SUMMARY
Whole-exome sequencing (WES) studies have demonstrated the contribution of de novo loss-of-function single nucleotide variants to autism spectrum disorders (ASD). However, challenges in the reliable detection of de novo insertions and deletions (indels) have limited inclusion of these variants in prior analyses. Through the application of a robust indel detection method to WES data from 787 ASD families (2,963 individuals), we demonstrate that de novo frameshift indels contribute to ASD risk (OR=1.6; 95%CI=1.0-2.7; p=0.03), are more common in female probands (p=0.02), are enriched among genes encoding FMRP targets (p=6×10−9), and arise predominantly on the paternal chromosome (p<0.001). Based on mutation rates in probands versus unaffected siblings, de novo frameshift indels contribute to risk in approximately 3.0% of individuals with ASD. Finally, through observing clustering of mutations in unrelated probands, we report two novel ASD-associated genes: KMT2E (MLL5), a chromatin regulator, and RIMS1, a regulator of synaptic vesicle release.
INTRODUCTION
Autism spectrum disorder (ASD) is a highly heritable neurodevelopmental syndrome of unknown etiology. An excess of de novo copy number variants (CNVs) in affected individuals is well established (Levy et al., 2011; Sanders et al., 2011; Sebat et al., 2007). Moreover, whole-exome sequencing (WES) studies have demonstrated that de novo loss-of-function (LoF) single nucleotide variants (SNVs) also carry significant risk for ASD (Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012; Sanders et al., 2012). Importantly, the observation of multiple de novo events at the same locus provides a reliable and statistically rigorous method to identify specific variations associated with ASD (Sanders et al., 2011; Sanders et al., 2012; Willsey et al., 2013). This approach has highlighted the contribution of CNVs at 16p11.2, 15q11.2-13, 22q11.2, 7q11.23, and NRXN1, and, to date, SNVs in nine genes: ANK2, CHD8, CUL3, DYRK1A, GRIN2B, KATNAL2, POGZ, SCN2A, and TBR1.
While these and similar studies have been critically important in outlining the genomic architecture of ASD (Buxbaum et al., 2012), they have not provided a comprehensive view of de novo variation in ASD. For example, systematic analysis of de novo insertions and deletions (indels) in WES data has been hindered by technological limitations including mapping errors and ambiguities in annotation leading to low sensitivity or infeasible numbers of confirmations.
We have resolved the most pressing issues in the detection of de novo indels by combining a family-based local realignment approach (Albers et al., 2011) with empirically derived quality metric thresholds to dramatically improve the accuracy of de novo indel prediction. We have applied this approach, followed by comprehensive de novo indel confirmation, to previously analyzed WES data from 2,963 individuals in 787 Simons Simplex Collection (SSC) families (Table S1), allowing a reliable analysis of the mutation rate in probands versus unaffected siblings. We identify 44 novel de novo coding indels and observe a significant excess of de novo frameshift indels in probands versus unaffected siblings with an odds ratio of 1.6, similar to that observed for de novo LoF SNVs. This additional data allows for a refinement of our prior analysis of the contribution of de novo disruptive events to ASD population risk. We now estimate that approximately 7% of affected individuals carry a de novo disruptive coding mutation contributing to ASD: 4% with a de novo LoF SNV and 3% with a de novo frameshift indel. Moreover, using our previously described approach to assessing the significance of clustering of de novo events at genomic loci (Sanders et al., 2011; Sanders et al., 2012; Willsey et al., 2013), we identify two novel ASD-associated genes: Lysine (K)-specific methyltransferase 2E (KMT2E, a.k.a Mixed-lineage leukemia 5 or MLL5) and Regulating synaptic exocytosis 1 (RIMS1), reinforcing prior findings highlighting a role for chromatin modification and synaptic function in the pathophysiology of ASD.
RESULTS
Identification and confirmation of de novo indels
To assess the burden of de novo indels in ASD, we analyzed WES data derived from whole-blood DNA from 787 families (602 quartets, 185 trios) in the SSC (Iossifov et al., 2012; O’Roak et al., 2012; Sanders et al., 2012; Willsey et al., 2013) (Table S1). Accurate prediction of indels is complicated by difficulties with alignment (Figure 1B) and multiple possible representations of the same indel in Variant Call File (VCF) format (Figure 1C). To overcome these difficulties, we developed an analysis pipeline optimized for de novo indel detection (Figure 1A) using Dindel local realignment (Albers et al., 2011) to correct alignment errors and the LeftAlignIndels tool from GATK (McKenna et al., 2010) to resolve problems with multiple representations of the same variant.
Figure 1. Experimental overview.
A) Indels were predicted in 787 families from the SSC using Dindel. Throughout the analytical pipeline, probands and siblings are treated equally to allow accurate assessment of de novo indel burden. Informative SNPs were used to establish the parent-of-origin of de novo indels. B) Alignment errors at the end of reads lead to indels being mis-called as SNVs. C) An indel can be represented in multiple ways in VCF format. See also Table S1.
Using this approach, we identified a total of 307 putative de novo indels (258 coding indels and 49 intronic) in cases and controls. All 307 were submitted for confirmation by PCR amplification and Sanger sequencing, blinded to affected status. High quality confirmation data were generated for 284 indels (93%), 146 of which were confirmed as being de novo (119 in coding regions and 27 in intronic regions), reflecting an overall confirmation rate of 51% (Table S2). While a 78% confirmation rate was achieved with more stringent detection thresholds, there was a corresponding 18% reduction in indel detection, so we elected to use the less stringent thresholds to maximize sensitivity.
To further assess the pipeline, we first evaluated our ability to detect 54 previously confirmed de novo indels within our current dataset (Iossifov et al., 2012; O’Roak et al., 2012). We correctly identified 52 (96%) of these; the remaining two indels were not detected by Dindel in the first step of our pipeline. In addition we detected and confirmed 6 (11%) novel de novo indels. Furthermore, using the latest iteration of GATK resulted in an 8% reduction in indel detection with no new de novo indels detected (Table S3). While the absence of a gold standard precludes accurate estimation of sensitivity, these results suggest that the method outlined in this manuscript is currently one of the most sensitive.
In addition to the 59 previously confirmed de novo coding indels in the SSC (Table S2) we confirmed an additional 16 previously predicted de novo coding indels and identified and confirmed 44 novel de novo coding indels.
Increased burden of de novo frameshift indels in ASD probands
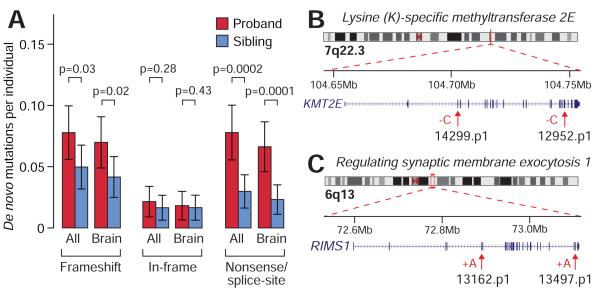
In total, we observed and confirmed 119 de novo coding indels: 79 in 787 probands and 40 in 602 unaffected siblings. To assess the burden of de novo indels in cases versus controls, we relied solely on the 100 indels detected in 602 quartet families that included both a proband and an unaffected sibling. We found 47 confirmed de novo indels that alter the reading frame (frameshift) in probands (0.078 per sample) compared to 30 (0.050 per sample) in siblings (OR: 1.6, 95% confidence interval: 1.0-2.7; p=0.03, one-sided Wilcoxon paired test; Figure 2A; Table S2); considering only brain-expressed genes resulted in a higher odds ratio of 1.7 (95% CI: 1.0-3.0; p=0.02; Figure 2A; Table S2). For de novo indels that do not alter the reading frame (in-frame) no such excess was observed with 13 (0.022 per sample) in probands and 10 (0.017 per sample) in siblings (OR: 1.3, 95% CI: 0.5-3.2; p=0.28, one-sided Wilcoxon paired test; Figure 2A). Similarly, no excess of intronic de novo indels was observed in ASD probands versus unaffected sibling controls (Figure S1). A similar burden of frameshift de novo indels was observed through the application of increasingly stringent quality metrics to the 258 putative de novo coding indels, instead of visualization and confirmation (Figure S2).
Figure 2. De novo indel burden and genes with multiple hits.
(A) The rate of de novo indels and SNVs is shown for 602 probands (red) and matched unaffected siblings (blue). “All” refers to all RefSeq genes in hg19. “Brain” refers to the subset of genes that are brain-expressed. “Nonsense” refers to single nucleotide substitutions that result in a premature stop codon; “splice-site” refers to single nucleotide substitutions that disrupt the canonical splice-site. Error bars represent the 95% confidence intervals and p-values are calculated with a one-sided paired Wilcoxon test. (B) Two de novo frameshift indels in independent samples are shown in the gene KMT2E. Both indels are likely to induce nonsense-mediated decay (Nagy and Maquat, 1998). (C) Two de novo frameshift indels in independent samples are shown in the gene RIMS1. Both indels are likely to induce nonsense-mediated decay (Nagy and Maquat, 1998). See also Figures S1 and S2.
As expected, these results mirror the previously reported burden of de novo LoF (nonsense or canonical splice-site) SNVs (OR: 2.4, 95% CI: 1.3-4.3; p=0.0002, one-sided Wilcoxon paired test; Figure 2A), while de novo missense SNVs show a trend toward overrepresentation in cases (OR: 1.1, 95% CI: 0.9-1.4; p=0.07) (Iossifov et al., 2012; O’Roak et al., 2012; Sanders et al., 2012; Willsey et al., 2013).
Two genes show multiple independent de novo LoF mutations
Given both the similar functional impact of frameshift indels and LoF SNVs, as well as the similarity between the observed odds ratio and frequency in ASD cases (Figure 2A), we concluded that these mutations could be treated as a single class of LoF mutations when considering the implications of observing multiple de novo disruptive mutations in the same gene. Using a permutation test (Sanders et al., 2012) that simulated de novo LoF mutations based on gene size and GC content at the rate observed in siblings (0.083 per sample), a gene with a single disruptive de novo mutation was found to have a 50.4% probability of being associated with ASD (q=0.496), while a gene with at least two disruptive de novo mutations has a 97.6% (q=0.024) probability of association with ASD.
Using this approach, we identified two ASD-associated genes (Table 1): Lysine (K)-specific methyltransferase 2E (KMT2E, also called Mixed-lineage leukemia 5 or MLL5, Figure 2B) and Regulating synaptic exocytosis 1 (RIMS1, Figure 2C).
Table 1. Novel de novo indels in genes with previously reported de novo non-synonymous mutations.
See also Table S2.
| Gene | Sample | hg19 Location | Variant | Effect | Source |
|---|---|---|---|---|---|
| CHD2 | 10C100480 | chr15:93518170 | C->T | Missense | (Neale et al., 2012) |
| 13618.p1 | chr15:93524060 | −AAAG | Frameshift | New | |
|
| |||||
| KMT2E | 14299.p1 | chr7:104702706 | −C | Frameshift | New |
| 12952.p1 | chr7:104748101 | −C | Frameshift | (Iossifov et al., 2012) | |
|
| |||||
| PHF3 | 14133.p1 | chr6:64413433 | −CG | Frameshift | New |
| 14110.p1 | chr6:64423242 | C->T | Missense | (Sanders et al., 2012) | |
|
| |||||
| RIMS1 | 13162.p1 | chr6:72889392 | +A | Frameshift | (Iossifov et al., 2012) |
| 13497.p1 | chr6:73102488 | +A | Frameshift | New | |
De novo frameshift indels support a role for FMRP targets in the pathophysiology of ASD
The identification of genes overtly reflecting chromatin modification and synaptic function in ASD led us to evaluate the putative functions of all 62 unique genes carrying de novo frameshift indels in the 787 probands (Table S2). We first assessed enrichment in gene ontology categories (GO) and KEGG pathways, as well as for connectivity of protein-protein interaction networks (DAPPLE). We found no significant results after correction for multiple comparisons.
We then turned to an assessment of mRNA targets of Fragile X Mental Retardation Protein (FMRP) in light of a recent analysis showing enrichment of de novo SNVs in this set of genes among affected individuals in the SSC (Iossifov et al., 2012). We assessed the intersection of genes in this study with 842 FMRP targets identified in mouse brain (Darnell et al., 2011) and 939 FMRP targets identified in embryonic kidney cells (HEK-293) (Ascano et al., 2012); 178 of these targets are present in both tissue types.
To ensure that factors known to influence de novo mutation rates did not confound the analysis, we used a generalized linear model of exome coverage, gene size and GC content, brain-expression, and identification as an FMRP target as predictors of genes carrying a de novo frameshift indel. We observed a strong signal for FMRP-targets identified in mouse brain but not human embryonic kidney (p = 6 × 10−9, mouse brain; p = 0.13, HEK-293; p = 1 × 10−6, combined list). No enrichment was observed for the 29 unique genes with frameshift de novo indels in siblings (p = 0.55 and p = 0.43, mouse brain and HEK-293 respectively).
We then considered our findings in light of the ASD-associated spatio-temporal co-expression networks recently reported by our group (Willsey et al., 2013). Since the prior work relied on overlapping sequencing data, including previously reported de novo indels, we focused only on the intersection of 18 newly identified frameshift indels detected in probands. The gene RIMS1 was found to be present in an ASD-associated network in the cerebellum and mediodorsal nucleus of the thalamus in early post-natal life.
Female probands have a greater burden of de novo frameshift indels
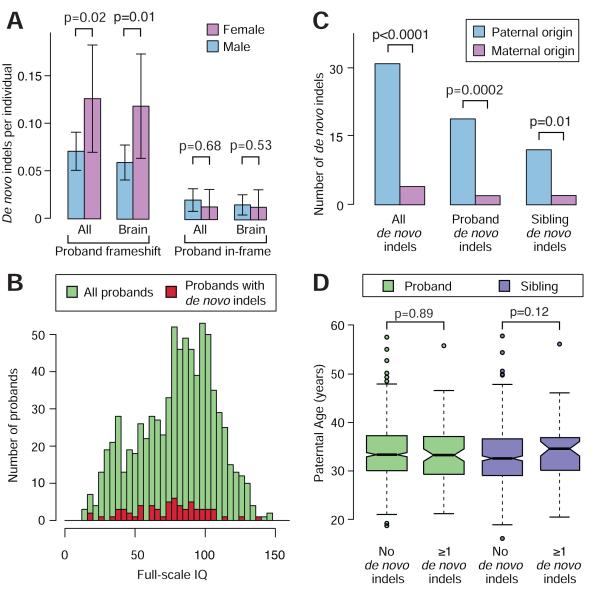
Female probands have previously been noted to have a higher burden of de novo CNVs than their male counterparts (Levy et al., 2011; Sanders et al., 2011), therefore we assessed the de novo indel burden by sex. A similar pattern was observed for de novo frameshift indels in probands, with 0.126 per sample in the 151 female cases compared to 0.071 per sample in the 636 male cases (OR: 1.9, 95% CI: 1.0-3.4; p=0.02, one-sided Wilcoxon unpaired test; Figure 3A). This sex-related burden was not observed for the de novo in-frame indels (OR: 0.6, 95% CI: 0.1-3.0; p=0.68, one-sided Wilcoxon unpaired test; Figure 3A).
Figure 3. Sex difference, parent-of-origin and parental age.
(A) A consistently higher rate of de novo frameshift indels was observed in female probands (pink) compared to male probands (blue), but this difference was not observed in unaffected siblings. “All” describes all de novo frameshift indels; “Brain” includes only those expressed in the brain. Error bars represent the 95% confidence intervals and p-values are calculated with a one-sided paired Wilcoxon test. (B) Histogram of full-scale IQ in all probands (green) and probands with a de novo frameshift indel (red). (C) The majority of de novo indels for which parent-of-origin could be resolved were found to be on the paternal (blue) rather than the maternal (pink) chromosome (p<0.001; Binomial). This result was observed in both probands and siblings separately. (D) No clear relationship between the presence of a de novo indel and increased paternal age was observed for probands (green) or siblings (purple). P-values were estimated with a Poisson regression.
De novo frameshift indels are associated with lower IQ
Given the significant clinical overlap between intellectual disability and ASD, and longstanding interest in the relative contribution of genetic risk to social versus intellectual disability (Skuse, 2007), we evaluated the relationship of IQ to mutation status. The presence of a de novo frameshift indel was associated with a 6.3 point decrement in proband full-scale IQ (FSIQ) (p<0.0001, Mann-Whitney U test) compared with probands with no known de novo LoF indel or SNV. However, de novo frameshift indels only explained a small fraction of variance in FSIQ (R2 = 0.004) and 43% of probands with de novo frameshift indels had FSIQ measures greater than the proband mean of 80.2 (Figure 3B). The current absence of FSIQ data for the parents prevents an analysis of the genetic deviation in FSIQ due to de novo mutations, as was recently performed for IQ in individuals with 16p11.2 CNVs (Zufferey et al., 2012) and for head circumference in the SSC (Chaste et al., 2013).
De novo indels arise predominantly from the paternal chromosome
Given the observation that the majority of de novo SNVs arise on the paternal chromosome (Kong et al., 2012; O’Roak et al., 2012) we assessed the parent-of-origin for the de novo indels. Informative SNPs (i.e., those unique to one parent and transmitted to the child) within 1,000bp of de novo indels were identified in WES data. The regions were amplified with PCR and sequenced on an Illumina MiSeq; visual inspection of the data allowed determination of parent-of-origin.
We observed a significant excess of de novo indels arising from the paternal chromosome (31 paternal vs. 4 maternal; p<0.001; binomial exact test; Figure 3C) as has been observed for de novo SNVs.
Correlation between parental age and de novo indels
Multiple prior studies, including our own (Kong et al., 2012; O’Roak et al., 2012; Sanders et al., 2012), have demonstrated a robust correlation of paternal age with the rate of de novo SNVs. Consequently, we tested for this relationship with regard to de novo indels by fitting a linear model with paternal age (yrs) at the child’s birth as a predictor for the presence of a de novo indel. Surprisingly, we found no association with paternal age (slope b=0.01, standard error ±0.01, p=0.33, regression). This result was not altered by considering maternal age (b=0.01, p=0.41), probands only (b=0.00, p=0.89; Figure 3D), siblings only (b=0.03, p=0.12; Figure 3D), or excluding frameshift indels (b=0.02, p=0.34). In comparison, applying the same model to the de novo SNVs continued to show a robust association for paternal age (b=0.02, standard error ±0.01, p=0.0002) equivalent to an extra 0.2 de novo coding mutations per decade of the father’s age.
The contribution of de novo indels and SNVs to ASD population risk
Based on the observed difference in de novo mutation burden between cases and controls (Figure 2A), we predict that 3% of affected individuals carry de novo risk frameshift indels in addition to 4% with de novo risk LoF SNVs. Should ASD association be demonstrated for de novo missense and de novo in-frame mutations (as is likely with increased power), they would potentially account for a further 7% of ASD individuals.
DISCUSSION
Analysis of 787 ASD families from the SSC, including 602 unaffected sibling controls, demonstrates the association of de novo frameshift indels with ASD. Furthermore, the similarity in odds ratio and mutation rate to that observed for de novo LoF SNVs, as well as the overlap in the functional consequences, fits with the assumption that de novo frameshift indels and de novo LoF SNVs can be considered as a single group of highly disruptive mutations. Overall, these disruptive mutations are predicted to contribute to risk in 7% of the ASD population.
The present re-analysis of WES data from the SSC cohort, using a more sensitive and reliable approach to de novo indel discovery, identifies two new ASD genes: KMT2E (MLL5) and RIMS1. KMT2E is a chromatin regulator recruited to methylated histones, specifically H3K4me3, found at the promoter of actively expressed genes. It was initially identified as a tumor suppressor gene and its role in hematopoietic stem cell homeostasis and self-renewal has been well documented. However, the gene is highly pleiotropic, with roles in cytokinesis, response to DNA damage, and genome maintenance (Ali et al., 2013). While KMT2E has not previously been associated with neurological disorders, chromatin regulation in fetal development has been identified as a key risk factor for ASD (O’Roak et al., 2012; Willsey et al., 2013), and the gene is highly expressed throughout the brain, especially during fetal development (Kang et al., 2011).
RIMS1 is a RAS signaling gene that is essential for multiple aspects of neurotransmitter release. It plays a role in presynaptic plasticity (Kaeser et al., 2012), with mouse knockouts showing deficits in learning and memory (Powell et al., 2004) and increased seizure frequency following induced status epilepticus (Pitsch et al., 2012). RIMS1 is expressed throughout the human brain, with levels increasing throughout development and reaching a plateau in the third trimester that persists throughout adulthood (Kang et al., 2011). The gene is present in an ASD-associated postnatal co-expression network in the cerebellum and mediodorsal nucleus of the thalamus (8-10 MD-CBC) due to its co-expression with the ASD gene SCN2A (Willsey et al., 2013).
FSIQ is below the proband average of 81 in the SSC (range 46-74) in all four individuals with mutations in KMT2E and RIMS1. Several scales within the Child Behavior Checklist (CBCL) were elevated for both individuals with RIMS1 mutations only, possibly indicating a degree of anxiety or depression. Inconsistent results were observed for other phenotypic measures, including seizures and head circumference.
While the ASD-associated de novo indels do not form a highly connected protein-protein interaction network or show enrichment for gene ontology terms, we do confirm the previously documented enrichment of FMRP target genes carrying de novo LoF mutations (Iossifov et al., 2012). In light of the strength and reproducibility of this relationship, the identification of mRNAs targeted by FMRP in the developing human brain is likely to be a valuable resource for ASD gene discovery.
Given the observed similarities between de novo frameshift indels and de novo LoF SNVs, a marked over-representation of mutations on the paternal allele might have been anticipated. However, we did not observe the expected correlation between these paternally enriched de novo mutations and paternal age. Given the relatively small number of indels it is likely that this negative result reflects inadequate statistical power. We will test this hypothesis as substantially larger WES datasets from ASD families become available in the near future (Buxbaum et al., 2012).
Finally, we investigated the relationship between de novo frameshift indels and IQ. Given the association of many established ASD mutations with decrements in cognitive functioning and the frequent phenotypic overlap seen in clinical samples, there has been speculation that de novo disruptive mutations may only carry risk for intellectual disability (ID), and not for the core social deficits that define ASD. Our data do not support this hypothesis. Though we observe lower IQ among probands that carry de novo frameshift indels, compared to probands without any de novo LoF mutations, the difference is small (6.3 IQ points), accounts for only a fraction of the variance in IQ (R2=0.004), and the distribution of IQ is similar to that of other probands (Figure 3B). Moreover, given an emerging picture of shared risks for de novo SNVs among a wide range of neurodevelopmental syndromes (Allen et al., 2013; Fromer et al., 2014; Moreno-De-Luca et al., 2014), the most parsimonious explanation is that a subset of highly disruptive risk mutations are associated with a range of phenotypic outcomes that includes, but is not limited to, ID, ASD, schizophrenia, and epilepsy.
Based on current estimates, detection of de novo frameshift indels and LoF SNVs has the capacity to identify a genetic contribution in approximately 7% of affected individuals, rivaling the contribution of de novo CNVs (Sanders et al., 2011). Moreover, in addition to confirming important recent observations regarding the genomic architecture of ASD, including the paternal origin of the majority of small de novo mutations, the approach is yielding a growing list of ASD risk genes, pointing to chromatin modification, synaptic functioning, and binding to FMRP as key pathophysiological mechanisms.
EXPERIMENTAL PROCEDURES
Sample collection and initial data processing
Whole-exome data for 2,963 samples from 787 families (602 quartets and 185 trios) in the SSC were obtained (Table S1). Exome capture had been performed using a NimbleGen custom array (N5210) or NimbleGen EZExomeV2.0 (N5718) followed by sequencing on the Illumina GAIIx or HiSeq2000 instruments. Reads were aligned to hg19 with BWA.
Family-based de novo indel detection
Indels were predicted in children using Dindel (Albers et al., 2011) followed by Dindel local realignment for all family members. The LeftAlignIndels tool from GATK (McKenna et al., 2010) was applied to all the resulting BAM files, and indels were assessed in the realigned files. Rare inherited heterozygous indels were used to set appropriate quality filters to identify rare de novo indels, including: ≥10 unique reads in all family members; indel not observed in other SSC families; and <5% of reads with an indel in either parent.
Realigned BAM files for the resulting 522 putative de novo coding indels (0.39 per sample in probands, 0.37 per sample in siblings) were visualized using Integrative Genome Viewer (IGV) (Thorvaldsdóttir et al., 2013) by two independent researchers who were blinded to affected status. High concordance between the two researchers was observed (kappa coefficient = 0.94) and any indel that was potentially de novo according to either researcher was submitted for confirmation. In total 258 indels (50%, 0.27 per sample in probands, 0.16 per sample in siblings) were selected. In addition, the 49 intronic de novo indels with the best indel quality scores were submitted for confirmation as an additional control, to give a total of 307 confirmations.
Indel confirmations
Indels were confirmed using PCR amplification of whole-blood DNA and Sanger sequencing. Of the 307 putative de novo indels, high quality confirmation data were generated for 284 (96%). Of these, no indel was observed in the child for 44 (15%), while an inherited indel was observed in 93 (33%). One confirmed indel was observed in both children, but not in either parent, suggesting germline mosaicism. This left 146 confirmed de novo indels and a confirmation rate of 51%.
Identifying parent-of-origin
Informative SNPs within 1,000bp of a confirmed de novo indel were identified in WES data. The regions were amplified from whole-blood DNA of the index child and both parents using PCR. Amplified DNA was normalized using PicoGreen quantitation and pooled separately for children, fathers, and mothers. Each pool underwent indexed library preparation and was run on an Illumina MiSeq with 250bp paired-end reads. The aligned sequence data were assessed in IGV.
Supplementary Material
HIGHLIGHTS.
De novo frameshift indels are associated with ASD with an odds ratio of 1.6
Multiple de novo indels in KMT2E and RIMS1 implicate these genes in ASD
88% of de novo indels arise on the paternal chromosome
Synaptic function, chromatin modification, and FMRP targets play key roles in ASD
ACKNOWLEDGEMENTS
We are grateful to the families participating in the Simons Foundation Autism Research Initiative (SFARI) Simplex Collection (SSC). This work was supported by a grant from the Simons Foundation (to MWS), the CIHR (DRA to AJW), the HHMI (International Student Research Fellowship to SJS), the NIMH (R37 MH057881 to KR and BD), and the National Center for Research Resources (NCRR, UL1 TR000142 and KL2 TR000140 to A.G.E.). LW was supported by National Natural Science Foundation of China (No. 31025014) and Ministry of Science and Technology of China (No. 2012CB837600). We would like to thank the SSC principal investigators A. L. Beaudet, R. Bernier, J. Constantino, E. H. Cook Jr, E. Fombonne, D. Geschwind, D. E. Grice, A. Klin, D. H. Ledbetter, C. Lord, C. L. Martin, D. M. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. W. State, W. Stone, J. S. Sutcliffe, C. A. Walsh and E. Wijsman and the coordinators and staff at the SSC clinical sites; the SFARI staff; the Rutgers University Cell and DNA repository for accessing biomaterials; N. Buenaventura and L. Chow for their help in administering the project at UCSF; and T. Brooks-Boone, N. Wright-Davis, and M. Wojciechowski for their help in administering the project at Yale.
Footnotes
AUTHOR CONTRIBUTIONS S.D., M.W.S, L.W. and S.J.S. designed the study. S.D., N.J.C., A.J.W., A.Y.Y., V.H.B., J.F.K., N.A.T., J.G. and S.J.S. developed analysis methods and analyzed the data. S.D., M.F.W., M.D., Z.W., L.E.G., J.D.D., S.F., J.D., J.D.M., C.A.S., N.M.D., R.O.K., Z.Y., S.M.S., A.G.E., A.R.G., S.M.M., M.S. and A.I.B confirmed the indels. S.D., K.R., B.D., M.W.S, L.W. and S.J.S. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Rincón-Arano H, Zhao W, Rothbart SB, Tong Q, Parkhurst SM, Strahl BD, Deng LW, Groudine M, Kutateladze TG. Molecular basis for chromatin binding and regulation of MLL5. Proc Natl Acad Sci U S A. 2013;110:11296–11301. doi: 10.1073/pnas.1310156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, Goldstein DB, Han Y, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW, Consortium TAS. The Autism Sequencing Consortium: Large-Scale, High-Throughput Sequencing in Autism Spectrum Disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, et al. Adjusting Head Circumference for Covariates in Autism: Clinical Correlates of a Highly Heritable Continuous Trait. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee Y.-h., Narzisi G, Leotta A, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeser PS, Deng L, Fan M, Südhof TC. RIM genes differentially contribute to organizing presynaptic release sites. Proc Natl Acad Sci U S A. 2012;109:11830–11835. doi: 10.1073/pnas.1209318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Wong WS, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee Y-H, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare De Novo and Transmitted Copy-Number Variation in Autistic Spectrum Disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Moreno-De-Luca A, Cubells JF, Sanders SJ. Cross-Disorder Comparison of Four Neuropsychiatric CNV Loci. Current Genetic Medicine Reports. 2014;2:1–11. [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012 doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012 doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsch J, Opitz T, Borm V, Woitecki A, Staniek M, Beck H, Becker AJ, Schoch S. The presynaptic active zone protein RIM1α controls epileptogenesis following status epilepticus. J Neurosci. 2012;32:12384–12395. doi: 10.1523/JNEUROSCI.0223-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, Südhof TC, Nestler EJ. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/s0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH. Rethinking the nature of genetic vulnerability to autistic spectrum disorders. Trends Genet. 2007;23:387–395. doi: 10.1016/j.tig.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey F, Sherr EH, Beckmann ND, Hanson E, Maillard AM, Hippolyte L, Macé A, Ferrari C, Kutalik Z, Andrieux J, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.