinically relevant variants are steadily improving as algorithmic advances more intelligently filter the large amounts of sequence data. For example, variants can be prioritized by considering heritability or variant association in populations61,78, correcting for gene-specific mutation rates10, accounting for evolutionary conservation79–81, and providing network context through systems biology approaches82–84. Beyond strictly biological applications, sequencing is also becoming an analytical tool for more esoteric questions, such as recording fluctuations in ion concentrations85 and even potentially detecting dark matter in astrophysics86. All these sequencing studies, however, are limited by the accuracy of the underlying sequencing experiments.
Here we have identified sources of sequencing error and presented a method for addressing the stochastic effects. Additional approaches to address other sources of error, such as experimental bias and software limitations, are also essential. These approaches include identifying erroneous SNPs exhibiting Hardy-Weinberg disequilibrium11, masking poor quality bases87, phasing and imputing variants in difficult-to-sequence regions or uncalled regions55 and improved methods for calling of structural variants, CNVs and indels. In conjunction with these computational approaches, the wise use of replicate genome sequencing will play an increasingly important role in reducing the noise in data processing and downstream analyses.
Supplementary Material
Acknowledgements
We offer a posthumous acknowledgement and sincere thank you to Dr. Tara Gianoulis for her feedback and inspiration. We would like to acknowledge Dr. Josée Dupuis, Professor of Biostatistics at Boston University, for her encouragement and feedback during the nascent stages of replicate analysis. We additionally would like to thank Dr. Wendell Jones, Global Head of Genomic Bioinformatics, Quintiles, and also Erik Aronesty, author of the popular ea-utils fastq processing package, for critical review of the manuscript. Some of this work was supported by the National Institutes of Health grant P50HG005550.
Glossary
- sequencing error
errors seen in the base call of the short reads from next-generation technology.
- sequencing read depth
the number of reads contributing to the variant call at a single location, a.k.a. read depth, fold coverage, depth of coverage. This term can also be used to refer to the average read depth across the entire targeted sequence area.
- short read
a short sequence of nucleotide bases and their respective quality scores, obtained via next-generation sequencing from a longer target sequence.
- misalignment
The alignment of a sequencing read to an incorrect location on a reference genome. This can occur when reads align equally well to multiple genomic locations due to indels, repeats, and low-complexity regions of the genome.
- multiple displacement amplification
(MDA) a technique used for amplifying DNA sequence by synthesizing DNA from random hexamer primers.
- barcode
a known DNA sequence appended to the ends of DNA fragments prior to sequencing for the purpose of pooling samples together to reduce cost.
- substitution error
when one base is substituted for another during sequencing.
- indel
a variant that is created by either the insertion or deletion of nucleotides with respect to a matching reference.
- homopolymer
a sequence of two or more consecutive, identical nucleotides.
- somatic mosaicism
genetic diversity among cells of a single organism.
- batch effect
the statistical bias of indeterminate cause observed in samples processed together with the same sample preparation, same library preparation and same sequencing experiment.
- base call
the identification of the nitrogenous base (A,G,C or T) added to the short read during sequencing.
- variant call error
an accumulation of misaligned reads, or of reads with base call errors over a particular locus, resulting in that locus being called variant when it truly matches reference, and vice-versa.
- read clipping
removal of adapter and barcode sequences or low quality bases near read ends following sequencing.
Biographies
Kimberly Robasky received her Ph.D. in bioinformatics from Boston University, with a research appointment in the Church Lab at the Department of Genetics at Harvard Medical School. Kimberly is currently Associate Director of Bioinformatics for Expression Analysis, a Quintiles Company, in Durham, NC.
Nathan E. Lewis obtained his Ph.D. in bioengineering at the University of California, San Diego, and has a degree in biochemistry from Brigham Young University. As a postdoctoral fellow at Harvard Medical School, he focused on using systems biology techniques to integrate disparate data types to discover functions of post-translational modifications and to infer gene regulatory networks in cell differentiation. He now is an assistant adjunct professor in the Division of Pediatric Pharmacology & Drug Discovery at the University of California, San Diego School of Medicine.
George Church is professor of genetics at the Harvard Medical School and the Wyss Institute for Biologically-Inspired Engineering. He also initiated the open-access Personal Genome Project, and co-developed many genomic sequencing, synthesis and computational technologies.
REFERENCES
- 1.O'Rawe J, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kircher M, Heyn P, Kelso J. Addressing challenges in the production and analysis of illumina sequencing data. BMC Genomics. 2011;12:382. doi: 10.1186/1471-2164-12-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. This review details many current sequencing technologies, including their strengths and limitations.
- 4.Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. 2011;12:125. doi: 10.1186/gb-2011-12-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ratan A, et al. Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PLoS One. 2013;8:e55089. doi: 10.1371/journal.pone.0055089. A thorough study of current error modes, coverage profiles and GC-biases of Next-generation technologies.
- 6. Peters BA, et al. Accurate whole-genome sequencing and haplotyping from 10 to 20 human cells. Nature. 2012;487:190–195. doi: 10.1038/nature11236. Highly accurate sequencing and haplotyping was achieved by fragmenting DNA from a few cells and separating fragments into hundreds of sequencing wells.
- 7.Williams C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yost SE, et al. Identification of high-confidence somatic mutations in whole genome sequence of formalin-fixed breast cancer specimens. Nucleic Acids Res. 2012;40:e107. doi: 10.1093/nar/gks299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE. Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn. 2005;7:36–39. doi: 10.1016/s1525-1578(10)60006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. A new approach to filter out variants in cancer that were likely non-causal
- 11.Leal SM. Detection of genotyping errors and pseudo-SNPs via deviations from Hardy-Weinberg equilibrium. Genet Epidemiol. 2005;29:204–214. doi: 10.1002/gepi.20086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 13.Hutchison CA, 3rd, Smith HO, Pfannkoch C, Venter JC. Cell-free cloning using phi29 DNA polymerase. Proc Natl Acad Sci U S A. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 15.Aird D, et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bystrykh LV. Generalized DNA barcode design based on Hamming codes. PLoS One. 2012;7:e36852. doi: 10.1371/journal.pone.0036852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform. 2010;11:484–498. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xuan J, Yu Y, Qing T, Guo L, Shi L. Next-generation sequencing in the clinic: Promises and challenges. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura K, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27:1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 21. Roberts RJ, Carneiro MO, Schatz MC. The advantages of SMRT sequencing. Genome Biol. 2013;14:405. doi: 10.1186/gb-2013-14-7-405. A detailed description of errors and strengths in the SMRT sequencing platform
- 22. Yang X, Chockalingam SP, Aluru S. A survey of error-correction methods for next-generation sequencing. Brief Bioinform. 2013;14:56–66. doi: 10.1093/bib/bbs015. A detailed review of error correction methods for sequencing data.
- 23.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurie CC, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C, Tsementzi D, Kyrpides N, Read T, Konstantinidis KT. Direct comparisons of Illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One. 2012;7:e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. Details the error model for a widely-used genotyper.
- 28. Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–1505. doi: 10.1101/gr.123638.111. Presents experimental and analytical methods for discerning adequate coverage.
- 29.Meynert AM, Bicknell LS, Hurles ME, Jackson AP, Taylor MS. Quantifying single nucleotide variant detection sensitivity in exome sequencing. BMC Bioinformatics. 2013;14:195. doi: 10.1186/1471-2105-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–739. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranzini SE, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reumers J, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat Biotechnol. 2012;30:61–68. doi: 10.1038/nbt.2053. Replicate sequencing was used to identify the optimal set of filters to remove false positives.
- 33. Lam HY, et al. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. The authors compared Illumina and Complete Genomics sequencing and variant calling accuracy for these platforms.
- 34.Jung H, Bleazard T, Lee J, Hong D. Systematic investigation of cancer-associated somatic point mutations in SNP databases. Nat Biotechnol. 2013;31:787–789. doi: 10.1038/nbt.2681. [DOI] [PubMed] [Google Scholar]
- 35.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 36.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelak K, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 40.Ball MP, et al. A public resource facilitating clinical use of genomes. Proc Natl Acad Sci U S A. 2012;109:11920–11927. doi: 10.1073/pnas.1201904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurie CC, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindgreen S. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes. 2012;5:337. doi: 10.1186/1756-0500-5-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degner JF, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genovese G, et al. Using population admixture to help complete maps of the human genome. Nat Genet. 2013;45:406–414. doi: 10.1038/ng.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Church DM, et al. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. doi: 10.1371/journal.pbio.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rusk N. One genome, two haplotypes. Nat Methods. 2011;8:107. doi: 10.1038/nmeth0211-107. [DOI] [PubMed] [Google Scholar]
- 53.Fan HC, Wang J, Potanina A, Quake SR. Whole-genome molecular haplotyping of single cells. Nat Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitzman JO, et al. Haplotype-resolved genome sequencing of a Gujarati Indian individual. Nat Biotechnol. 2011;29:59–63. doi: 10.1038/nbt.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bansal V, Bafna V. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics. 2008;24:i153–i159. doi: 10.1093/bioinformatics/btn298. [DOI] [PubMed] [Google Scholar]
- 57.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lupski JR, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 61.Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- 62.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 64.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robins WP, Faruque SM, Mekalanos JJ. Coupling mutagenesis and parallel deep sequencing to probe essential residues in a genome or gene. Proc Natl Acad Sci U S A. 2013;110:E848–E857. doi: 10.1073/pnas.1222538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conrad TM, Lewis NE, Palsson BO. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol. 2011;7:509. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shendure J, et al. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 68.Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet. 2013 doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X, et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lewis NE, et al. Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol. 2013;31:759–765. doi: 10.1038/nbt.2624. [DOI] [PubMed] [Google Scholar]
- 71.Brinkrolf K, et al. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol. 2013;31:694–695. doi: 10.1038/nbt.2645. [DOI] [PubMed] [Google Scholar]
- 72.Becker J, et al. Unraveling the Chinese hamster ovary cell line transcriptome by next-generation sequencing. J Biotechnol. 2011;156:227–235. doi: 10.1016/j.jbiotec.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 73.Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. The emerging CHO systems biology era: harnessing the 'omics revolution for biotechnology. Curr Opin Biotechnol. 2013 doi: 10.1016/j.copbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 74.Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meaburn E, Schulz R. Next generation sequencing in epigenetics: insights and challenges. Semin Cell Dev Biol. 2012;23:192–199. doi: 10.1016/j.semcdb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 76.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum Mol Genet. 2010;19:4313–4318. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schneeberger K, et al. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 79.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez-Perez A, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods. 2013;10:723–729. doi: 10.1038/nmeth.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis NE, Abdel-Haleem AM. The evolution of genome-scale models of cancer metabolism. Front Physiol. 2013;4:237. doi: 10.3389/fphys.2013.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ala-Korpela M, Kangas AJ, Inouye M. Genome-wide association studies and systems biology: together at last. Trends Genet. 2011;27:493–498. doi: 10.1016/j.tig.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 84.Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nat Rev Genet. 2012;13:523–536. doi: 10.1038/nrg3253. [DOI] [PubMed] [Google Scholar]
- 85.Zamft BM, et al. Measuring cation dependent DNA polymerase fidelity landscapes by deep sequencing. PLoS One. 2012;7:e43876. doi: 10.1371/journal.pone.0043876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Freese K, Lisanti M, Savage C. Annual modulation of dark matter: a review. arXiv preprint arXiv:1209.3339. 2012 [Google Scholar]
- 87.Hubisz MJ, Lin MF, Kellis M, Siepel A. Error and error mitigation in low-coverage genome assemblies. PLoS One. 2011;6:e17034. doi: 10.1371/journal.pone.0017034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Macabeo-Ong M, et al. Effect of duration of fixation on quantitative reverse transcription polymerase chain reaction analyses. Mod Pathol. 2002;15:979–987. doi: 10.1097/01.MP.0000026054.62220.FC. [DOI] [PubMed] [Google Scholar]
- 89.Kerick M, et al. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68. doi: 10.1186/1755-8794-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin MT, et al. Quantifying the relative amount of mouse and human DNA in cancer xenografts using species-specific variation in gene length. Biotechniques. 2010;48:211–218. doi: 10.2144/000113363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Innis MA, Gelfand DH, Sninsky JJ, White TJ. PCR protocols: a guide to methods and applications. Academic press; 1990. [Google Scholar]
- 92.Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kanagawa T. Bias and artifacts in multitemplate polymerase chain reactions (PCR) J Biosci Bioeng. 2003;96:317–323. doi: 10.1016/S1389-1723(03)90130-7. [DOI] [PubMed] [Google Scholar]
- 94.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pont-Kingdon G, et al. Design and analytical validation of clinical DNA sequencing assays. Arch Pathol Lab Med. 2012;136:41–46. doi: 10.5858/arpa.2010-0623-OA. [DOI] [PubMed] [Google Scholar]
- 96.Gogol-Doring A, Chen W. An overview of the analysis of next generation sequencing data. Methods Mol Biol. 2012;802:249–257. doi: 10.1007/978-1-61779-400-1_16. [DOI] [PubMed] [Google Scholar]
- 97.Whiteford N, et al. Swift: primary data analysis for the Illumina Solexa sequencing platform. Bioinformatics. 2009;25:2194–2199. doi: 10.1093/bioinformatics/btp383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 99.Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.