Abstract
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β superfamily, which utilize BMP receptors and intracellular SMADs to transduce their signals to regulate cell differentiation, proliferation, and apoptosis. Because mutations in BMP receptor type IA (BMPRIA) and SMAD4 are found in the germline of patients with the colon cancer predisposition syndrome juvenile polyposis, and because the contribution of BMP in colon cancers is largely unknown, we examined colon cancer cells and tissues for evidence of BMP signaling and determined its growth effects. We determined the presence and functionality of BMPR1A by examining BMP-induced phosphorylation and nuclear translocation of SMAD1; transcriptional activity via a BMP-specific luciferase reporter; and growth characteristics by cell cycle analysis, cell growth, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide metabolic as-says. These assays were also performed after transfection with a dominant negative (DN) BMPR1A construct. In SMAD4-null SW480 cells, we examined BMP effects on cellular wound assays as well as BMP-induced transcription in the presence of transfected SMAD4. We also determined the expression of BMPR1A, BMP ligands, and phospho-SMAD1 in primary human colon cancer specimens. We found intact BMP signaling and modest growth suppression in HCT116 and two derivative cell lines and, surprisingly, growth suppression in SMAD4-null SW480 cells. BMP-induced SMAD signaling and BMPR1A-mediated growth suppression were reversed with DN BMPR1A transfection. BMP2 slowed wound closure, and transfection of SMAD4 into SW480 cells did not change BMP-specific transcriptional activity over controls due to receptor stimulation by endogenously produced ligand. We found no cell cycle alterations with BMP treatment in the HCT116 and derivative cell lines, but there was an increased G1 fraction in SW480 cells that was not due to increased p21 transcription. In human colon cancer specimens, BMP2 and BMP7 ligands, BMPRIA, and phospho-SMAD1 were expressed. In conclusion, BMP signaling is intact and growth suppressive in human colon cancer cells. In addition to SMADs, BMP may utilize SMAD4-independent pathways for growth suppression in colon cancers.
Keywords: transforming growth factor-β, tumor suppressor
Bone Morphogenetic Protein (BMP) is a member of the transforming growth factor (TGF)-β superfamily known to regulate cell proliferation, apoptosis, and differentiation and to participate in the mesenchymal development of most tissues and organs in vertebrates. However, its role in epithelial growth regulation is not well understood. As part of the TGF-β superfamily, BMP utilizes a similar signaling cascade to that of TGF-β. BMP ligands use serine-threonine kinase receptors, type IA (BMPRIA), type IB (BMPRIB), and type II (BMPRII), to transmit their signal in the cell (7, 25, 36). BMP ligands bind either cooperatively to preformed receptor complexes or first to a BMPRI receptor that then recruits BMPRII (23). Additionally, BMPs can bind to activin type II (ACVR2) and type IIB (ACVR2B) receptors (29, 34, 38). BMP7 appears to preferentially bind to ACVRII and ACVRI but also has affinity for BMPRII, BMPRIA, and BMPRIB, whereas BMP2 and BMP4 appear to bind to BMPRIA and BMPRII preferentially (11, 30, 38). Upon ligand binding, BMPRII phosphorylates BMPRI, which, in turn, phosphorylates intracellular SMAD1, -5, or -8 at their COOH terminus (28). The phosphorylated (p)SMADs associate with SMAD4, and the complex translocates to the nucleus to regulate the expression of various genes that control cell proliferation, cell differentiation, and apoptosis (2).
BMPs and their receptors have been linked to the pathogenesis of some solid tumors. BMPRIA, BMPRII, and BMP2 mRNA levels have been found upregulated in pancreatic cancers, 55% of which have biallelic loss of SMAD4 (32, 39). BMP2 might promote pancreatic cancer progression by enhancing the growth of pancreatic cancer cells (6, 24). mRNAs for BMP receptors and BMP ligands are greater in cells with high metastatic potential than in cells with less metastatic potential (1). However, BMP2 inhibits the proliferation of breast cancer cell lines that express both SMAD1 and SMAD4 (CAMA-1, MCF7, MDA-MB-231, T-47D, and ZR-75-1) and upregulates the cyclin-dependent kinase inhibitor p21WAF1 in these cells (33). BMP2 has also been shown to inhibit proliferation and cause cell cycle arrest in the G1 phase of MKN74 gastric cells (37). Growth inhibitory effects of BMP2 have also been seen in the androgen-sensitive prostate cancer cell line LNCAP, but no effect was found in androgen-insensitive PC-3 and DU145 cell lines (3).
Recently, Hardwick et al. (14) found that BMP2 inhibits normal colonic epithelial cell growth by promoting apoptosis and differentiation and inhibiting proliferation. They also found that BMP2, BMPRIA, BMPRIB, BMPRII, pSMAD1, and SMAD4 are expressed predominantly in mature colonocytes at the epithelial surface in normal adult human and mouse colon tissue samples (14).
Juvenile polyposis syndrome (JP) is an autosomal dominant gastrointestinal hamartomatous polyposis syndrome that increases the afflicted patient's risk for developing colon cancer by ~12-fold. Germline mutations in the tumor suppressor SMAD4 and BMPRIA have been described in JP patients (18, 40). A small percentage of JP kindreds have also shown germline mutations in PTEN (20). Four common BMPRIA germline mutations have been described in JP leading to premature stop codons and subsequent inactivation of BMPRIA (17). Recently, Howe et al. (19) found that of 77 JP cases, germline SMAD4 mutations accounted for 18.2%, whereas germline BMPRIA mutations accounted for 20.8%, of the JP cases. Additionally, Haramis et al. (13) found that inhibition of BMP signaling by conditional knockout of BMPRIA resulted in the formation of numerous ectopic crypts that mimic the intestinal histopathology of JP.
Colon cancer develops as a result of uncontrolled cellular proliferation and dysregulation of cell death mechanisms, and inactivation of TGF-β superfamily signaling appears to play a key role (4). Inactivation of TGF-β signaling occurs in ~80% of colon cancers (10). Inactivation of activin signaling via mutations in ACVR2, another TGF-β superfamily receptor, occurs in the majority of colon tumors with microsatellite instability (MSI) (16, 22). Because patients with the colon cancer predisposition syndrome JP develop germline mutations in key BMP signaling molecules, and because the effects of BMP signaling in colon cancer are largely unknown, we aimed to answer two questions: 1) is BMP signaling disrupted in colon cancer like its TFG-β and activin counterparts; and 2) if not, does BMP signaling confer growth control in colon cancer cells? We found that BMP signaling is intact in human colon cancer specimens and in several cell lines and is moderately growth suppressive.
MATERIALS AND METHODS
Cell lines
We utilized the cell lines listed in Table 1 to investigate whether the BMP pathway was intact. These include the HCT116 cell line, which is a MSI cell line due to biallelic mutations in human mutL homolog 1 (hMLH1) and has mutations in TGF-β receptor type 2 (TGFBR2) and ACVR2 (5). We also utilized HCT116 cells complemented with chromosome 3 (HCT116 + chr3), which are microsatellite stable (MSS) at single base mispairs due to hMLH1 complementation and have wild-type TGFBR2 restored (5, 26). We also used HCT116 cells complemented by chromosome 2 transfer (HCT116 + chr2), which demonstrate MSI and have ACVR2 restored as well as an additional copy of BMPRII (21, 26). Additionally, we used SW480 cells, which are MSS and are SMAD4 null (8, 12).
Table 1.
Genetic characteristics of the various cell lines used in this study
| Cell Line | Microsatellite Status | Key Affected Genes | Key Restored Genes |
|---|---|---|---|
| HCT116 | MSI-H | ACVR2, TGFBR2, hMLH1 | |
| HCT116 + chr3 | MSS | ACVR2 | TGFBR2, hMLH1 |
| HCT116 + chr2 | MSI-H | TGFBR2, hMLH1 | ACVR2 |
| SW480 | MSS | SMAD4 |
MSI-H, high microsatellite instability; MSS, microsatellite stable; ACVR2, activin receptor 2; TGFBR2, transforming growth factor (TGF)-β receptor type 2; hMLH1, human mlh homolog 1.
Cell culture
The HCT116 cell line, its derivatives, and the SW480 cell line were maintained in Iscove's modified Dulbecco's medium (IMDM; Invitrogen; Carlsbad, CA) with 10% FBS and penicillin G-streptomycin (Invitrogen). G418 was added to cultures of HCT116 + chr2 and HCT116 + chr3 to maintain the transferred chromosome.
Nuclear/cytoplasmic fractionation, immunoprecipitation, and immunoblot analysis
We separated the nuclear and cytoplasmic fractions with hypotonic lysis buffer [10 mM Tris·HCl (pH 7.4), 3 mM MgCl2, 0.2% Nonidet P-40 (NP-40), 0.1 mM EDTA, 40 mM NaF, 5 mM glycerophosphate, 10 μg/ml aprotinin, 5 μg/ml leupeptin, 1 mM sodium orthovanadate, and 0.5 mM PMSF] and RIPA buffer [1% NP-40, 0.1% SDS, 1% deoxycholic acid, 0.15 M NaCl, 50 mM Tris·HCl (pH 7.2), and 1 mM PMSF]. Immunoprecipitation of the receptors was performed by overnight incubation by rotary rotation with BMPRIA antibody with 500 μg of cell extract at 4°C. Protein agarose A beads (Upstate; Lake Placid, NY) were added, and the mixture was further rotated for 3 h at 4°C. After the mixture was washed with PBS, the antibody-protein complex was denatured at 100°C for 5 min, and the protein was loaded onto an 8.5% polyacryl-amide gel. After electrophoresis, proteins were transferred onto a nylon membrane, blocked for 1 h with 5% milk, and probed overnight with primary antibodies at 4°C. Blotting was done with antibodies to BMPRIA (1:150), total SMAD1 (1:300), SMAD4 (1:400), histone (1:1,000; SC 5676, SC 7965, SC 7966, and SC 8030, respectively, Santa Cruz Biotechnology; Santa Cruz, CA), and pSMAD1 (1:400; 06-702, Upstate). The next day, several 0.1% PBS-Tween (PBS-T) washes were performed along with the appropriate secondary antibody incubation. Blotted proteins were detected with horseradish peroxidase-linked secondary antibodies (Sigma; St. Louis, MO) followed by enhance chemiluminescence detection (Amersham; Little Chalfont, UK).
Luciferase assays
Transient transfection of colon cancer cells with the BMP-responsive element (BRE)-luciferase (Luc) plasmid (a gift from Dr. Peter ten Dijke, Netherlands Cancer Institute, Amsterdam, The Netherlands) was done to assess the effects of BMP on BMP-specific transactivation. The pWWP-luc plasmid (a gift from Burt Vogelstein, Johns Hopkins Univerisity, Baltimore, MD) was transfected to assess the effects of BMP on p21WAF1 transactivation. Reporter vectors (0.75 μg/ml) and the pRL-TK vector (20 ng/ml) were transiently delivered by Transfectin (Promega; Madison, WI) in 12-well plates with a ratio of 3:1 of vector to transfection reagent in OPTI-MEM reduced serum-free media (GIBCO; Carlsbad, CA). Two hours posttransfection, 1 ml of complete media was added per well, and, 12–16 h posttransfection, cells were treated with 50 ng/ml BMP2 or BMP7. Luc activity was measured by a dual-Luc kit (Promega) 20–24 h after the treatment, and normalization was performed using the Renilla Luc activity expressed by the cotransfected pRL-TK vector.
SMAD4 and dominant negative and constitutively active BMPR1A transfections
SMAD4, dominant negative (DN) BMPR1A, or constitutively active (CA) BMPR1A were transiently delivered by Transfectin (Promega) at a ratio of 3:1 of vector to transfection reagent using 1 μg/ml SMAD4 vector (a generous gift from Dr. Masayuki Funaba, Azabu University School of Vetinary Medicine, Azabu, Japan) or 3 μg/ml DN BMPR1A or CA BMPR1A in OPTI-MEM reduced serum-free media (GIBCO). After 2–3 h, IMDM with FBS and penicillin G-streptomycin was added to the transfected cells. At 2 h posttransfection, complete media were added and later used in the experiments.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
The effect of BMP treatment on cell growth was assessed by using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)-based metabolic assay. Cells were seeded in 48-well plates at a density of 10–20,000 cells/well in 0.4 ml culture medium supplemented with BMP2 or BMP7 (100 ng/ml). Metabolic activity, corresponding to growth, was assayed after 2 or 4 days of incubation at fixed intervals for MTT-dependent absorbency. For this, cells were stained for 3 h with MTT dye, the reaction product released by lysis with SDS, and absorbency was detected at a wavelength of 570 nm using a Beckman-Coulter DU640B spectrophotometer (Beckman-Coulter; Fullerton, CA).
Cell counting for growth
Cells were seeded at a density of 10,000 cells/well and, 24 h later, were treated with 100 ng/ml BMP2 or BMP7 in the presence of FBS. After 48 and 96 h, cells were lysed in 0.5 ml of 0.05% trypsin and counted using a hemacytometer.
Wound closure assay
Cells were plated in six-well plates and grown to 95% confluency. A wound was then created in the shape of a cross with a plastic pipet tip. Media were replaced with fresh media, and cells were photographed using a Carl Zeiss Axiovert 200 microscope (Carl Zeiss; Thornwood, NY). Cells were then treated with 100 ng/ml BMP2 or BMP7 or 300 ng/ml Noggin. Every 18 h thereafter, the media were refreshed, and cells were photographed and retreated. All pictures were taken from the same location on the plates at the top of the intersection of the cross.
Total RNA extraction and semiquantitative RT-PCR
Total RNA extraction was performed using TRIzol reagent (Invitrogen). Cells grown on six-well plates were lysed with TRIzol (1 ml/well), combined with chloroform, and mixed. Supernatants were then precipitated with isopropanol, and RNA pellets were washed with 75% ethanol, air dried, and then resuspended in water. Two micrograms of total RNA were converted into cDNA by reverse transcriptase and amplification of BMP2 and BMP7 (SuperScript II, Invitrogen). Briefly, after inactivation at 65°C for 10 min, 1 μl of the reaction mixture was incubated in buffer containing 0.2 mM concentrations of dATP, dCTP, dGTP, and dTTP; 0.2 μM concentrations of each of the oligonucleotide primers; 3 mM MgCl2; and 10× buffer consisting of 200 mM Tris·HCl (pH 8.0), 500 mM KCl, and 1 unit Taq polymerase. The following primers were designed to amplify BMP2 and BMP7: BMP2, forward 5′-CCCAGCGTGAAAAGAGAGAC-3′ and reverse 5′-GAGACCGCAGTCCGTCTAAG-3′; and BMP7, forward 5′TCGTGGAACATGACAAGGAA-3′ and reverse 5′-CTGATCCGGAACGTCTCATT-3′. Primers for p21/Waf1 were as follows: forward 5′-CAGGGGACAGCAGAGGAAGA-3′ and reverse 5′TTAGGGCTTCCTCTTGGAGAA-3′. GAPDH served as a loading control (forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse 5′-TCCACCACCCTGTTGCTGTA-3′). PCR was performed as follows: denaturation at 95°C for 3 min and 35 cycles of 94°C for 30 s, 55°C for 30 s, and 74°C for 4 min for BMP7 and GAPDH; denaturation at 95°C for 3 min and 40 cycles of 94°C for 30 s, 57°C for 30 s, and 74°C for 4 min for BMP2; and denaturation at 95°C for 3 min and 40 cycles of 94°C for 30 s, 55°C for 30 s, and 74°C for 4 min for p21.
Cell cycle analysis
Cells were grown on 10-cm dishes until 50% confluent. After 24 h of serum starvation, 10% serum was added containing 50 ng/ml BMP2 or BMP7 for 48 h. Cells were harvested using 0.05% trypsin, washed with PBS, and resuspended in 0.6 ml of PBS and 1.0 ml of 100% ethanol. Cells were fixed overnight at 4°C. The next day, cells were centrifuged, washed in PBS, and centrifuged again for 5 min. The pellet was resuspended in 0.5 ml of PBS solution containing 40 units/ml RNase A and 50 μg/ml propidium iodide, incubated at 37°C for 1 h, and then placed on ice until analysis with a Beckman-Coulter Elite Flow Cytometer Multicycle (Phoenix Flow; San Diego, CA).
Propidium iodide viability
For viability assays, cells were serum starved 30 min and then treated with 50 ng/ml ligand for 48 h. Cells were then harvested with 0.05% trypsin, centrifuged at 1,500 rpm for 5 min, and resuspended in 0.3 ml PBS. Propidium iodide (10 μg/ml) was added to the cells, and cells were placed on ice until analysis with a Beckman-Coulter Elite Flow Cytometer (Phoenix Flow).
Immunohistochemical analysis
Under institutional review board approval (University of California-San Diego Protocol No. 050958XT), 10 random MSS and 3 random MSI slides from a population-based study (35) containing colon cancer tissue were deparaffinized in xylene and rehydrated in graded alcohols to water. Slides were immersed in sodium citrate buffer (pH 6.0) and heated in a microwave for 4 min for four times for antigen retrieval. Slides were then processed using a DAKO Signal Catalyzed Amplification System (DAKO; Carpinteria, CA). Endogenous peroxidase activity was blocked by incubation with 3% H2O2. Goat serum (10%) was added for 15 min to block nonspecific protein binding. Slides were incubated overnight with primary antibody [BMPRIA (1:150, SC 5676, Santa Cruz Biotechnology), BMP2 (4.5 μg/ml), BMP7 (25 μg/ml, AF 355 and AF354, R&D Systems; Minneapolis MN), pSMAD1 (1:100, AB 3848, Chemicon; Temecula CA)] and then rinsed with 0.1% PBS-T. Biotinylated secondary antibody was added for 15 min, followed by an incubation with peroxidase-labeled streptavidin for 15 min at room temperature. Sections were washed with PBS-T, incubated with diaminobenzidine and H2O2 for 1 min, lightly counterstained with hematoxylin, dehydrated in graded alcohols, cleared in xylene, and coverslipped. All sections and immunostains were reviewed by a single gastrointestinal pathologist.
Statistical analysis
Statistical significance was determined using either the Student's t-test or two-factor without replication ANOVA. P values of <0.05 were considered to be significant.
RESULTS
The BMPRIA receptor is present and SMAD signaling is intact in the majority of colon cancer cells
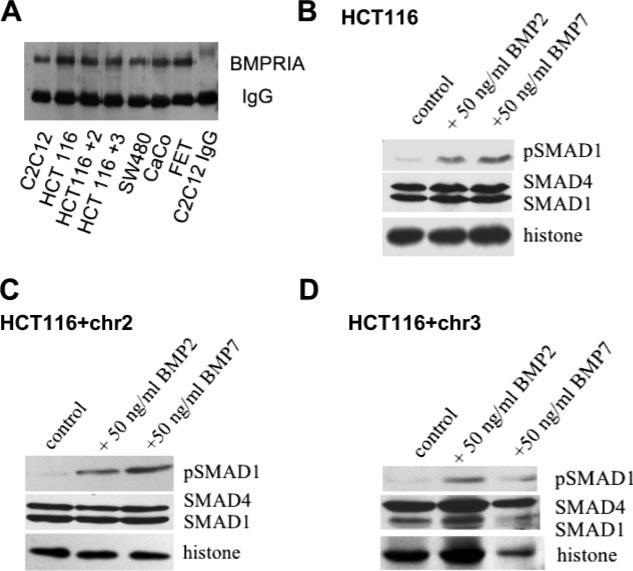
To investigate whether the BMP pathway is perturbed in colon cancer cell lines, we first sought to examine whether the BMPRIA receptor was present, because this receptor is often mutated in colon cancer-prone JP patients. All of the colon cancer cell lines examined expressed BMPRIA at the protein level (Fig. 1A). We next examined whether the receptor was functional. We treated cells with BMP2 or BMP7 and examined SMAD1 phosphorylation using a phospho-specific SMAD1 antibody after nuclear and cytoplasmic fractionation. In response to BMP ligand, the HCT116, HCT116 + chr2, and HCT116 + chr3 cancer cell lines phospohorylated and translocated pSMAD1 to the nucleus. Total SMAD1 and SMAD4 were also present in both the cytoplasm and nucleus (Fig. 1, B–D).
Fig. 1.
Bone morphogenetic protein (BMP) receptor type IA (BMPRIA) and SMAD protein expression. A: immunoprecipitation of BMPRIA in various colon cancer cell lines. The C2C12 mouse cell line is used as a positive control. FET and CaCo are additional colon cancer cell lines. B–D: nuclear extracts of HCT116 (B), HCT116 + chr2 (C), and HCT116 + chr3 (D) cell lines treated with BMP2 or BMP7. Membranes were blotted for phosphorylated (p)SMAD1, SMAD1, and SMAD4. Histone was used as a loading control for nuclear fractionations.
BMP-induced specific transcription is intact in colon cancer cells
Transcriptional activation due to BMP2 or BMP7 stimulation was determined with the use of the BMP-specific SMAD-induced Luc reporter BRE-Luc (a generous gift from Dr. Peter ten Dijke) (27). Transcriptional activation by BMP increased over untreated controls but varied among the cell lines: in HCT116 and HCT116 + chr3 cells, transcriptional activity increased 3- to 5-fold, whereas in HCT116 + chr2 cells, transcriptional activity increased 8- to 13-fold (Fig. 2, A and B). Nontransfected and mock-transfected SMAD4-null SW480 cells did not exhibit increased transcriptional activity to BMP2 or BMP7 over untreated controls (Fig. 2C). To determine the effect of reconstituting the SMAD pathway on BMP signaling, we transfected SMAD4 into SW480 cells. SMAD4-transfected SW480 cells showed a modest increase in BRE-Luc activation when treated with ligand over control; however, there was strong basal transcriptional activation of SMAD signaling in the absence of exogenous BMP, suggesting an autocrine system in these cells with stimulation of BMP receptors (Fig. 2C). When SMAD4-transfected SW480 cells were treated with 100 ng/ml Noggin, a BMP-specific inhibitor, there was reversal of BMP-induced transcriptional activity in these cells, confirming the high endogenous BMP activity. To further confirm the endogenous production of BMP2 and BMP7, we treated SW480 cells with BMP2, BMP7, and Noggin for 1, 6, 24, and 48 h and then performed RT-PCR on SW480 cells. SW480 cells produced both endogenous BMP2 and endogenous BMP7 at the transcriptional level (Fig. 2D). We did not observe transcriptional changes in endogenous BMP2 and BMP7 expression with ligand or inhibitor (Noggin) treatment.
Fig. 2.
BMP-induced transcriptional activity in colon cancer cells. A and B: fold induction of BMP2 (A) or BMP7 (B) treatment over no treatment (control, CNT) of BMP-induced SMAD transcriptional activity in HCT116 cell line derivatives. C: relative amount of SMAD-induced transcriptional activity in SW480 cells with and without SMAD4 transfection. D: RT-PCR of SW480 cells for basal (endogenous) BMP2 and BMP7 expression and after treatment with BMP2, BMP7, or Noggin. GAPDH was used as a control.
BMP ligands are moderately growth suppressive in colon cancer cells
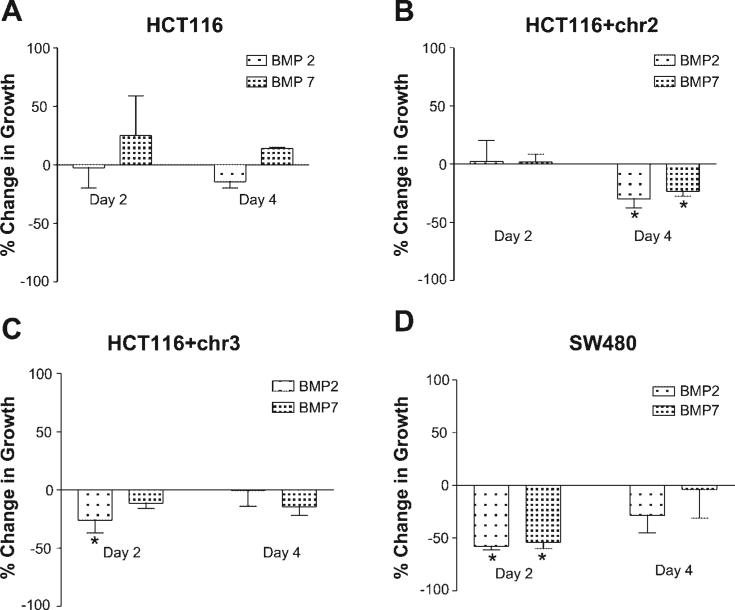
Cell growth was indirectly evaluated by MTT assay and directly by cell counting. HCT116 + chr2 and HCT116 + chr3 cells demonstrated modest but significant decreases in growth when treated with 100 ng/ml BMP2 or BMP7 for 48 h, as assessed by the MTT assay (Fig. 3). HCT116 cells also demonstrated significant decreases in growth when treated with BMP7 but not with BMP2. Although transcriptional activity was unchanged by exogenous BMP treatment, SW480 cells also showed significant growth suppression with BMP without the presence of SMAD4 (Fig. 3). Direct cell counting of the cell lines after BMP treatment showed decreased growth of BMP-treated cells compared with untreated cells. We observed inhibition after 2 days of growth in HCT116 + chr3 and SW480 cells with either BMP2 or BMP7 ligand but in not HCT116 or HCT116 + chr2 cells. HCT116 cells after BMP2 and HCT116 + chr2 cells after BMP2 and BMP7 showed decreased cell growth after 4 days of ligand treatment (Fig. 4, A–D). Only HCT116 cells treated with BMP7 had some modest growth enhancement, which was reduced by day 4. Overall, our data were consistent with the MTT results.
Fig. 3.
Effect of BMP2 or BMP7 treatment on cell growth as assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in HCT116, HCT116 + chr2, HCT116 + chr3, and SW480 cells. Two-factor with replications ANOVA was used to determine P values: *P < 0.05 and **P < 0.01.
Fig. 4.
Effect of BMP2 or BMP7 treatment on cell growth as assessed by cell counting in HCT116 (A), HCT116 + chr2 (B), HCT116 + chr3 (C), and SW480 (D) cells after 2 and 4 days of BMP2 and BMP7 treatment.
DN BMPR1A reverses transcriptional activity and growth inhibitory effects of BMP ligands
We transfected HCT116 + chr2, HCT116 + chr3, and SW480 cells with a DN BMPRIA construct and compared the results with a transfected CA BMPR1A vector or mock vector (15). Posttransfection, we treated the cells with BMP2 or BMP7 and assayed transcription via the BRE-Luc assay. Transcriptionally, DN BMPR1A reduced the effect of BMP ligands compared with mock and CA BMPR1A transfections in HCT116 + chr2 and HCT116 + chr3 cell lines (Fig. 5, A and B). In SMAD4-null SW480 cells, DN BMPR1A transfection demonstrated no difference in transcriptional activity over mock transfection (Fig. 5C). However, when DN BMPR1A was cotransfected with SMAD4, BMP2-induced transcriptional activity was reduced by 50% compared with SMAD4 transfectants alone, although it did not reduce to the levels seen with Noggin treatment. Thus DN BMPR1A trasfection reduces BMP-SMAD-mediated transcription in our cell models.
Fig. 5.
Relative amount of SMAD-induced transcriptional activity after BMP ligand treatment and the effect of dominant negative (DN) BMPR1A transfection. In both HCT116 + chr2 (A) and HCT116 + chr3 (B) cells, DN BMPR1A transfection reduced BMP-induced SMAD transcriptional activity. Constitutively active (CA) BMPR1A is shown as a positive control in the absence of ligand treatment. In SMAD4-null SW480 cells (C), DN BMPR1A reduced endogenous (CNT) and BMP2-induced SMAD transcriptional activity when cotransfected with SMAD4.
DN BMPR1A reversed BMP-induced growth-suppressive effects in our cell models. In both HCT116 + chr2 and HCT116 + chr3 cells, CA BMPR1A was more effective in reducing growth than exogenous BMP2 or BMP7 ligand treatment. However, the presence of DN BMPR1A attenuated or reversed BMP-induced growth suppression in both of these cell lines (Fig. 6, A and B). In SMAD4-null SW480 cells, the presence of DN BMPR1A completely reversed BMP2-induced growth suppression to levels similar with cells treated with the BMP inhibitor Noggin (Fig. 6C). Thus we showed reversal of BMP-induced growth suppression when signaling through BMPR1A was impaired, indicating that this receptor is a gateway for growth suppression.
Fig. 6.
DN BMPR1A transfection reversed BMP-induced growth suppression as assessed by MTT assay in HCT116 + chr2 (A), HCT116 + chr3 (B), and SW480 (C) cells. Two-factor with replications ANOVA was used to determine P values: *P < 0.05 and **P < 0.01.
BMP2 decreases wound closure by SW480 cells
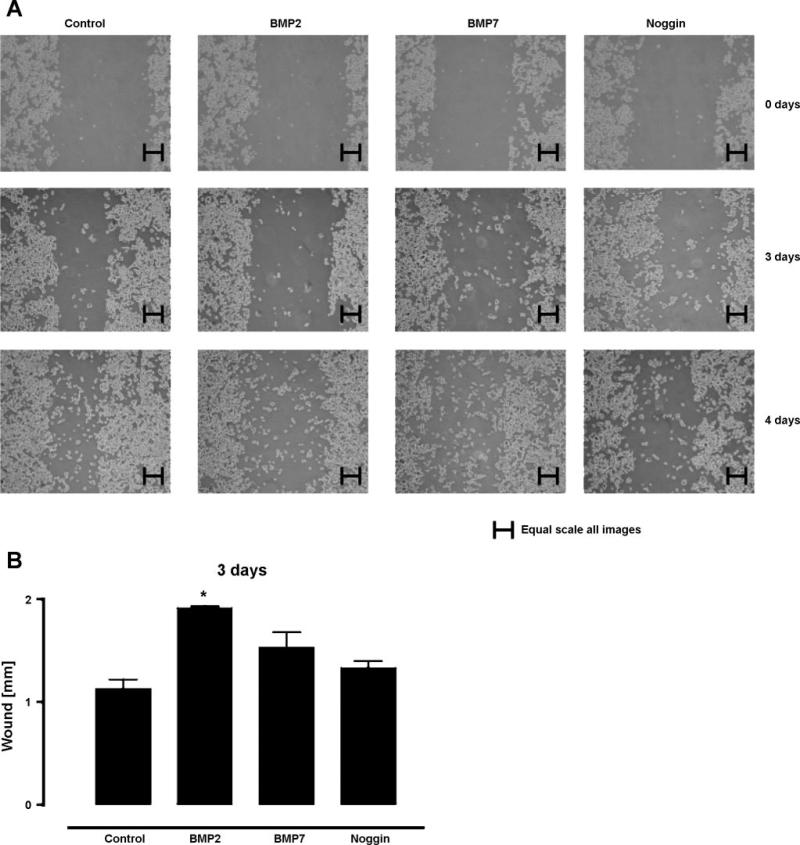
We performed a wound closure assay on SW480 cells treated with BMP2, BMP7, and Noggin. We found that untreated cells were able to fill into the wound gap, whereas BMP2 inhibited wound closure by SW480 cells. BMP7 also inhibited wound closure by SW480 cells but to a lesser extent than BMP2 did. Noggin, a BMP ligand inhibitor, did not inhibit wound closure by SW480 cells to any significant extent (Fig. 7, A and B).
Fig. 7.
Wound closure assay of untreated (control) SW480 cells and those treated with BMP2, BMP7, or Noggin. A: representative pictures for days 0, 3, and 4. B: bar graph of SW480 wound closure at day 3.
BMP2 and BMP7 and alteration of the cell cycle
We examined the functional effects of BMP treatment on the cell cycle in colon cancer cell lines. Despite intact BMP signaling and growth suppression, none of the HCT116 cell lines examined demonstrated significant alteration of the cell cycle (Table 2). However, SW480 cells demonstrated an increase in the percentage of cells in the G1 phase when treated with BMP2 and a significant increase when treated with BMP7. Because p21 has been shown to be upregulated by BMPs in some model systems, we further examined whether p21WAF1 was transcriptionally activated by BMP in cell lines by transfecting pWWP-Luc, a p21WAF1-specific reporter plasmid. We found no up-regulation in transcriptional activity of p21WAF1 upon stimulation with BMP2 or BMP7 over controls in HCT116, HCT 116 + chr2, HCT116 + chr3, and SW480 cells. Indeed, in particular, SW480 cells showed a slight reduction in p21 transcription over baseline with BMP7 treatment but no increase, as would be expected by the degree of growth suppression (Fig. 8A). We also examined endogenous p21 mRNA expression by RT-PCR after BMP2 and BMP7 treatement and found no change in p21 mRNA levels in any of the cell lines examined (Fig. 8, B–E). In SW480 cells, Noggin may have increased p21 expression in this semiquantitative experiment. Additionally, neither BMP2 nor BMP7 stimulation caused significant changes in cell survival as examined by propidium iodide viability in any of the cell lines examined (data not shown).
Table 2.
Cell cycle analysis from flow cytometric data on various colon cancer cell lines
| Cell Cycle Phase |
|||
|---|---|---|---|
| Cell Line | G1 | S | G2 |
| HCT 116 | |||
| Control | 66.17 | 18.03 ± 0.97 | 15.8 ± 0.6 |
| +BMP2 | 69.93 | 16.3 ± 0.93 | 13.73 ± 0.53 |
| +BMP7 | 70.33 | 16.63 ± 0.87 | 13.1 ± 0.57 |
| +TGF-β | 68.7 | 18.2 ± 0.9 | 13.1 ± 0.5 |
| HCT 116 + chr2 | |||
| Control | 78.57 | 8.7 ± 0.77 | 12.7 ± 0.67 |
| + BMP2 | 79.13 | 8.86 ± 1 | 11.96 ± 0.63 |
| + BMP7 | 78.97 | 8.53 ± 0.63 | 12.56 ± 0.43 |
| +TGF-β | 79.80 | 7.4 ± 1.3 | 12.7 ± 0.9 |
| HCT 116 + chr3 | |||
| Control | 69.10 | 14.83 ± 1.43 | 12.73 ± 0.93 |
| + BMP2 | 68.53 | 16.83 ± 1.4 | 14.6 ± 1.03 |
| + BMP7 | 69.43 | 18.23 ± 1.13 | 12.33 ± 0.7 |
| +TGF-β | 79.8* | 7.4 ± 1.3* | 12.7 ± 0.9 |
| SW480 | |||
| Control | 41.73 | 43.06 ± 1.43 | 15.23 ± 0.7 |
| + BMP2 | 44.67 | 42.6 ± 1.93 | 12.73 ± 0.9 |
| + BMP7 | 48.03* | 41.53 ± 1.6* | 10.43 ± 0.83* |
| +TGF-β | 47.4* | 42.9 ± 1.4 | 9.7 ± 0.6* |
Values are means ± SE expressed as the percentage of cell in a given cell cycle phase. BMP, bone morphogenetic protein.
P < 0.01.
Fig. 8.
Effect of BMPs on p21 transcription. A: results of p21 luciferase assay using pWWP-luc plasmid on HCT116, HCT116 + chr2, HCT116 + chr3, and SW480 cell lines with BMP2 and BMP7 treatment. B–E: results of p21 mRNA expression in HCT116 (B), HCT116 + chr2 (C), HCT116 + chr3 (D), and SW480 (E) cells after treatment with BMP2, BMP7, and Noggin.
Human colon cancer specimens exhibit intact BMP signaling components
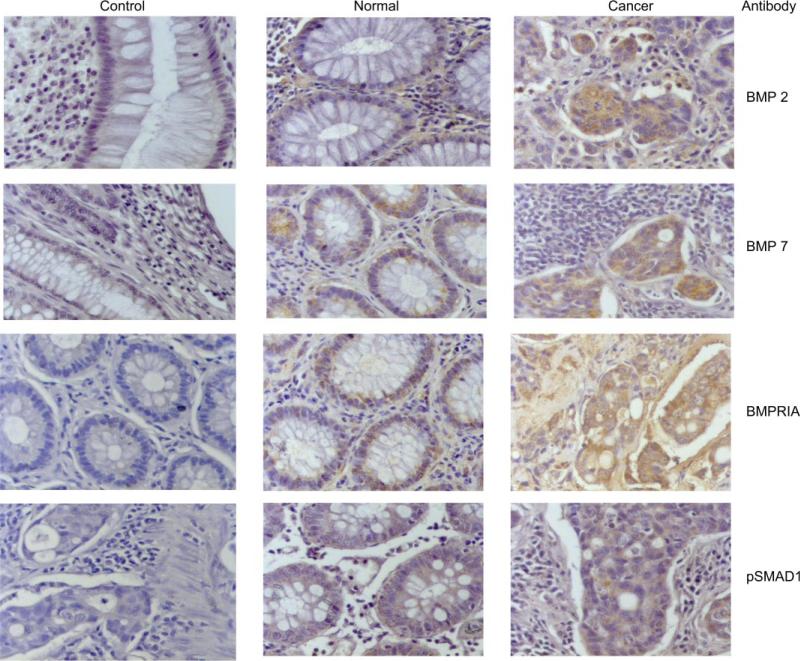
To assess the status of BMP signaling in primary human colon cancer specimens, we utilized immunohistochemistry to examine the presence or absence of signaling molecules. We examined 13 primary colon cancer tissues: 3 high-MSI (MSI-H) and 10 MSS tissues. All of the cancers (and paired normal colonic tissue) expressed pSMAD1, BMPRIA, BMP2, and BMP7. Representative pictures of the immunohistochemical stains are shown in Fig. 9.
Fig. 9.
Representative pictures of immunohistochemistry on 4 paired human colon cancer specimens stained for BMP2, BMP7, BMPRIA, and pSMAD1. Controls represent stained samples without primary antibody. Intestinal hamartomatous polyps with and without mutations in BMPR1A were used as additional controls (not shown). Magnification: ×40.
DISCUSSION
BMPs are known to play a role in tissue development, but, until recently, little work has been done examining the significance of BMP signaling in cancer. Work by several groups has previously shown BMP signaling to affect epithelial cell growth (3, 14, 33). TGF-β and activin ligands from the same superfamily as BMP are known growth suppressors and have shown to be inactivated in subsets of colon cancers (9, 16, 22, 31). BMP signaling can be inactivated by a germline mutation of BMPRIA in the colon cancer predisposition syndrome JP (17, 40). Taken together, these facts led us to hypothesize that the BMP pathway might be affected in colon cancer cell lines and human colon cancer specimens without JP. In this study, we investigated several colon cancer cell lines for the presence of intact BMP2 and BMP7 signaling and whether growth was affected by ligand treatment.
We determined that the BMPRIA receptor was present in all of the colon cancer cell lines and tissues examined. Thus BMPRIA is expressed in colon cancers as opposed to some polyps from JP patients. To further evaluate whether the BMPRIA receptor was active, we found that SMAD1 was phosphorylated and was transported to the nucleus in the colon cancer tissues and the HCT116 cell line and its derivatives. Furthermore, we utilized a Luc reporter vector specific for BMP-induced SMAD signaling to confirm that BMP signaling could induce transcription. While we found severalfold increases in transcriptional activity induced with ligand treatment in both the HCT116 (TGFBR2 and ACVR2 mutated) and HCT116 + chr3 (TGFBR2 reconstituted) cell lines, the HCT116 + chr2 cell line, which contains an extra copy of BMPRII and a functional copy of ACVR2, had a higher transcriptional activity with ligand treatment over controls (21). We propose that the higher levels of BMP-induced transcription in HCT116 + chr2 cells is a direct result of either one or both of the reconstituted receptors being present, with the ability to transduce the effects of BMP. Using DN BMPR1A transfection, we showed inhibition of BMP-induced SMAD-dependent transcription, further indicating that this receptor is necessary for transducing the effects of the BMP ligands. The reduction in transcriptional activity as a result of DN BMPR1A transfection appeared to be greater in HCT116 + chr2 cells compared with HCT116 + chr3 cells, again perhaps due to the presence of additional receptors encoded on chromosome 2 that are involved in BMP signaling. Additionally, we found that upon stimulation with BMP2 and BMP7, HCT116, HCT116 + chr2, and HCT116 + chr3 cells exhibited modest but significant growth suppression as assayed by the MTT assay. The growth suppression was verified in our models with the use of DN BMPR1A to impair signaling through this receptor. Indeed, DN BMPR1A reverse the BMP-induced growth suppression in our cell models. These results are consistent with an intact and functional BMP pathway in these colon cancer cells, with the manifestation of growth suppression.
We also investigated whether BMP2 or BMP7 stimulation induced changes in cell viability or cell cycle progression. We did not find BMP2 and BMP7 stimulation to induce changes in cell viability (data not shown) in our system, in contrast to what has been previously reported by other groups (14). Our cell cycle analysis did not identify any changes in cell cycle progression except for an increase in the G1 phase of SW480 cells, but our analysis of p21WAF1 transcriptional activity did not demonstrate differences with BMP stimulation. Thus the moderate growth suppression seen in these cells does not appear to be directly related to activation of p21WAF1, consistent with results reported for breast cancer cell lines (18).
SW480 is a SMAD4-null cell line, and, upon reconstitution of SMAD4, there was strong transcriptional activation of BMP-regulated SMAD signaling in the absence of exogenous BMP ligands, indicating high autocrine stimulation of the BMP receptors. This was verified by treating SMAD4-transfected cells with Noggin (which binds free ligand). A subsequent decrease in transcriptional activity was demonstrated as well as the presence of BMP2 and BMP7 ligand transcripts, which can make protein available for autocrine or paracrine activity upon BMP receptors. Furthermore, nontransfected SW480 cells exhibited significant decreases in growth when treated with BMP2 and BMP7, and BMP2-induced growth suppression was reversed with transfection with the DN BMPR1A vector. Taken together, these findings indicate that BMP can induce SMAD4-independent growth effects in SW480 cells by an as-yet-uncharacterized pathway. The SMAD4-independent growth-suppressive effect might be partly through nonligand saturable mechanisms given the high endogenous production of ligand in SW480 cells and little increase in BMP-specific transcriptional activity with exogenous ligand treatment.
Because of their more dramatic effect on growth inhibition with BMP, SW480 cells were also analyzed for their ability to close a wound scratched through the center of confluent cells. In particular, BMP2 ligand treatment inhibited the ability of SW480 cells to close the wound. SW480 cells treated with Noggin were able to close the wound to near the same extent as untreated SW480 cells. Thus BMP2 may slow the growth and/or inhibit the migratory ability of BMP-sensitive colon cancer cells. We also evaluated HCT116 + chr2 cells for their ability to close a wound in response to BMP ligands and observed no changes from controls (data not shown). This may be due to the less-dramatic growth effect of BMPs upon HCT116 + ch2 cells or possibly that cells without SMAD4 (such as SW480 cells, which lack SMAD4-dependent signaling) have unbalanced SMAD4-independent signaling that allows increased growth suppression.
While it was not a main focus of this study, we found no difference in BMP signaling between MSI-H and MSS tissues, as was expected because the BMPR1A receptor does not contain a coding microsatellite. This is in contrast to other TGF-β super-family members, TFG-β and activin, which have their signaling inactivated by mutations within coding microsatellites in key surface receptors (10, 22). Thus we have no evidence that BMP would have differential effects on MSI-H tumors compared with the TGF-β and activin pathways. BMP signaling could represent a potential pharmacological target that may lead to moderate growth suppression in both MSI-H and MSS tumors.
In conclusion, unlike other TGF-β superfamily member signaling (such as TFG-β and activin signaling), which is often inactivated in subsets of colon cancer, BMP signaling appears to be intact in these tumors. To our knowledge, this is the first examination of BMP7-induced signaling in sporadic colon cancers. In juvenile polyps from patients with JP, loss of BMP signaling might contribute to the high risk of colon cancer observed over the lifetime of these patients. Given that BMP signaling appears to be growth suppressive, there are a few possibilities as to why BMP signaling remains intact in colon cancers. First, the degree of tumor suppression may be moderate and thus overcome by other genetic events that occur in colon cancers. Second, other signaling pathways may adversely affect BMP growth-suppressive signaling, such that traditional SMAD signaling is abrogated without mutation of the components. Finally, some growth suppression by BMP may be induced via SMAD4-independent pathways, as suggested by our results in SW480 cells. This uncharacterized signaling pathway(s) could be redundant or affected by genetic events that occur during the formation and progression of colon cancers. These findings indicate further exploration is necessary of the pathways, and potential cross-talk between the pathways should be investigated.
ACKNOWLEDGMENTS
We thank Dr. Stephen Harris (University of Texas Health Science Center, San Antonio, TX) for the DN and CA BMPR1A vectors, Dr. Peter ten Dijke (Netherlands Cancer Institute, Amsterdam, The Netherlands) for the BRE-Luc construct, Dr. Masayuki Funaba (Azabu University School of Veterinary Medicine, Azabu, Japan) for the SMAD4 construct, and Dr. Burt Vogelstein (Johns Hopkins University, Baltimore, MD) for the pWWP construct. We also thank Dr. Katsumi Miyai (Department of Pathology, Univeristy of California, San Diego, CA) for the pathological expertise.
A portion of this work was presented in abstract form at the 2004 annual meeting of the American Association for Cancer Research in Orlando, FL, and the 2005 annual meeting of the American Gastroenterolgical Association in Chicago, IL.
GRANTS
This work was supported by National Institutes of Health Grants T32-HL-07212 (to S. E. Beck), DK-064560 (to S. C. Huang), and CA-90231 and DK-067287 (to J. M. Carethers).
REFERENCES
- 1.Arnold SF, Tims E, McGrath BE. Identification of bone morpho-genetic proteins and their receptors in human breast cancer cell lines: importance of BMP2. Cytokine. 1999;11:1031–1037. doi: 10.1006/cyto.1999.0508. [DOI] [PubMed] [Google Scholar]
- 2.Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- 3.Brubaker KD, Corey E, Brown LG, Vessella RL. Bone morpho-genetic protein signaling in prostate cancer cell lines. J Cell Biochem. 2004;91:151–160. doi: 10.1002/jcb.10679. [DOI] [PubMed] [Google Scholar]
- 4.Carethers JM. Biology of colorectal carcinoma. In: Rustgi A, Crawford J, editors. Gastrointestinal Cancers: Biology and Clinical Management. 1st ed. Saunders; Philadelphia, PA: 2003. pp. 407–419. [Google Scholar]
- 5.Carethers JM, Pham TT. Mutations of transforming growth factor beta 1 type II receptor, BAX, and insulin-like growth factor II receptor genes in microsatellite unstable cell lines. In Vivo. 2000;14:13–20. [PubMed] [Google Scholar]
- 6.Dicuonzo G, Angeletti S, Garcia-Foncillas J, Brugarolas A, Okrouzhnov Y, Santini D, Tonini G, Lorino G, De Cesaris M, Baldi A. Colorectal carcinomas and PTEN/MMAC1 gene mutations. Clin Cancer Res. 2001;7:4049–4053. [PubMed] [Google Scholar]
- 7.Gilboa L, Nohe A, Geissendorfer T, Sebald W, Henis YI, Knaus P. Bone morphogenetic protein receptor complexes on the surface of live cells: a new oligomerization mode for serine/threonine kinase receptors. Mol Biol Cell. 2000;11:1023–1035. doi: 10.1091/mbc.11.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyette MC, Cho K, Fasching CL, Levy DB, Kinzler KW, Paraskeva C, Vogelstein B, Stanbridge EJ. Progression of colorectal cancer is associated with multiple tumor suppressor gene defects but inhibition of tumorigenicity is accomplished by correction of any single defect via chromosome transfer. Mol Cell Biol. 1992;12:1387–1395. doi: 10.1128/mcb.12.3.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grady WM, Myeroff LL, Swinler SE, Rajput A, Thiagalingam S, Lutterbaugh JD, Neumann A, Brattain MG, Chang J, Kim SJ, Kinzler KW, Vogelstein B, Willson JK, Markowitz S. Mutational inactivation of transforming growth factor beta receptor type II in micro-satellite stable colon cancers. Cancer Res. 1999;59:320–324. [PubMed] [Google Scholar]
- 10.Grady WM, Rajput A, Myeroff L, Liu DF, Kwon K, Willis J, Markowitz S. Mutation of the type II transforming growth factor-beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res. 1998;58:3101–3104. [PubMed] [Google Scholar]
- 11.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 12.Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromo-some 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 13.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 14.Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, Offerhaus GJ, Van Deventer SJ, Peppelen-bosch MP. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126:111–121. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- 15.He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Mishina Y, Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 16.Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK, Yeo CJ, Hruban RH, Kern SE. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994–999. [PubMed] [Google Scholar]
- 17.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 18.Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, Stone EM, Aaltonen LA. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 19.Howe JR, Sayed MG, Ahmed AF, Ringold J, Larsen-Haidle J, Merg A, Mitros FA, Vaccaro CA, Petersen GM, Giardiello FM, Tinley ST, Aaltonen LA, Lynch HT. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet. 2004;41:484–491. doi: 10.1136/jmg.2004.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang SC, Chen CR, Lavine JE, Taylor SF, Newbury RO, Pham TT, Ricciardiello L, Carethers JM. Genetic heterogeneity in familial juvenile polyposis. Cancer Res. 2000;60:6882–6885. [PubMed] [Google Scholar]
- 21.Jung B, Fiorino A, Doctolero RT, Smith EJ, Bocanegra M, Cabrera BL, Carethers JM. Functional correlation of growth suppression with activin treatment in microsatellite unstable, activin type 2 receptor (ACVR2)-restored colorectal cancer cells. Gastroenterology. 2004;126:A–264. [Google Scholar]
- 22.Jung B, Doctolero RT, Tajima A, Nguyen AK, Keku T, Sandler RS, Carethers JM. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654–659. doi: 10.1053/j.gastro.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2-BRIA ectodomain complex. Nat Struct Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 24.Kleeff J, Maruyama H, Ishiwata T, Sawhney H, Friess H, Buchler MW, Korc M. Bone morphogenetic protein 2 exerts diverse effects on cell growth in vitro and is expressed in human pancreatic cancer in vivo. Gastroenterology. 1999;116:1202–1216. doi: 10.1016/s0016-5085(99)70024-7. [DOI] [PubMed] [Google Scholar]
- 25.Koenig BB, Cook JS, Wolsing DH, Ting J, Tiesman JP, Correa PE, Olson CA, Pecquet AL, Ventura F, Grant RA, Chen G-X, Wrane JL, Massague J, Rosenbaum JS. Characterization and cloning of a receptor for BMP-2 and BMP-4 from NIH 3T3 cells. Mol Cell Biol. 1994;14:5961–5974. doi: 10.1128/mcb.14.9.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N′-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 27.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 28.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 29.Liu F, Ventura F, Doody J, Massague J. Human type II receptor for bone morphogenic proteins (BMPs): extension of the two-kinase receptor model to the BMPs. Mol Cell Biol. 1995;15:3479–3486. doi: 10.1128/mcb.15.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J Biol Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 31.Markowitz SD, Roberts AB. Tumor suppressor activity of the TGF-beta pathway in human cancers. Cytokine Growth Factor Rev. 1996;7:93–102. doi: 10.1016/1359-6101(96)00001-9. [DOI] [PubMed] [Google Scholar]
- 32.Moskaluk CA, Hruban RH, Schutte M, Lietman AS, Smyrk T, Fusaro L, Fusaro R, Lynch J, Yeo CJ, Jackson CE, Lynch HT, Kern SE. Genomic sequencing of DPC4 in the analysis of familial pancreatic carcinoma. Diagn Mol Pathol. 1997;6:85–90. doi: 10.1097/00019606-199704000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Pouliot F, Labrie C. Role of Smad1 and Smad4 proteins in the induction of p21WAF1,Cip1 during bone morphogenetic protein-induced growth arrest in human breast cancer cells. J Endocrinol. 2002;172:187–198. doi: 10.1677/joe.0.1720187. [DOI] [PubMed] [Google Scholar]
- 34.Rosenzweig BL, Imamura T, Okadome T, Cox GN, Yamashita H, ten Dijke P, Heldin CH, Miyazono K. Cloning and characterization of a human type II receptor for bone morphogenetic proteins. Proc Natl Acad Sci USA. 1995;92:7632–7636. doi: 10.1073/pnas.92.17.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satia JA, Keku T, Galanko JA, Martin C, Doctolero RT, Tajima A, Sandler RS, Carethers JM. Diet, lifestyle, and genomic instability in the North Carolina Colon Cancer Study. Cancer Epidemiol Biomarkers Prev. 2005;14:429–436. doi: 10.1158/1055-9965.EPI-04-0486. [DOI] [PubMed] [Google Scholar]
- 36.ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin CH, Miyazono K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J Biol Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- 37.Wen XZ, Miyake S, Akiyama Y, Yuasa Y. BMP-2 modulates the proliferation and differentiation of normal and cancerous gastric cells. Biochem Biophys Res Commun. 2004;316:100–106. doi: 10.1016/j.bbrc.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita H, ten Dijke P, Huylebroeck D, Sampath TK, Andries M, Smith JC, Heldin CH, Miyazono K. Osteogenic protein-1 binds to activin type II receptors and induces certain activin-like effects. J Cell Biol. 1995;130:217–226. doi: 10.1083/jcb.130.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 40.Zhou XP, Woodford-Richens K, Lehtonen R, Kurose K, Aldred M, Hampel H, Launonen V, Virta S, Pilarski R, Salovaara R, Bodmer WF, Conrad BA, Dunlop M, Hodgson SV, Iwama T, Jarvinen H, Kellokumpu I, Kim JC, Leggett B, Markie D, Mecklin JP, Neale K, Phillips R, Piris J, Rozen P, Houlston RS, Aaltonen LA, Tomlinson IP, Eng C. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am J Hum Genet. 2001;69:704–711. doi: 10.1086/323703. [DOI] [PMC free article] [PubMed] [Google Scholar]