Abstract
A heterogeneous population of inhibitory neurons controls the flow of information through a neural circuit1–3. Inhibitory synapses that form on pyramidal neuron dendrites modulate the summation of excitatory synaptic potentials4–6 and prevent the generation of dendritic calcium spikes7,8. Precisely timed somatic inhibition limits both the number of action potentials and the time window during which firing can occur8,9. The activity-dependent transcription factor NPAS4 regulates inhibitory synapse number and function in cell culture10, but how this transcription factor affects the inhibitory inputs that form on distinct domains of a neuron in vivo was unclear. Here we show that in the mouse hippocampus behaviourally driven expression of NPAS4 coordinates the redistribution of inhibitory synapses made onto a CA1 pyramidal neuron, simultaneously increasing inhibitory synapse number on the cell body while decreasing the number of inhibitory synapses on the apical dendrites. This rearrangement of inhibition is mediated in part by the NPAS4 target gene brain derived neurotrophic factor (Bdnf), which specifically regulates somatic, and not dendritic, inhibition. These findings indicate that sensory stimuli, by inducing NPAS4 and its target genes, differentially control spatial features of neuronal inhibition in a way that restricts the output of the neuron while creating a dendritic environment that is permissive for plasticity.
We assessed NPAS4 expression in the hippocampus by immunofluorescence microscopy. We found that in hippocampi from mice maintained in standard housing, NPAS4 protein is undetectable by immunostaining (Fig. 1a). By contrast, exposure of mice to an enriched environment induces NPAS4 protein expression in neurons in the pyramidal layer, whereas no NPAS4-positive nuclei are observed in the neuropil (Fig. 1b). To obtain more widespread induction of NPAS4, mice were injected intraperitoneally with kainic acid to trigger seizure activity. This treatment led to NPAS4 expression in nearly all excitatory neurons in the pyramidal layer and a minority of inhibitory neurons in the neuropil (Fig. 1c).
Figure 1.
Exposure of mice to increased circuit activity reveals an NPAS4-dependent regulation of inhibition in vivo. a–c, Hippocampal sections from wild-type mice in standard housing (a), enriched environment (EE) (b) or after kainic acid (KA) injection (c). Sections were immunostained for NPAS4 (red) and NeuN (blue) protein. Quantification is shown in b and c. Data are shown as mean ± s.e.m. For each condition n=3 animals with 3–5 sections per animal. *P<0.01. d, Wide-field (left) and fluorescence (right) image of hippocampal slice from an Npas4f/f mouse injected with AAVCre–GFP. e, Experimental configuration. f–h, mIPSC frequency (top) and amplitude (bottom) recorded from neighbouring NPAS4-WT and NPAS4-KO neurons from mice maintained in standard housing (f) (n=14 pairs), enriched environment (g) (n=23 pairs, freq: P<0.05) or after kainic acid injection (h) (n=26 pairs, freq: P,0.01, amp: P<0.05). Open circles represent NPAS4-KO/NPAS4-WT pairs. Red circles indicate mean ± s.e.m.
We next asked whether the loss of NPAS4 affects inhibitory synapse function in vivo. Adeno-associated virus encoding a Cre–GFP fusion protein (AAV-Cre–GFP)11 was injected into the CA1 region of the hippocampus in Npas4 conditional mice (Npas4f/f) (Fig. 1d, Extended Data Fig. 1). Post-operative animals were transferred to an enriched environment or maintained in standard housing for 7 to 12 days. Subsequently acute hippocampal slices were prepared, whole-cell voltage clamp recordings were made from neighbouring NPAS4 knockout and wild-type neurons (referred to as NPAS4-KO and NPAS4-WT, respectively) and pharmacologically isolated miniature inhibitory postsynaptic currents (mIPSCs) were recorded (Fig. 1e).
We found that when mice were exposed to an enriched environment significantly less frequent and slightly smaller amplitude mIPSCs were recorded from NPAS4-KO neurons compared to the neighbouring NPAS4-WT neurons (Fig. 1g, Supplementary Table 1). Similar results were obtained from mice injected with kainic acid to induce NPAS4 (Fig. 1h). By contrast, no significant difference in mIPSC frequency or amplitude was observed between neighbouring NPAS4-WT and NPAS4-KO neurons in slices acquired from mice maintained in standard housing (Fig. 1f). Next we measured mIPSCs in wild-type mice and detected no differences between AAV-Cre–GFP infected and uninfected neurons (Extended Data Fig. 2a, b) indicating that the observed changes in mIPSCs in Npas4f/f neurons are due to Npas4 excision. Finally, we crossed EMX-Cre and Npas4f/f mice to generate animals that lack NPAS4 postnatally and specifically in excitatory neurons (Extended Data Fig. 2c, d). Both mIPSC frequency and amplitude were significantly reduced in NPAS4-KO neurons from animals injected with kainic acid compared to mice maintained in standard housing (Extended Data Fig. 2e). From these analyses we conclude that disruption of behaviourally induced NPAS4 expression leads to a decrease in mIPSC frequency recorded from CA1 pyramidal neurons.
This decrease in mIPSC frequency could be a consequence of homeostatic regulation of all inhibitory synapses or the modification of inhibitory synapses that form on discrete regions of the pyramidal neuron. Local inhibitory neuron subtypes have axons that elaborate preferentially in one or more layers of the hippocampus2,3. Thus, stimulation of axons in the different laminar domains of the hippocampus and analysis of the resulting inhibitory currents has the potential to reveal whether the NPAS4-dependent effects on inhibition are due to cell wide or domain-specific regulation of inhibitory synapses.
We sparsely excised Npas4 and made direct comparisons of layer specific, pharmacologically isolated, monosynaptic evoked inhibitory postsynaptic currents (eIPSCs) recorded simultaneously from neighbouring NPAS4-KO and NPAS4-WT CA1 neurons (Fig. 2a). As predicted by cable properties, eIPSC rise times correlated with distance of the stimulating electrode from the recording electrode (Extended Data Fig. 3, Supplementary Table 2) indicating that stimulation of axons within a given layer is predominately activating synapses within that layer.
Figure 2.
Behaviourally induced NPAS4 differentially regulates inhibitory synapse function across the somato-dendritic axis of pyramidal neurons. a, Experimental configuration. L, lacunosum; R, radiatum; P, pyramidale; O, oriens. b–e, eIPSCs measured from mice maintained in standard housing. Top shows average eIPSC, normalized pairwise to the wild-type neuron, measured from NPAS4-WT (black) and NPAS4-KO (green) neurons in response to stimulation in lacunosum (b), radiatum (c), pyramidale (d) or oriens (e). Scale bars indicate per cent change from wild type. Bottom shows eIPSC amplitude measured from pairs of neighbouring NPAS4-KO and NPAS4-WT neurons in response to stimulation of axons in lacunosum (b) (n=14 pairs), radiatum (c) (n=13 pairs), pyramidale (d) (n=17 pairs) or oriens (e) (n=13 pairs). Open circles represent NPAS4-KO/NPAS4-WT pairs. Red circles indicate mean ± s.e.m. f–i, eIPSCs measured from mice exposed to an enriched environment. Data are displayed as in b–e. L, n=16 pairs; R, n=16 pairs, P<0.05; P, n=14 pairs, P<0.01; O, n=18 pairs.
We found that in slices obtained from mice housed in a standard environment, equivalent eIPSCs were recorded from NPAS4-WT and NPAS4-KO neurons in response to stimulation of axons in any of the hippocampal layers (Fig. 2b–e, Supplementary Table 2). However, in slices obtained from mice exposed to an enriched environment, stimulation of axons in the pyramidal layer generated smaller eIPSCs in NPAS4-KO neurons than in the neighbouring NPAS4-WT neurons (Fig. 2h). By contrast, stimulation of axons in stratum radiatum generated significantly larger eIPSCs in NPAS4-KO neurons than in neighbouring NPAS4-WT neurons (Fig. 2g), whereas no difference was detected between NPAS4-WT and NPAS4-KO neurons upon stimulation of axons in oriens or lacunosum (Fig. 2f, i). Similar results were obtained from mice injected with kainic acid to induce NPAS4 (Extended Data Fig. 4). These findings indicate that NPAS4 expression does not result in uniform, homeostatic regulation of inhibitory synapses. Rather, NPAS4 functions in an unprecedented manner, simultaneously decreasing inhibition within the proximal apical dendrites while increasing inhibition at the soma.
Extracellular stimulation of axons could result in the depolarization of axons of passage that preferentially form synapses on NPAS4-WT or NPAS4-KO neurons and in regions other than the intended layer. To address this possibility we compared the rise times of the eIPSCs recorded from NPAS4-WT and NPAS4-KO neurons and found them to be indistinguishable (Extended Data Fig. 3). This indicates that the redistribution of inhibition observed is not due to a bias in synaptic transmission associated with axons of passage onto NPAS4-WT or NPAS4-KO neurons. Additionally, the hippocampus contains several interneuron subtypes that form synapses within multiple layers (that is, bistratified and trilaminar cells). If NPAS4 regulates synapses made by these cell types, it is probable that only boutons that synapse on the apical, and not basal, dendrites are affected by loss of NPAS4.
We next investigated the cellular mechanisms by which NPAS4 differentially regulates inhibition, including possible effects on dendritic growth, changes in the probability of inhibitory synaptic vesicle release or changes in inhibitory synapse number. We first asked whether loss of NPAS4 results in altered dendritic complexity which could indirectly lead to larger eIPSCs in response to stimulation of axons in radiatum. However, this seems unlikely because Sholl analysis revealed that the disruption of NPAS4 function has no effect on dendrite number or length (Fig. 3a, b, Extended Data Fig. 5a, b). In addition, the capacitance of neurons from mice housed under standard conditions, in an enriched environment or exposed to kainic acid are equivalent (Supplementary Table 3).
Figure 3.
Behaviourally induced NPAS4 regulates inhibitory synapse number. a, Standard housing example NPAS4-WT and NPAS4-KO CA1 pyramidal neurons (left) and quantification of NPAS4-WT (black, n=10 neurons) and NPAS4-KO (red, n=10 neurons) dendrites (right). b, Enriched environment Sholl analysis, as in a. NPAS4-WT, n=7neurons; NPAS4-KO, n=10 neurons. c–e, PPRs measured from NPAS4-WT and NPAS4-KO neurons in response to stimulation in radiatum (c), pyramidale (d) or oriens (e). Standard housing (black, R, n=9 neurons; P, n=10 neurons; O, n=8 neurons) or enriched environment (green, R, n=16 neurons; P, n=11 neurons; O, n=16 neurons). Examples are from standard housing (top) and are shown normalized to peak current recorded from the NPAS4-WT neuron. Scale bars indicate per cent change from wild type. ISI, interstimulus interval. f, Example image of dendrites in radiatum. g, Quantification of dendritic GFP–gephyrin puncta from NPAS4-WT (black, standard housing, n=15 sections/63 dendritic segments; enriched environment, n=14 sections/40 dendritic segments) and NPAS4-KO (red, standard housing, n=18 sections/172 dendritic segments; enriched environment n=18 sections/167 dendritic segments) neurons. Enriched environment P<0.01. h, Example image of somata in pyramidale. i, Quantification of somatic GFP–gephyrin puncta from NPAS4-WT (black, standard housing, n=14 sections/32 soma; enriched environment, n=11 sections/20 soma) and NPAS4-KO (red, standard housing, n=17sections/94 soma, enriched environment: n=21sections/75 soma) neurons. Enriched environment P<0.05. All data are shown as mean ± s.e.m.
We next asked whether disruption of Npas4 leads to changes in the presynaptic release of neurotransmitter at axons that synapse onto different regions of the pyramidal neuron. We measured the paired pulse ratios (PPRs) upon stimulation of axons in each layer, and compared NPAS4-WT and NPAS4-KO neurons in slices obtained from mice housed under standard conditions, in an enriched environment, or injected with kainic acid. We found no difference between the PPRs of NPAS4-WT and NPAS4-KO neurons under any of these conditions (Fig. 3c–e, Extended Data Fig. 5c–e, Supplementary Table 2) indicating that the NPAS4-dependent changes in eIPSCs are not likely due to changes in the probability of presynaptic vesicle release.
Finally we asked whetherNPAS4 differentially regulates the number of inhibitory synapses on the soma or apical dendrites. To quantify inhibitory synapses we co-injected AAVs encoding mCherry-IRESCre and Cre-dependent GFP–gephyrin fusion protein into the CA1 region of the hippocampus in wild-type and Npas4f/f mice. GFP–gephyrin fusion proteins when expressed at low levels localize to inhibitory synapses without significantly altering neurotransmission12,13. Cre was sparsely expressed and GFP–gephyrin puncta quantified on NPAS4-WT andNPAS4-KOneurons. This analysis revealed that when mice were kept in standard housing, NPAS4-WT and NPAS4-KO hippocampal pyramidal neurons have equivalent densities of GFP–gephyrin puncta on the soma and apical dendrites. However, after exposure to an enriched environment, NPAS4-KO pyramidal neurons have significantly fewer somatic and more dendritic GFP–gephyrin puncta than NPAS4-WT neurons (Fig. 3f–h) consistent with the electrophysiology data described above. Strikingly, comparison of the number of GFP–gephyrin puncta measured on wild-type neurons revealed that exposure to an enriched environment leads to an increase in the number of somatic and a decrease in the number of dendritic inhibitory synapses. In both domains of the neuron, the behaviourally triggered changes in inhibition are NPAS4 dependent.
NPAS4 is a transcription factor and thus to understand how it mediates sensory-dependent changes in inhibitory synapse number we sought to identify NPAS4 target genes and assess their function. We focused our attention on putative NPAS4 targets that satisfy three criteria: (1) by deep sequencing NPAS4-bound DNA (ChIP-seq), NPAS4 is shown to be bound within 10 kilobases of the putative target gene; (2) by RNA-seq, the mRNA transcribed near the NPAS4 peak is induced greater than fivefold (Extended Data Table 1)14; (3) upon short hairpin RNA (shRNA)-mediated knockdown of the putative target gene there is a significant change in mIPSC frequency or amplitude. Through this screen we identified 16 putative NPAS4 targets that display a change in mIPSC frequency or amplitude upon knockdown with gene-specific shRNAs in organotypic hippocampal slices, five of which we further validated (Extended Data Figs 6–8).
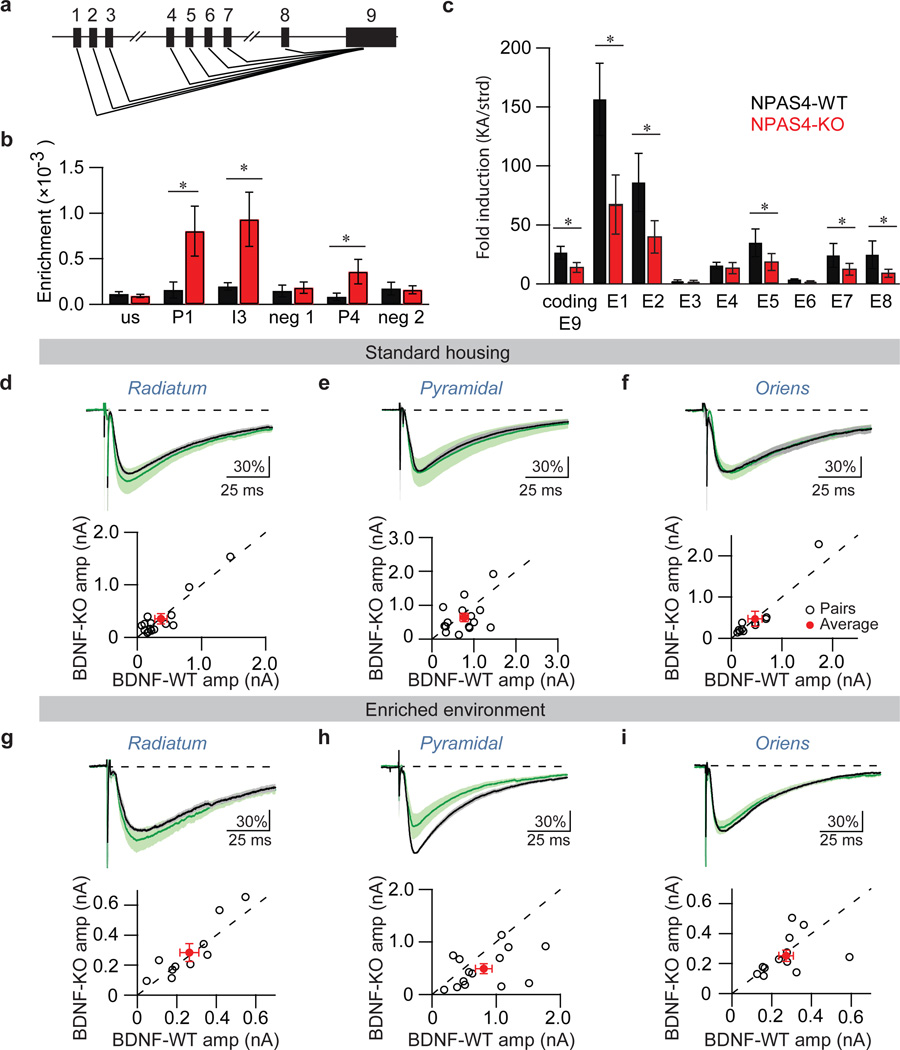
Of the putative NPAS4 target genes identified by the screen, Bdnf seemed a possible candidate to control the number of inhibitory synapses that form on a specific region of an excitatory neuron in response to sensory stimulation. BDNF expression is regulated by neuronal activity in vivo15–18 and it regulates inhibitory synapse number and function19–21. In addition, Bdnf mRNAs can be targeted to discrete regions of the neuron22, and BDNF-containing vesicles are known to be secreted from the axon, soma or dendrites of pyramidal neurons23,24. Furthermore, we confirmed by ChIP and quantitative PCR with reverse transcription (qRT–PCR) that NPAS4 binds to three sites within the Bdnf gene (Fig. 4a, b), and that induction of the transcription of multiple Bdnf isoforms is significantly reduced in the hippocampus of Npas4 knockout (Npas4−/−) compared to wild-type (C57BL/6J) mice (Fig. 4c).
Figure 4.
Behaviourally induced BDNF regulates somatic but not dendritic inhibition. a, Schematic of Bdnf gene. Exons 1–9 are indicated. b, qPCR of NPAS4 bound DNA. ChIP samples are from wild-type mice in standard conditions (black) or after kainic acid injection (red). us, upstream region; P1, promoter 1; I3, intron 3; neg 1, control region; P4, promoter 4; neg 2, control region. Enrichment relative to input DNA. N=3. *P<0.05. c, Induction of Bdnf exons in wild-type (black, standard housing, n=5 animals; kainic acid, N=6 animals) or Npas4−/− animals (red, standard housing, n=5 animals; kainic acid, n=6 animals) mice as measured by qRT–PCR with exon specific primers, E1–9.*P<0.05. Data in b and c are shown as mean ± s.e.m. d–f, eIPSCs measured from mice maintained in standard housing. Top shows average eIPSC, normalized pairwise to the wild-type neuron, measured from BDNF-WT (black) and BDNF-KO (green) neurons in response to stimulation in radiatum(d), pyramidale (e) or oriens (f). Scale bars indicate per cent change from wild type. Bottom shows eIPSC amplitude measured from pairs of neighbouring BDNF-KO and BDNF-WT neurons in response to stimulation of axons in radiatum (d) (n=14 pairs), pyramidale (e) (n=15 pairs) or oriens (f) (n=11 pairs). Open circles represent BDNF-KO/BDNF-WT pairs. Red circles indicate mean ± s.e.m. g–i, eIPSC measured from mice housed in an enriched environment. Data are shown as in d–f. R, n=10 pairs; P, n=14 pairs; P<0.05; O, n=14 pairs.
To determine if BDNF is a target of NPAS4 that selectively regulates somatic and/or dendritic inhibition of pyramidal neurons in vivo, we injected AAV-Cre–GFP into BDNF conditional knockout mice (Bdnff/f) mice and compared eIPSCs between neighbouring Bdnf knockout and Bdnf wild-type neurons (BDNF-KO and BDNF-WT, respectively) in response to stimulation of axons in the somatic or dendritic layers. In mice housed under standard conditions, we found no differences in eIPSC amplitude measured from neighbouring BDNF-KO and BDNF-WT neurons upon stimulation of axons in any of the layers of the hippocampus (Fig. 4d–f, Supplementary Table 2).
However, when we exposed mice to an enriched environment to induce NPAS4 as well as BDNF expression and secretion, stimulation of axons in the pyramidal layer results in a significantly smaller amplitude eIPSC in BDNF-KO neurons relative to neighbouring BDNF-WT neurons (Fig. 4h). The disruption of Bdnf function did not have a significant effect on eIPSC amplitude when axons in radiatum or oriens were stimulated (Fig. 4g, i). Similar results were observed when BDNF expression was induced with kainic acid (Extended Data Fig. 9). Taken together these findings indicate that the differential effects of NPAS4 on pyramidal neuron inhibition are mediated, at least in part, through the induction of BDNF that functions selectively and locally to promote inhibition at the soma. Given that the disruption of BDNF function does not affect eIPSCs in radiatum, the ability of NPAS4 to relieve inhibition on the apical dendrites of CA1 pyramidal neurons is probably mediated by NPAS4 targets other than BDNF.
These findings indicate that when a mouse engages with its environment, the neuronal activity-regulated transcription factor NPAS4 is expressed in pyramidal neurons of the hippocampus where it promotes an increase in the number of inhibitory synapses on the cell soma and a decrease in the number of inhibitory synapses on the apical dendrites. In light of recent reports indicating that inhibition within these cellular domains can have distinct functions25–28, we speculate that this redistribution of inhibition will have significant effects on information processing in the postsynaptic neuron. For example, the opposing regulation of somatic and apical dendritic inhibition may allow integration or plasticity of excitatory events in the apical dendrites while still limiting the generation of somatic action potentials and thus the propagation of information to downstream neurons. This is in contrast to homeostatic scaling of inhibitory synapses—a process whereby the action potential output of a neuron is regulated by cell-wide scaling of synaptic strengths that preserves the relative strengths among synapses. The NPAS4-dependent redistribution of inhibitory circuitry may underlie recent findings that associate NPAS4 function with contextual or stimulus dependent fear conditioning29,30.
Methods
Animal husbandry and handling
Animals were handled according to protocols approved by the Harvard University Standing Committee on Animal Care and the University of California San Diego Institutional Animal Care and Use Committee, and were in accordance with federal guidelines. The following animal lines were used: Npas4f/f, NPAS4 knockout (Npas4−/−) mice10, wild type (C57BL/6J, JAX 000664), Bdnff/f (Bdnf < tm3Jae>/J, JAX 004339) and EMX-Cre (B6.129S2-EMX1TM1(cre)Krj/J, JAX 005628). Both male and female mice were used. Electrophysiology, Sholl analysis and GFP–gephyrin experiments were performed on animals between postnatal day 21 and 28 (P21–P28). Immunostaining, ChIP and qRT-PCR experiments were done on samples collected from P25–P35 animals.
For experiments in which mice were exposed to an enriched environment, three days after virus injection (P16–P18) animals (dam and pups) were moved to a larger cage that contained a running wheel, hut, tunnel and several other novel objects. To maximize novelty, the objects in the environment were rearranged and new objects introduced every second day. Mice were housed in the enriched environment for 4–10 days. Although we detect a limited number of NPAS4 positive excitatory neurons at a given time point, exploration of a novel environment increases NPAS4 immunoreactivity to an extent similar to that observed for other activity inducible genes such as Fos and Arc. Moreover, behaviourally triggered NPAS4 induction appears to occur iteratively, and in distinct subpopulations of neurons as the animal engages and explores novel features of the environment, such that the number of NPAS4 immunopositive neurons detected at any given time point significantly underestimates the number of neurons that express NPAS4 over a period of several days.
For experiments in which seizures were induced, kainic acid (2.5–10 mg per kg) was injected intraperitoneally. Mice were euthanized 2 h after the first visible seizure for ChIP, at 3 hours for immunohistochemistry, at 6 h for qRT–PCR or 24 h later for electrophysiology experiments. These time points were selected to allow sufficient time for expression of NPAS4 protein, the execution of an NPAS4-dependent program of gene expression and potential synaptic regulation but before detectable seizure-related cell death.
Sterotaxically guided surgery
All surgeries were performed according to protocols approved by the Harvard University Standing Committee on Animal Care and the University of California San Diego Institutional Animal Care and Use Committee, and were in accordance with federal guidelines. Surgeries were performed on mice between P13 and P15. Animals were deeply anaesthetized by inhalation of isoflurane (initially 3–5% in O2, maintained with 1–2%) and secured in the stereotaxic apparatus (Kopf). Animal temperature was maintained at 37 °C. The fur was shaved and scalp cleaned with betadine and 100% ethanol three times before an incision was made to expose the skull. A small hole was drilled through the skull and the CA1 region of hippocampus was specifically targeted (medial/lateral: ± 2.9 mm; anterior/posterior: −2.5 mm; dorsal/ventral: 2.8 mm below the dura) and virus was injected (250–300 nl; 150 nl min−1). Five minutes post-injection, the needle was retracted, the scalp sutured and the mouse returned to its home cage. All animals were monitored for at least one hour post-surgery and at 12-h intervals for the next 5 days. Post-operatively, analgesic (flunixin, 2.5 mg per kg) was administered at 12-h intervals for 72 h.
Virus production
AAV-Cre–GFP was produced by the Harvard Gene Therapy Initiative or UNC Vector Core with a plasmid provided by M. During (Ohio State University). The plasmids containing the genome for AAV-EF1a-YFP-2A-Cre, AAV-mCherry-IRES-Cre and AAV-FLEX-GFP–gephyrin were generated using standard molecular cloning techniques and were produced by the UNC Vector Core.
Acute slice preparation
Transverse hippocampal slices were prepared from Npas4f/f, EMX-Cre; Npas4f/f, Bdnff/f or C57BL/6 mice (P21–P28). Animals were anaesthetized by inhalation of isoflurane. The cerebral hemispheres were quickly removed and placed into ice-cold choline-based artificial cerebrospinal fluid (choline-ACSF) consisting of (in mM): 110 choline-Cl, 25 NaHCO3, 1.25 Na2HPO4, 2.5 KCl, 7 MgCl, 25 glucose, 0.5 CaCl2, 11.6 ascorbic acid, 3.1 pyruvic acid and equilibrated with 95% O2/5% CO2. Tissue was blocked and transferred to a slicing chamber containing choline-ACSF. Slices (300 µm) were cut with a LeicaVT1000 s vibratome (Leica Instruments) and transferred to a holding chamber containing ACSF consisting of (in mM): 127 NaCl, 25 NaHCO3, 1.25 Na2HPO4, 2.5 KCl, 2 CaCl2, 1 MgCl2, 25 glucose, and saturated with 95% O2/5% CO2. Slices were incubated at 30 °C for 30–40 min and then kept at room temperature for no more than 6 h until recordings were performed. For mice injected with AAV-Cre–GFP, slices showing infection of between 20% to 40% in CA1, as determined by GFP fluorescence, were used for recording. Slices showing greater than ~40% infected neurons or infection outside of CA1 were discarded.
Organotypic slice preparation and transfection
Hippocampi were rapidly dissected from wild-type mice at P7 in ice cold dissection media consisting of (in mM): 1 CaCl2, 5 MgCl2, 10 glucose, 4 KCl, 26 NaHCO3, 218 sucrose, 1.3 NaH2PO4·H2O, 30 HEPES. Tissue was transferred to a tissue chopper (McIliwan Ted Pella) and 400-µm thick sections cut. Sections were transferred to tissue culture plates and grown on PTFE inserts (Milicell Organotypic insert, EMD Millipore) in slice culture media consisting of (in mM or percentage) 1× MEM, 20% horse serum, 1 l-glutamine, 0.125% ascorbic acid, 1 CaCl2, 2 MgCl2, 12.8 glucose, 5.25 NaHCO3, 30 HEPES, pH 7.4, 320 mOsm) for 7–10 days. On the second day in vitro, cultures were biolistically (Gene Gun, Biorad) co-transfected with 1-µm gold particles coated with GFP and shRNAs targeting the gene of interest (particles prepared with GFP 15 µg, 3–9 µg of shRNA, pcDNA3 to a final DNA mass of 50 µg as per the manufacturer’s instructions). The shRNA sequences are indexed in Supplementary Table 4.
Electrophysiology
Whole-cell voltage clamp recordings were obtained from CA1 pyramidal neurons visualized with infrared, differential interference contrast microscopy. Neurons were held at −70 mV except for experiments measuring evoked responses from stimulation of axons in stratum lacunosum, during which neurons were held at 0 mV. Patch pipettes (open pipette resistance 2–4 MΩ) were filled with an internal solution consisting of (in mM) 147 CsCl, 5 Na2-phosphocreatine, 10 HEPES, 2 MgATP, 0.3 Na2GTP and 2 EGTA. Osmolarity and pH were adjusted to 300 mOsm and 7.3 with double distilled water and CsOH, respectively. In experiments in which mIPSCs were recorded, currents were pharmacologically isolated with bath application of 0.5 µM tetrodotoxin citrate (Tocris Bioscience), 10 µM (R)-CPP (Tocris Bioscience) and 10 µM NBQX disodium salt (Tocris Bioscience). In experiments in which eIPSCs were recorded, inhibitory currents were pharmacologically isolated with bath application of CPP and NBQX. Additionally, 5 mM QX-314 (Sigma) was added to the internal solution. Extracellular stimulation of local axons within specific lamina of the hippocampus was delivered by current injection through a theta glass stimulating electrode that was placed in the centre of the relevant layer (along the somato-dendritic axis of the CA1 neuron) and within 100–200-µm laterally of the patched pair. The stimulus strength was the minimum required to generate an eIPSC in both NPAS4-KO and NPAS4-WT neurons.
Data acquisition and analysis
Electrophysiology data were acquired using pClamp software, either a Multiclamp 700B or Axoclamp 200B amplifier, and digitized with a DigiData 1440 data acquisition board (Axon Instruments). Data were sampled at 10 kHz and filtered at 4 or 6 kHz except for experiments with stimulation of axons in lacunosum which were filtered at 1 kHz. Off-line data analysis was performed using custom software written in Igor Pro by B.L.B. (Wavemetrics).
Experiments were discarded if the holding current was greater than −500 pA or if the series resistance was greater than 25 MΩ. In experiments in which direct comparisons were made between two neurons, recordings were discarded if the series resistance differed by more than 25% between the two recordings. All recordings were performed at room temperature (~20–22 °C).
Threshold for mIPSC detection was determined independently for each neuron and was based on the average root mean square (r.m.s.) of the first 150 ms of the recording. Amplitude threshold was set to 1.5 times r.m.s. for that cell and recordings were discarded if the r.m.s. was greater than 6 pA. This strategy resulted in fewer missed events and fewer erroneously called events than applying a single amplitude threshold.
The amplitude of eIPSCs was calculated by averaging the amplitude 0.5 ms before to 2 ms after the peak of the current. Data are shown as positive values for clarity. Slopes of the rise times of the eIPSCs were measured by normalizing the eIPSC, then measuring the slope between 10–90% of the peak. Paired pulse ratios (PPR) were calculated by recording a template eIPSC for each cell, subtracting the template wave from the first pulse, and then measuring the corrected amplitude of the second peak.
Immunohistochemistry, Sholl analysis and GFP–gephyrin quantification
Animals were anaesthetized by inhalation of isoflurane. Hippocampi were rapidly dissected in ice-cold dissection media as described above and drop fixed in 4% paraformaldehyde in PBS at 4 °C for 1.5–3 h followed by overnight incubation in 30% sucrose in PBS. Cryoprotected tissue was stored in Tissue-Tec O.C.T. at −80 °C and subsequently sectioned at a thickness of 40–45-µm (Leica CM1950 cryostat) for immunostaining or GFP–gephyrin puncta quantification, or at a thickness of 100-µm for Sholl analysis (Leica Instruments).
For NPAS4 immunostaining, hippocampal sections were blocked in 2% goat serum and 0.2% Triton X-100 in PBS for 1 h at room temperature. Sections were incubated in primary antibody overnight at 4 °C, washed three times in PBS, incubated in species-matched fluorescently conjugated secondary for 1 h at room temperature and washed again in PBS. Finally, sections were mounted on slides with Fluoromount-G (SouthernBiotech). The following antibodies were used: rabbit anti-NPAS4 (1:1,000, made in house), mouse anti-NeuN (1:1,000, Millipore) and rabbit anti-GFP (1:500, Invitrogen). The secondary antibodies were Alexa-488 or Alexa-555 against the appropriate species (1:500, Invitrogen). Sections were imaged on a Zeiss LSM5 Pascal confocal microscope with a ×20 objective. In cases in which the tissue was too large, multiple images were taken and stitched together post-hoc in Adobe Photoshop. Images for DAPI and NeuN quantification were obtained on an Olympus BX61VS microscope with a ×20 objective and subsequently process and exported into JPEG format with the manufacturer’s software. When necessary, nuclei were labelled by adding Hoechst 33582 dye (Invitrogen) to the final PBS (1:5,000). Nuclei positive for the relevant marker were counted and quantified using ImageJ. In Npas4f/f mice that have been sparsely infected with AAV-Cre–GFP and then administered kainic acid, we see dendritic NPAS4 immunoreactivity in the NPAS4-WT but not in the NPAS4-KO neurons. This indicates to us that the low levels of dendritic staining we observe with our NPAS4 antibody is unlikely due to cross-reactivity with another protein.
The neurons used for Sholl analysis were from animals injected with the AAV-EF1a-YFP-2A-Cre. Tissue was fixed as described above. To maximize signal, GFP signal was amplified by staining with rabbit anti-GFP antibodies (1:1,000, Invitrogen). The secondary antibody was anti-rabbit Alexa-488 (1:500, Invitrogen). Neurons selected for analysis if there were few or no other infected neurons nearby to facilitate reconstruction of dendritic morphology. After imaging, the neurons were traced and skeletonized using the NeuronJ plugin for ImageJ and Sholl intersections were subsequently quantified and significance determined by ANOVA.
The neurons used for GFP–gephyrin puncta quantification were from animals injected with AAV-mCherry-IRES-Cre and AAV-FLEX-GFP–gephyrin. GFP was not amplified by immunostaining. Images were acquired on a Leica SP5 confocal with resonant scanner through a ×63 oil immersion objective. Fluorophores were excited using Argon 488 and HeNe 594 lasers and emitted photons detected through GFP or Alexa 594 filter cubes with Leica hybrid detectors (HyD3). Images were acquired at 16 bit resolution, 1024 × 1024 pixels and each frame the average of 8 scans. Images were analysed with ImageJ. Maximum intensity projections were made of each channel. A Yen threshold was applied to the green channel and puncta identified using the particle counter. Our reported puncta densities are lower than expected by electron microscopy. This is probably due to detection limitations of our microscope, incomplete or faint labelling of synapses by GFP–gephyrin, and the thickness of our sections.
Chromatin immunoprecipitation
Hippocampi from wild-type mice maintained in standard housing or injected with kainic acid (2.5–10 mg per kg, 2–3 h prior) were rapidly dissected in ice-cold dissection media (as above). The tissue was homogenized and DNA and protein were cross linked for 11 min (in mM: 10 HEPES-NaOH pH 7.5, 100 NaCl., 1 EDTA. 1 EGTA, 1% formaldehyde, in PBS). Formaldehyde was quenched with 2 M glycine in PBS and the sample incubated on a rocker at room temperature for 5 min. Tissue was pelleted (2,000 r.m.p., 5 min, 4 °C), washed with PBS plus PMSF, and re-pelleted (2,000 r.p.m., 5 min, 4 °C). The supernatant was removed and the pellet resuspended in 5 ml L1 buffer (in mM: 50 HEPES-NaOH 7.5, 140 NaCl, 1 EDTA, 1 EGTA, 0.25% Triton X-100, 0.5% NP40, 10% glycerol, 1M BGP, 0.2M NaVO4, 0.5M NaF, 1× complete protease inhibitor cocktail without EDTA (Roche)). The sample was the further homogenized by ten strokes with a tight pestle in a Dounce homogenizer, pelleted (2,000 r.p.m., 5 min, 4 °C), washed in L1, pelleted and then resuspended in buffer L2 (in mM: 10 Tris-HCl pH 8.0, 100 NaCl, 1M BGP, 0.2 M NaVO4, 0.5 M NaF, 1× complete protease inhibitor cocktail without EDTA (Roche)). Samples were placed on a rotator at room temperature for 10 min, pelleted and resuspended in L3 buffer (in mM 10 Tris-HCl pH 8.0, 1 EDTA, 1 EGTA, 1M BGP, 0.2M NaVO4, 0.5M NaF, 1× complete protease inhibitor cocktail without EDTA (Roche)) to a final concentration of 10 million nuclei per ml. Twenty million nuclei were sonicated using a Misonix 3000 for a total time of 7 min, power 7.5 and at 4 °C. NPAS4 immunoprecipitation was done using protein G Dyna Beads (Life Technologies) according to the manufacturer’s instructions using 4 µg of NPAS4 antibody10. NPAS4-bound DNA was quantified using qPCR. A list of putative NPAS4 target genes was determined by analysing ChIP and RNA-seq data published under Gene Expression Omnibus accession number GSE21161.
qRT–PCR
qRT–PCR was on cDNA samples made from RNA icollected from wild-type of NPAS4-KO hippocampi from mice that been housed in standard conditions or injected with kainic acid. qRT–PCR was carried out with SYBR green (Life Technologies) and primers with the following sequences for NPAS4 ChIP samples probed at the Bdnf locus:
| us | 5′-TGGTGAAAACACTTGGGCATA-3′; 5′-TGATGAGCTGGGAACTCTGC-3′; |
| P1 | 5′-GTCCGCTGGAGACCCTTAGT-3′, 5′-CTGAGCCAGTTACGTGACCA-3′; |
| I3 | 5′-CTTCCCAGATGGTGCTGT-3′, 5′-AATCTCCCAGTTCTGCGTTC-3′; |
| P4 | 5′-CCCTGGAACGGAATTCTTCT-3′, 5′-TGCACGAATTACCAGAATCA-3′; |
| Neg1 | 5′-CATTCAGCACCTTGGACAGA-3′, 5′-GCTTGACAGCGAGGAAAAGA-3′; |
| Neg2 | 5′-GGCCTGAAGTTCAAGGATGG-3′,5′-GCCTGCCACTGAAGCTTGTA-3′. |
In cDNA samples probed for various Bdnf isoforms, Npas4, PCSK1, Adcyap1 and Penk1, the primers used were:
| Bdnf exon1 | 5′-CACTGAGCAAAGCCGAACTTCTC-3′, 5′-TCACCTGGTGGAACATTGTGGC-3′; |
| Bdnf exon 2 | 5′-AGCGGTGTAGGCTGGAATAGACTC-3′, 5′-GGTGGAACTTCTTTGCGGCTTAC-3′; |
| Bdnf exon 3 | 5′-TACCCCTTTCTATCATCCCTCCCCG-3′, 5′-GAAGCATCCGGCCCGACAGTTCCAC-3′; |
| Bdnf exon 4 | 5′-CGCCATGCAATTTCCACTATCAATAATTTAAC-3′, 5′-CGCCTTCATGCAACCGAAGTATG-3′; |
| Bdnf exon 5 | 5′-CCATAACCCCGCACACTCTGTGTAG-3′, 5′-CTTCCCGCACCTTCACAGTTCCAC-3′; |
| Bdnf exon 6 | 5′-GATCCGAGAGCTTTGTGTGGAC-3′, 5′-GCCTTCATGCAACCGAAGTATG-3′; |
| Bdnf exon 7 | 5′-GGTCCAAGGTCAACGTTTA-3′, 5′-TAAACGTTGACCTTGGACC-3′, |
| Bdnf exon 8 | 5′-GAACAAACTGATTGCTGAA-3′, 5′-TTCAGCAATCAGTTTGTTC-3′; |
| Bdnf exon9/coding exon | 5′-GATGCCGCAAACATGTCTATGA-3′, 5′-TAATACTGTCACACACGCTCAGCTC-3′; |
| Npas4: | 5′-AGGGTTTGCTGATGAGTTGC-3′, 5′-CCCCTCCACTTCCATCTTC-3′; |
| PCSK1: | 5′-TGGAGTTGCATATAATTCCAAAGTT-3′, 5′-AGCCTCAATGGCATCAGTTAC-3′; |
| Adcyap1 | 5′-GAGAATCTGGGGGCAAGTCT-3′, 5′-CACCAGCACCTGATCTGTCA-3′; |
| Penk | 5′-CCCAGGCGACATCAATTT-3′, 5′-TCTCCCAGATTTTGAAAGAAGG-3′. |
Western blot analysis
Myc-tagged cDNA encoding NPAS4, BDNF, Pcsk1, Adcyap1 or PENK1 was transfected into 293T cells using Lipofectamine. Twenty-four hours later cells were lysed in 2× Laemmli buffer and boiled for 5 min. Lysates were resolved by SDS–PAGE and immunoblotted with antibodies targeting Myc (1:1,000, Abcam) or actin (1:2,500, Abcam).
Statistics
All data are shown as the mean ± s.e.m. unless otherwise noted. Data for which a specific P value is not indicated are not significant (P > 0.05).
For electrophysiology experiments, significance was determined by paired two-tailed t-test for direct comparisons of neighbouring neurons, two-tailed t-test for comparisons between populations, one-way ANOVA (Dunnett’s test) for PPRs, and Wilcoxon for mIPSCs in the screen of putative NPAS4 target genes. All data sets were acquired from at least 3 animals, 2 slices from each animal.
For imaging experiments, significance was determined by ANOVA or two-tailed t-test. All data sets were acquired from at least 3 animals, 3–5 images per animal.
For qRT–PCR experiments, significance was determined by two tailed t-test.
Extended Data
Extended Data Figure 1.
Infection with AAV-Cre-GFP effectively excises Npas4. a, Schematic of the AAV-Cre–GFP virus genome. b, Representative hippocampal section from an AAV-Cre–GFP; Npas4f/f animal 3 h after kainic acid injection. Sections were imaged for native GFP fluorescence (green) and immunostained NPAS4 (red) and NeuN (blue) protein. c, Quantification of neurons with overlapping GFP and NPAS4 in the sections represented in b. n = 14 neurons, *P < 0.01.
Extended Data Figure 2.
NPAS4-dependent changes in inhibition require loss of NPAS4 in excitatory neurons. a, mIPSCs frequency (left) and amplitude (right) were measured from pairs of neighbouring GFP+ and GFP− neurons from wild-type (C57BL/6) mice maintained in standard housing. n = 17 pairs. b, mIPSCs frequency (left) and amplitude (right) were measured from pairs of neighbouring GFP+ and GFP− neurons from wild-type (C57BL/6) mice 24 h after injection of kainic acid. n = 17 pairs. Open circles indicate individual GFP+/GFP− pairs. Red circles indicate mean ± s.e.m. c, Representative hippocampal CA1 section from an EMX-Cre;Npas4f/+ and EMX-Cre;Npas4f/f mouse 3 h after kainic acid injection. Sections were immunostained for NPAS4 (red) protein. d, Quantification of NPAS4 immunoreactivity. n = 9 sections from 3 animals. *P < 0.05. e, mIPSCs frequency and amplitude measured from neurons in EMX-Cre;Npas4f/f mice maintained in standard housing (n = 14 neurons) or 24 h after kainic acid injection (n = 16 neurons). freq: P < 0.01; amp: P < 0.05.
Extended Data Figure 3.
Stimulation of axons in different hippocampal layers generates eIPSCs with distinct rise times. a, Cartoon of a CA1 pyramidal neuron and example eIPSCs generated in response to stimulation of axons in lacunosum (L, blue), radiatum (R, yellow), pyramidale (P, red) and oriens (O, green). Slopes (10–90% of the peak eIPSC) are indicated by coloured lines. b, Standard housing summary of the slopes of the eIPSC rise times recorded from neighbouring NPAS4-WT and NPAS4-KO neurons in response to stimulation of axons in each of the four hippocampal layers. Measurements are from eIPSCs in Fig. 2b–e. c, Enriched environment summary of the slopes as in b. Measurements are from eIPSCs in Fig. 2f–i. d, Kainic acid summary of the slopes as in (b). Measurements are from eIPSCs in Extended Data Fig. 4b–e.
Extended Data Figure 4.
NPAS4 differentially regulates inhibitory synapses across the somato-dendritic axis of pyramidal neurons in response to kainic acid. a, Experimental configuration. L, lacunosum; R, radiatum; P, pyramidale; O, oriens. b–e, eIPSCs measured from mice injected with kainic acid. Top shows average eIPSC, normalized pairwise to the wild-type neuron, measured from NPAS4-WT (black) and NPAS4-KO (green) neurons in response to stimulation in lacunosum (b), radiatum (c), pyramidale (d) or oriens (e). Scale bars indicate per cent change from wild type. Bottom shows eIPSC amplitude measured from pairs of neighbouring NPAS4-KO and NPAS4-WT neurons in response to stimulation of axons in lacunosum (b) (n = 13 pairs); radiatum (c) (n = 17 pairs, P < 0.05); pyramidale (d) (n = 18 pairs, P < 0.01); or oriens (e) (n = 15 pairs, P < 0.05). Open circles represent NPAS4-KO/NPAS4-WT pairs. Red circles indicate mean ± s.e.m.
Extended Data Figure 5.
Sholl analysis and paired pulse ratios for NPAS4-WT and NPAS4-KO neurons from mice injected with kainic acid. a, Schematic of the AAV-YFP-2A-Cre virus genome used for Sholl analysis. b, Kainic acid example NPAS4-WT and NPAS4-KO CA1 pyramidal neurons (left) and quantification of NPAS4-WT (black, n = 10 neurons) and NPAS4-KO (red, n = 13 neurons) dendrites (right). c–e, PPRs measured from NPAS4-WT and NPAS4-KO neurons in response to stimulation in radiatum (c), pyramidale (d) or oriens (e). Standard housing (black) or kainic acid (red). Standard housing data are re-plotted from Fig. 3c–e. R, n = 11 neurons; P, n = 13 neurons; O, n = 8 neurons. All data are shown as mean ± s.e.m.
Extended Data Figure 6.
Functional screen of putative NPAS4 target genes identifies many genes that regulate mIPSCs frequency or amplitude. a, b, mIPSCs were recorded from CA1 pyramidal neurons transfected with a pool of three shRNAs targeting a single putative NPAS4 target gene. mIPSC frequency (a) and amplitude (b) of those significantly different from control are shown. Knockdown of genes that result in fewer or smaller mIPSCs are shown on top (mustard); knockdown of genes that result in more or larger mIPSCs are shown on the bottom (blue). The dashed line represents the median value from control conditions. P < 0.05.
Extended Data Figure 7.
Validation of five putative NPAS4 target genes. a–e, Select putative NPAS4 target genes that influence mIPSC frequency and/or amplitude were subjected to additional validation: Npas4 (a), Bdnf (b), Pcsk1 (c), Penk1 (d) and Adycap1 (e). Top shows mIPSC frequency and amplitude recorded from CA1 pyramidal neurons that were transfected with individual shRNAs from the pool used in the primary screen. The mean ± s.e.m. mIPSC frequency and amplitude recorded from control neurons is indicated by the grey bar. Expression of individual shRNAs phenocopied expression of the pool. Bottom shows expression of target-specific shRNAs reduces expression of the target protein. f, qRT–PCR using primers specific to the target gene indicate that all five genes are induced in the hippocampus and their induction is misregulated in NPAS4 knockout neurons. Standard housing, n = 5 animals; kainic acid, n = 6 animals. All data are shown as mean ± s.e.m. *P < 0.05.
Extended Data Figure 8.
Many putative NPAS4 target genes do not regulate inhibitory synapses. a, b, mIPSCs were recorded from CA1 pyramidal neurons transfected with a pool of three shRNAs targeting a single putative NPAS4 target gene. mIPSC frequency (a) and amplitude (b) of those not significantly different from control are shown.
Extended Data Figure 9.
BDNF regulates somatic inhibitory synapses in response to kainic acid. a–c, eIPSCs were measured simultaneously from neighbouring BDNF-WT (black) and BDNF-KO (green) neurons from mice injected with kainic acid. Top shows average eIPSC, normalized pairwise to the wild-type neuron, measured from BDNF-WT (black) and BDNF-KO (green) neurons in response to stimulation of axons in radiatum (a), pyramidale (b) or oriens (c). Scale bars indicate per cent change from wild type. Bottom shows eIPSC amplitude measured from pairs of neighbouring BDNF-KO and BDNF-WT neurons in response to stimulation of axons in radiatum (a) (n = 14 pairs), pyramidale (b) (n = 12 pairs, P < 0.01), or oriens (c) (n = 15 pairs). Open circles represent BDNF-KO/BDNF-WT pairs. Red circles indicate mean ± s.e.m.
Extended Data Table 1.
Putative NPAS4 target genes
| gene | chr | pos | induction of gene | induction at peak |
|---|---|---|---|---|
| 4921504E06Rik | 2 | 19476120 | 8 | 6.2 |
| 9530008L14Rik | 13 | 41896890 | 8.6 | 21 |
| Abcd2 | 15 | 90983260 | 2.48429 | 5.5 |
| Adcyap1 | 17 | 93602815 | 4.33656 | 5.929 |
| Ankrd56 | 5 | 93483350 | 15.9545 | 11 |
| Arhgap29 | 3 | 121676880 | 3.56853 | 5.429 |
| Bdnf | 2 | 109532475 | 35.2919 | 205.333 |
| Bdnf | 2 | 109518455 | 35.2919 | 4.333 |
| Bdnf | 2 | 109514675 | 35.2919 | 128 |
| Cd6 | 19 | 10859575 | 2.11111 | 7.5 |
| Cpa4 | 6 | 30522930 | 5.70588 | 25.5 |
| Cpxm2 | 7 | 139350455 | 2.56693 | 6.286 |
| Crem | 18 | 3281155 | 5.14777 | 16.8 |
| Crem | 18 | 3281830 | 5.14777 | 22 |
| Cyr61 | 3 | 145314740 | 4.5102 | 6 |
| D16Ertd472e | 16 | 78576220 | 5.54462 | 12 |
| D3Bwg0562e | 3 | 117060470 | 4.03298 | 10 |
| Dbc1 | 4 | 68615695 | 4.20687 | 5.005 |
| Ddx3y | 21 | 622975.5 | 3.93276 | 10 |
| Dkk2 | 3 | 131749835 | 9.25532 | 10.5 |
| Dusp6 | 10 | 98786170 | 2.54083 | 9 |
| E430004N04Rik | 10 | 28532080 | 8.13043 | 13 |
| Fer1l3 | 19 | 38066355 | 5.39535 | 74 |
| Fer1l3 | 19 | 37982665 | 5.39535 | 9 |
| Fos | 12 | 86812870 | 33.5814 | 14 |
| Fosb | 7 | 19905495 | 53 | 5.333 |
| Fosb | 7 | 19894115 | 53 | 20 |
| Fosl2 | 5 | 32435940 | 5.29613 | 14 |
| Fosl2 | 5 | 32438065 | 5.29613 | 5.875 |
| Gabra4 | 5 | 72008145 | 3.34333 | 5.5 |
| Gabrb1 | 5 | 72460260 | 2.17759 | 6 |
| Gcnt2 | 13 | 41013390 | 3.71429 | 6.2 |
| Gpr22 | 12 | 32394835 | 5.68515 | 7.676 |
| Gpr3 | 4 | 132767740 | 10.1849 | 15 |
| Hcn1 | 13 | 118394055 | 2.98126 | 5.5 |
| Hdac9 | 12 | 35213780 | 8.31166 | 9.947 |
| Hdac9 | 12 | 35214265 | 8.31166 | 9.915 |
| Hdac9 | 12 | 35182840 | 8.31166 | 5 |
| Hdac9 | 12 | 35166755 | 8.31166 | 24.75 |
| Hdac9 | 12 | 35183850 | 8.31166 | 18 |
| Hdac9 | 12 | 35173085 | 8.31166 | 5.31 |
| Hspb3 | 13 | 114453860 | 11.2703 | 10.947 |
| Hunk | 16 | 90387145 | 2.93033 | 5 |
| Iqsec3 | 6 | 121423910 | 2.03163 | 7.009 |
| Junb | 8 | 87502555 | 18.6986 | 19.684 |
| Kcna4 | 2 | 107257495 | 3.2902 | 8.857 |
| Kcnj3 | 2 | 55289600 | 5.1252 | 6.675 |
| Klhl4 | 20 | 111587385 | 6.72727 | 5.889 |
| Maff | 15 | 79177910 | 10.8679 | 14 |
| Maff | 15 | 79178470 | 10.8679 | 6.5 |
| Maml3 | 3 | 51906195 | 3.28369 | 5.75 |
| Maml3 | 3 | 51907390 | 3.28369 | 11.333 |
| Mest | 6 | 30695130 | 4.66716 | 6.274 |
| Myo3b | 2 | 69952395 | 2.02105 | 5 |
| Nfil3 | 13 | 53078245 | 4.87197 | 12.32 |
| Npas4 | 19 | 4989805 | 73.8489 | 58.538 |
| Npas4 | 19 | 4993095 | 73.8489 | 19 |
| Nptx2 | 5 | 145308275 | 7.35033 | 9.568 |
| Nr4a1 | 15 | 101097330 | 36.0876 | 66.4 |
| Nr4a2 | 2 | 56969220 | 6.71864 | 10.25 |
| Nr4a3 | 4 | 48057945 | 12.8444 | 10.559 |
| Nr4a3 | 4 | 48056410 | 12.8444 | 11.375 |
| Nr4a3 | 4 | 48057290 | 12.8444 | 12.034 |
| Olfml2a | 2 | 38806110 | 2.34211 | 17 |
| Pacsin3 | 2 | 91095860 | 2.12644 | 15 |
| Pam | 1 | 99928620 | 3.52883 | 11.647 |
| Pappa | 4 | 64788065 | 2.88618 | 8 |
| Pcdh11x | 20 | 117723835 | 2.41197 | 9 |
| Pcsk1 | 13 | 75228705 | 37.6 | 13.1 |
| Pde4d | 13 | 110612880 | 2.31523 | 50 |
| Penk1 | 4 | 4065825 | 2.1 | 5.231 |
| Per1 | 11 | 68913080 | 3.159 | 7.8 |
| Rabggtb | 3 | 153574995 | 3.08718 | 6.065 |
| Rgs2 | 1 | 145852135 | 8.16283 | 10.181 |
| Rgs2 | 1 | 145851380 | 8.16283 | 7.207 |
| Rgs2 | 1 | 145850695 | 8.16283 | 8.76 |
| Rspo3 | 10 | 29259485 | 2.27746 | 6 |
| Scn1a | 2 | 66246895 | 2.98474 | 6.875 |
| Sertad1 | 7 | 28271930 | 9.30631 | 7.526 |
| Slc6a17 | 3 | 107319165 | 2.54003 | 5 |
| Snx7 | 3 | 117526220 | 2.07258 | 5.5 |
| Snx7 | 3 | 117529915 | 2.07258 | 6.5 |
| Sorcs3 | 19 | 48315650 | 2.76436 | 11 |
| Sorcs3 | 19 | 48659720 | 2.76436 | 5.5 |
| Sorcs3 | 19 | 48172075 | 2.76436 | 8 |
| Spry2 | 14 | 106297475 | 2.01437 | 14.75 |
| Stac | 9 | 111591890 | 3.65815 | 5.083 |
| sult2b1 | 13 | 96731800 | 4.05891 | 9 |
| Sv2c | 3 | 93251570 | 2 | 7 |
| Tchh | 8 | 66685070 | 6.68364 | 5.579 |
| Tll1 | 9 | 53914130 | 2.51701 | 5.75 |
| Tnfaip8l3 | 1 | 162079810 | 11.2174 | 13 |
| Tnn | 9 | 8544010 | 8.38462 | 17 |
| Trpc6 | 6 | 30871095 | 5.66667 | 6.75 |
| Tsga13 | 6 | 30871525 | 5.66667 | 6 |
| Tsga13 | 6 | 30858185 | 5.66667 | 12 |
| Tsga13 | 8 | 74194390 | 2.24115 | 5.667 |
| Unc13a | 5 | 137505265 | 13.7907 | 10.625 |
| Vgf | 5 | 137506375 | 13.7907 | 14.077 |
| Vgf | 9 | 119920665 | 13.7907 | 6.8 |
Supplementary Material
Acknowledgements
We thank members of the Greenberg laboratory for comments, suggestions and critical reading of the manuscript. We thank A. Mardinly for observations and comments on NPAS4 protein induction, P. Greer and S. Flavell for observations and comments on Npas4 mRNA induction in response to enriched environment and B. Cruz Moreno for help with sectioning. We thank M. During for the gift of the AAV-Cre–GFP plasmid. GFP–gephyrin images were collected at the UCSD Neuroscience Microscopy Shared Facility sponsored by grant P30 NS047101. This work was funded by Helen Hay Whitney Foundation, L’Oreal USA Fellowship for Women in Science and The Medical Foundation/Charles A. King Trust Postdoctoral Fellowship (B.L.B.), NSF graduate student research fellowship (N.S.) and National Institutes of Health grant NS028829 (M.E.G.).
Footnotes
Author Contributions Experiments were designed by B.L.B., N.S. and M.E.G. Experiments were conducted and analysed by B.L.B., N.S., H.A.B. and A.Z.T. The manuscript was written by B.L.B., N.S. and M.E.G.
The authors declare no competing financial interests.
REFERENCES
- 1.Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J. Physiol. (Lond.) 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danglot L, Triller A, Marty S. The development of hippocampal interneurons inrodents. Hippocampus. 2006;16:1032–1060. doi: 10.1002/hipo.20225. [DOI] [PubMed] [Google Scholar]
- 3.Freund TF, Buzsáki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 4.Turner DA. Feed-forward inhibitory potentials and excitatory interactions in guinea-pig hippocampal pyramidal cells. J. Physiol. (Lond.) 1990;422:333–350. doi: 10.1113/jphysiol.1990.sp017987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu G. Local structural balance and functional interaction of excitatory and inhibitory synapses in hippocampal dendrites. Nature Neurosci. 2004;7:373–379. doi: 10.1038/nn1206. [DOI] [PubMed] [Google Scholar]
- 6.Chiu CQ, et al. Compartmentalization of GABAergic inhibitionby dendritic spines. Science. 2013;340:759–762. doi: 10.1126/science.1234274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larkum ME, Zhu JJ, Sakmann B. A new cellular mechanism for coupling inputs arriving at different cortical layers. Nature. 1999;398:338–341. doi: 10.1038/18686. [DOI] [PubMed] [Google Scholar]
- 8.Miles R, Toth K, Gulyas AI, Hajos N, Freund TF. Differences between somatic and dendritic inhibition in the hippocampus. Neuron. 1996;16:815–823. doi: 10.1016/s0896-6273(00)80101-4. [DOI] [PubMed] [Google Scholar]
- 9.Pouille F, Scanziani M. Enforcement of temporal fidelity in pyramidal cells by somatic feed-forward inhibition. Science. 2001;293:1159–1163. doi: 10.1126/science.1060342. [DOI] [PubMed] [Google Scholar]
- 10.Lin Y, et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adesnik H, Li G, During MJ, Pleasure SJ, Nicoll RA. NMDA receptors inhibit synapse unsilencing during brain development. Proc. Natl Acad. Sci. USA. 2008;105:5597–5602. doi: 10.1073/pnas.0800946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JL, et al. Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron. 2012;74:361–373. doi: 10.1016/j.neuron.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Versendaal D, et al. Elimination of inhibitory synapses is a major component of adult ocular dominance plasticity. Neuron. 2012;74:374–383. doi: 10.1016/j.neuron.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rocamora N, Welker E, Pascual M, Soriano E. Upregulation of BDNF mRNA expression in the barrel cortex of adult mice after sensory stimulation. J. Neurosci. 1996;16:4411–4419. doi: 10.1523/JNEUROSCI.16-14-04411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castrén E, Zafra F, Thoenen H, Lindholm D. Light regulates expression of brain-derived neurotrophic factor mRNA in rat visual cortex. Proc. Natl Acad. Sci. USA. 1992;89:9444–9448. doi: 10.1073/pnas.89.20.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lein ES, Shatz CJ. Rapid regulation of brain-derived neurotrophic factor mRNA within eye-specific circuits during ocular dominance column formation. J. Neurosci. 2000;20:1470–1483. doi: 10.1523/JNEUROSCI.20-04-01470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falkenberg T, et al. Increased expression of brain-derived neurotrophic factor mRNA in rat hippocampus is associated with improved spatial memory and enriched environment. Neurosci. Lett. 1992;138:153–156. doi: 10.1016/0304-3940(92)90494-r. [DOI] [PubMed] [Google Scholar]
- 19.Marty S, Wehrle R, Sotelo C. Neuronal activity and brain-derived neurotrophic factor regulate the density of inhibitory synapses in organotypic slice cultures of postnatal hippocampus. J. Neurosci. 2000;20:8087–8095. doi: 10.1523/JNEUROSCI.20-21-08087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohara K, et al. A local reduction in cortical GABAergic synapses after a loss of endogenous brain-derived neurotrophic factor, as revealed by single-cell gene knock-out method. J. Neurosci. 2007;27:7234–7244. doi: 10.1523/JNEUROSCI.1943-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldelli P, Hernandez-Guijo J-M, Carabelli V, Carbone E. Brain-derived neurotrophic factor enhances GABA release probability and nonuniform distribution of N- and P/Q-type channels on release sites of hippocampal inhibitory Synapses. J. Neurosci. 2005;25:3358–3368. doi: 10.1523/JNEUROSCI.4227-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattabiraman PP, et al. Neuronal activity regulates the developmental expression and subcellular localization of cortical BDNF mRNA isoforms in vivo. Mol. Cell. Neurosci. 2005;28:556–570. doi: 10.1016/j.mcn.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Brigadski T, Hartmann M, Lessmann V. Differential vesicular targeting and time course of synaptic secretion of the mammalian neurotrophins. J. Neurosci. 2005;25:7601–7614. doi: 10.1523/JNEUROSCI.1776-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean C, et al. Distinct subsets of Syt-IV/BDNF vesicles are sorted to axons versus dendrites and recruited to synapses by activity. J. Neurosci. 2012;32:5398–5413. doi: 10.1523/JNEUROSCI.4515-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atallah BV, Bruns W, Carandini M, Scanziani M. Parvalbumin-expressing interneurons linearly transform cortical responses to visual stimuli. Neuron. 2012;73:159–170. doi: 10.1016/j.neuron.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovett-Barron M, et al. Regulation of neuronal input transformations by tunable dendritic inhibition. Nature Neurosci. 2012;15:423–430. doi: 10.1038/nn.3024. [DOI] [PubMed] [Google Scholar]
- 27.Royer S, et al. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nature Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, et al. Activation of specific interneurons improves V1 feature selectivity and visual perception. Nature. 2012;488:379–383. doi: 10.1038/nature11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ploski JE, Monsey MS, Nguyen T, DiLeone RJ, Schafe GE. The neuronal PAS domain protein 4 (Npas4) is required for new and reactivated fear memories. PLoS ONE. 2011;6:e23760. doi: 10.1371/journal.pone.0023760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramamoorthi K, et al. Npas4 regulates a transcriptional programin CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.