Abstract
Genetic and genomic studies have enhanced our understanding of complex neurodegenerative diseases that exert a devastating impact on individuals and society. One such disease, age-related macular degeneration (AMD), is a major cause of progressive and debilitating visual impairment. Since the pioneering discovery in 2005 of complement factor H (CFH) as a major AMD susceptibility gene, extensive investigations have confirmed 19 additional genetic risk loci, and more are anticipated. In addition to common variants identified by now-conventional genome-wide association studies, targeted genomic sequencing and exome-chip analyses are uncovering rare variant alleles of high impact. Here, we provide a critical review of the ongoing genetic studies and of common and rare risk variants at a total of 20 susceptibility loci, which together explain 40–60% of the disease heritability but provide limited power for diagnostic testing of disease risk. Identification of these susceptibility loci has begun to untangle the complex biological pathways underlying AMD pathophysiology, pointing to new testable paradigms for treatment.
Keywords: complex disease, genetic susceptibility, neurodegeneration, retina, blindness
INTRODUCTION
Impairment of sight and blindness are debilitating and are among the three most feared medical conditions, after cancer and cardiovascular disease. Age-related macular degeneration (AMD) afflicts almost 10 million individuals in the United States alone, twice the number of Alzheimer’s disease patients and roughly equal to the total of all cancer patients combined (32, 39, 43). At least 2 million Americans over the age of 50 suffer from late-stage AMD that severely affects the quality of their life and results in billions of dollars in health care costs (72).Worldwide, AMD is the third-largest cause of vision loss (76). The problem is expected to worsen as the population ages, and no effective treatment or cure is currently available for a majority of affected patients.
AMD is a multifactorial, late-onset human disease characterized by the formation of lipid-rich extracellular deposits, localized inflammation, and ultimately neurodegeneration in the central part of the retina (termed the macula). The etiology of AMD is complex, with advanced age and family history being major risk factors (52, 92). In addition, smoking and nutrition have a significant impact on disease progression (8, 86). Patients with AMD exhibit loss of central vision with a spectrum of clinical phenotypes.
Clinical examination of human retinas can reveal distinct hallmarks of AMD (Figure 1) that can be broadly divided into early/intermediate and late (advanced) stages (29). Early/intermediate AMD [Age-Related Eye Disease Study (AREDS) grades 2 and 3] is the most common and least severe form, characterized by pigmentary abnormalities in the macula and accumulation of extracellular aggregates (called drusen). Late AMD (AREDS grades 4 and 5) is usually subdivided into dry [geographic atrophy (GA)] and wet [choroidal neovascularization (CNV)] (29). Vision impairment in AMD is largely irreversible. CNV can be treated but not cured with inhibitors of vascular endothelial growth factor (VEGF) (81). Monthly injections of VEGF inhibitors are expensive to the health care system and burdensome to patients (20). Additionally, patients with CNV exhibit a great deal of variability in response to anti-VEGF treatments (65). GA is not yet treatable (67).
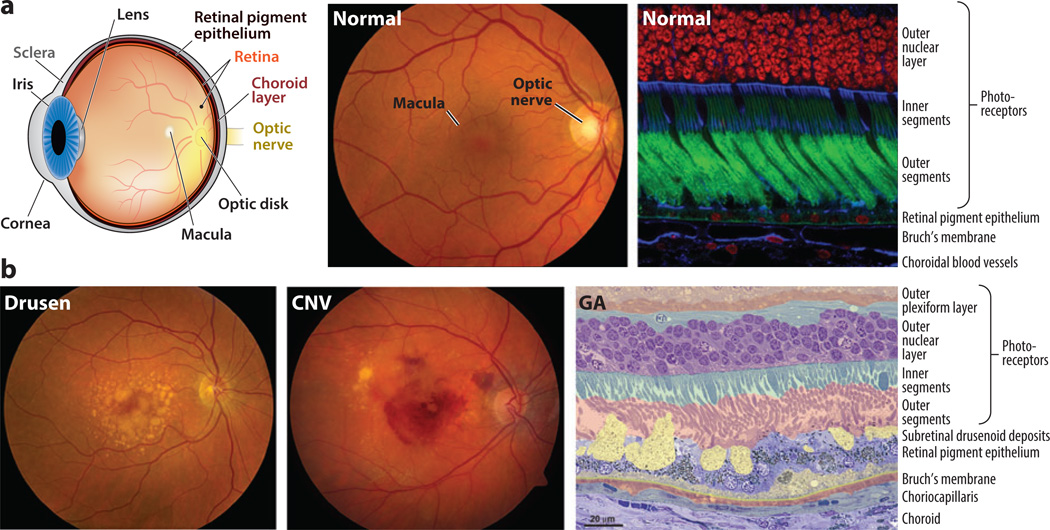
Figure 1.
(a) Normal retina. (Left) Anatomical depiction of the human eye viewed in sagittal section. Light entering the eye passes through the cornea and is focused by the lens onto the retina. The macula (Latin for “spot”) is a feature of human and most nonhuman primate retinas and lies in the central visual axis. The fovea (Latin for “pit”) is a small region at the center of the macula adapted for high-acuity vision. Two vascular networks—one in the neural retina and the other in the choroid—support the high metabolic demands of the retina. Rod and cone photoreceptors rely principally on the choroidal blood supply. Metabolites destined for the rods and cones are shuttled from the choroid to the photoreceptors by the retinal pigment epithelium. Neuronal impulses generated in the retina are conveyed to the brain by the optic nerve. (Center) Fundus photograph of the retina of a healthy individual. Using an ophthalmoscope, the retina can be evaluated as part of a routine eye exam. The optic nerve, macula, and retinal blood vessels provide ophthalmologists with clues about patients’ ocular health. (Right) Confocal microscopy image of a human retina labeled with fluorescent probes to show the close apposition of photoreceptors, retinal pigment epithelium, Bruch’s membrane, and choroidal blood vessels. Cell nuclei in the outer nuclear layer of photoreceptors are red, photoreceptor outer segments are green, and structures containing high concentrations of filamentous actin (cell–cell junctions and vessel walls) are blue. The inner segments of photoreceptors contain metabolic components, including numerous mitochondria. (b) AMD pathology. (Left,center) Fundus photographs depicting two distinct pathological features of AMD. The photograph on the left shows the characteristic accumulation of drusen, which appear as yellow splotches and flecks within the macula. The photograph in the center shows the unmistakable evidence of hemorrhage beneath the retina resulting from the pathological process of choroidal neovascularization (CNV). The decline in visual function associated with CNV hemorrhage is often rapid and irreversible. (Right) Micrograph showing pathological changes characteristic of AMD, outside the zone of geographic atrophy (GA). Layers of cells and extracellular lesions have been pseudocolored to highlight the extensive disorganization that has occurred at the interfaces between the photoreceptors, the retinal pigment epithelium, Bruch’s membrane, and the choroid. The outer plexiform layer includes synapses of photoreceptors with neurons in the inner retina. Lipid-rich deposits accumulate between the retinal pigment epithelium and Bruch’s membrane (dull yellow), and between the retinal pigment epithelium and photoreceptors (bright yellow). These deposits compromise the delivery of metabolites from the choroid to the photoreceptors and may initiate or exacerbate inflammatory processes, resulting in retinal pigment epithelium and photoreceptor cell death. Adapted in part from Reference 104.
We are ushering in an exciting era in medicine, with high expectations that genetic and genomic advances will transform patient care (10, 28, 58). The influences of the Human Genome Project, the International HapMap Consortium, and next-generation DNA sequencing are clearly evident in ophthalmology and vision research (105). Five years ago in this journal, we reviewed genetic studies of AMD and suggested a model of AMD progression, providing a context for retinal biology and the impact of aging (104). Since then, many articles have reviewed the clinical and biological aspects of AMD and discussed the relationship of inflammation, the complement pathway, and cholesterol transport to disease (2, 3,15, 30, 69, 74). Here, in addition to providing the current status of AMD genetics, we critically review the genetic data and place it in the context of our understanding of biological pathways and AMD pathogenesis. We also propose a multi-hit “threshold” model of AMD pathogenesis, in which aging and environmental factors synergistically augment the impact of genetic predisposition on disease onset and progression.
PHOTORECEPTORS–RETINAL PIGMENT EPITHELIUM–CHOROID: A LIFELONG PARTNERSHIP
AMD can be considered a multifactorial disease of the photoreceptor support system, which includes the retinal pigment epithelium (RPE), Bruch’s membrane (BrM), and the choroidal vasculature. The fundamental cause of vision loss in AMD is progressive damage to photoreceptors, which can be triggered by RPE dysfunction and atrophy; impaired transport of oxygen, nutrients, and metabolites between vessels and outer retinal cells; and leakage from choroidal capillaries that invade the retina through the RPE. We first explain the anatomical features of the eye involved in AMD to provide a context for understanding the impact of genetics on the disease biology.
Light entering the eye is focused on the retina, where exquisitely specialized rod and cone photoreceptors transduce the stimuli into chemical signals through a process termed phototransduction (Figure 1). A complex neural circuitry within the retina relays these signals to visual centers in the brain (62). Retinal photoreceptors are metabolically active neurons with oxygen requirements that are among the highest in the human body (116). The outer segments of rod and cone photoreceptors comprise stacks of flattened membranous discs containing light-sensitive photopigments (rhodopsin in rods and spectrally tuned red-, green-, or blue-sensitive opsins in cones) that must be regenerated after photon capture (77). In humans, rods and cones exhibit a distinct topography; the macula (6 mm in diameter) contains a cone-dominated fovea (0.8 mm in diameter) that is associated with high-acuity vision (19) (Figure 1a).
A unique support system serves the needs of photoreceptors by delivering the resources to carry out visual function. The photoreceptors have an intimate relationship with the RPE, which is critical for recycling the retinoids for phototransduction. The RPE comprises a single layer of polarized cells with extensive villi that interdigitate with the tips of rod and cone outer segments (Figure 1a). Approximately 10% of photoreceptor outer-segment discs are shed daily and phagocytosed by the RPE (59, 119). The RPE maintains a blood–retina barrier that isolates the photoreceptors on one side from the systemic circulation on the other side. Rod and cone photoreceptors depend on the RPE for delivery of oxygen and metabolites (101, 114). Photoreceptor support and choroidal maintenance functions are performed by the RPE for the entire life span without respite.
The choroid is an extensive vascular complex lining the posterior part of the eye. The innermost layer of choroidal vessels is a meshwork of capillaries that supply nutrients utilized by the retina and act as a conduit for the by-products of photoreceptor and RPE metabolism. Blood flow through the choroid sustains the high oxygen levels required by the mitochondria-packed photoreceptor inner segments. The inner aspect of the choroid, next to the RPE, is BrM, a laminar extracellular matrix (ECM) of collagen and elastin (16). BrM represents a crucial link in the supply chain through its transport of oxygen, glucose, and other metabolic components to the RPE and photoreceptors and its return of metabolic waste to the systemic circulation.
AGING-ASSOCIATED CHANGES IN THE RETINA
Age is the most significant risk factor for AMD. Disease prevalence for late AMD can peak near 10% in persons over age 80 (72). When genetic risk and environmental triggers such as smoking or diet act in concert with increasing age, the pathological changes of AMD become more likely.
In the human retina, advanced age is associated with an almost 30% loss of rod photoreceptors in the central macula, with a minimal effect on cone number (18). In addition, the RPE cells accumulate autofluorescent granules termed lipofuscin, which are remnants of retinoid metabolites from shed photoreceptor outer-segment membranes. A2E, an abundant component of lipofuscin (21, 25, 101), is hypothesized to exacerbate AMD through photooxidation and phototoxicity in RPE cells (96, 97). However, direct evidence for a role of A2E in vivo is lacking.
Age-related ocular changes also include increased BrM thickness (27), a potential mechanism for AMD onset (99). However, decreased thickness of the collagenous and elastin components of BrM has also been reported in some AMD patients (9). Another significant pathological change in BrM is the accumulation of lipoproteins that contain apolipoproteins B and E, cholesterol (17), and a toxic oxidized product termed 7-ketocholesterol (70). A preferential accumulation of these materials in the macula can act as a diffusion barrier and stimulus for inflammation.
AMD PATHOLOGY
AMD histopathological analysis has focused on human donor eyes, as a good animal model of AMD has yet to emerge. In early AMD, the macula has an abnormal RPE pigment distribution and drusen in the sub-RPE space between the basal lamina of RPE and the inner collagenous layer of BrM (85, 98), which may lead to a modest decline in visual acuity and function. Large or soft drusen with indistinct borders are a major risk factor for AMD progression (Figure 1). Drusen components include lipids (>40% of volume) and an array of proteins, including those involved in complement regulation, TIMP3, vitronectin, β-amyloid, and apolipoproteins (E, B, A-I, C-I, and C-II), plus zinc and iron ions (12, 14, 71). The abundance of a diffusely distributed and stereotypic thickening of RPE basal lamina, called basal laminar deposit, also serves as a marker of disease severity, likely reflecting RPE stress level (83). Other extracellular lesions between the photoreceptors and RPE, called subretinal drusenoid deposits, are observed in many patients (Figure 1b) and can confer risk for AMD progression independent of drusen (16, 122).
GA, a common manifestation of late AMD, includes the degeneration and death of RPE cells, photoreceptors, and choroidal capillaries (66). In contrast to the typical slow progression of GA, CNV can decrease vision acutely through the abrupt onset of edema and bleeding from new capillaries invading the RPE and neural retina.
THE GENETIC ARCHITECTURE OF AMD
AMD susceptibility is determined by a combination of aging, genetics, and environmental factors (104). The strongest nongenetic risk factors are advanced age and smoking (23, 108). Support for a genetic contribution is incontrovertible and comes from both classical epidemiological and twin studies (38, 50,51, 68, 87) and gene-mapping studies. In the early twenty-first century, family-based linkage studies suggested major susceptibility loci on chromosomes 1 and 10 in addition to weak linkage at many other genomic regions (31, 103). In a groundbreaking accomplishment, several groups reported and validated the identification of complement factor H (CFH) on chromosome 1q as a strong AMD susceptibility locus (24, 35,37, 56, 120), unleashing a flurry of research that successfully identified many additional loci within a short period (11, 34,48, 78, 118). Risk alleles have also been suggested in mitochondrial DNA, although the evidence remains inconsistent (6, 45,84, 110).
More recently, genome-wide association studies of large samples (7, 73) and subsequent meta-analyses have uncovered common risk variants for advanced AMD in 19 susceptibility loci (33) (Figure 2). In addition to the primary signals (Figure 3), independently associated common variants have been identified at several loci (33), including—for example—the CFH and C2/CFB genes (7, 24,34, 61). Targeted genomic resequencing of selected loci has identified nonsynonymous rare variants in four complement genes (CFH, CFI, C3, and a new AMD susceptibility locus, C9) (Figure 2). One nonsynonymous variant, CFH:p. Arg1210Cys, increases AMD risk by >20-fold and is virtually absent in control individuals (1 heterozygous carrier in 2,268 sequenced controls) (121). Three other nonsynonymous variants (CFI:p.Gly119Arg, C3:p.Lys155Gln, and C9:p.Pro167Ser) have allele frequencies between 0.1% and 1% in controls and are associated with odds ratios of 2–4 (40, 75,90, 113, 121). Future studies with larger sample sizes and deeper coverage of exomic regions might identify additional functional variants with larger effects.
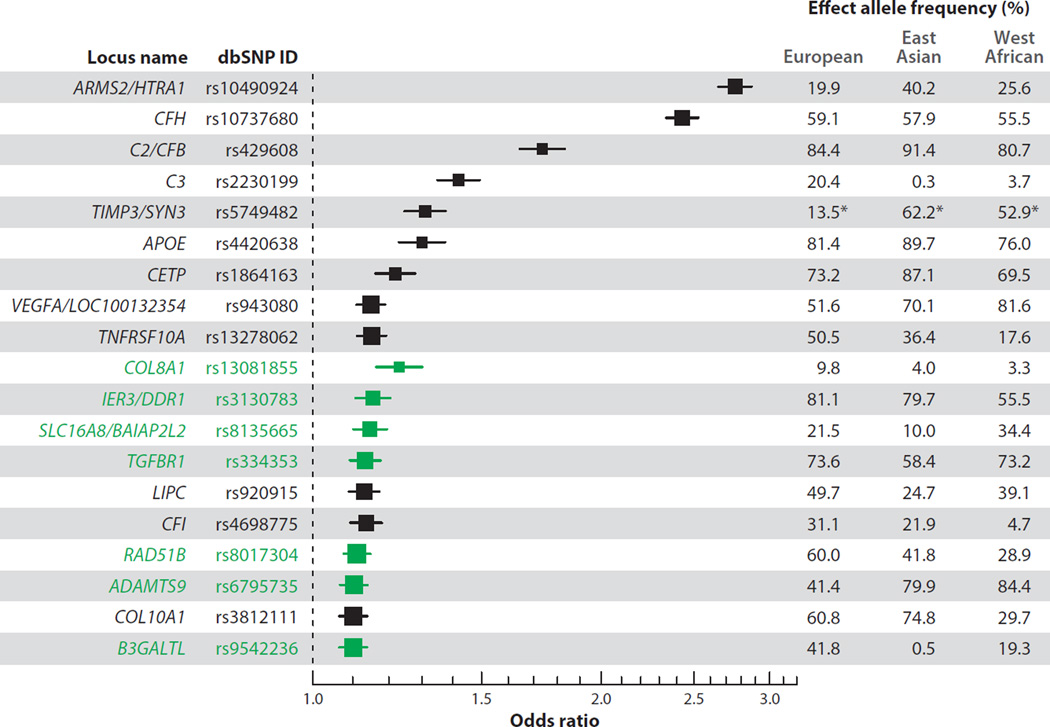
Figure 2.
Schematic of human chromosomes showing reported association signals of common (minor allele frequencies ≥1%) and rare variants (minor allele frequencies <1%) for 20 AMD susceptibility loci/genes. Locus names indicate genes that overlap or are close to the observed association signal but do not necessarily represent the underlying disease-causing gene (33, 40,75, 90, 113, 121). The visualization was created using quantsmooth R package version 1.24.0.
Figure 3.
Effect sizes and 95% confidence intervals for the 19 common AMD risk variants in the discovery study of the AMD Gene Consortium meta-analysis (33). Box sizes indicate the precision of effect estimates. The 7 loci that were reported with genome-wide significance (P < 5 × 10−8) for the first time are highlighted in green. Population-based effect allele frequencies are given for European (N = 379), East Asian (N = 286), and West African (N = 244) ancestry groups of the 1000 Genomes Project (phase I v3) (1). rs5749482 for TIMP3 was not available in the 1000 Genomes Project, and effect allele frequencies in this row (indicated by asterisks) are instead given for the highly correlated rs5754227 [r2 = 0.92 in CEU (Utah residents with ancestry from northern and western Europe) samples from the International HapMap Project].
The involvement of five complement genes in the AMD pathway, including CFH and CFI, has highlighted the role of the innate immune system in the development of AMD. Although pathways associated with the 15 other loci remain uncharacterized, each discovered novel AMD risk locus provides a much-needed starting point for functional downstream analyses that may ultimately uncover key players in AMD pathogenesis.
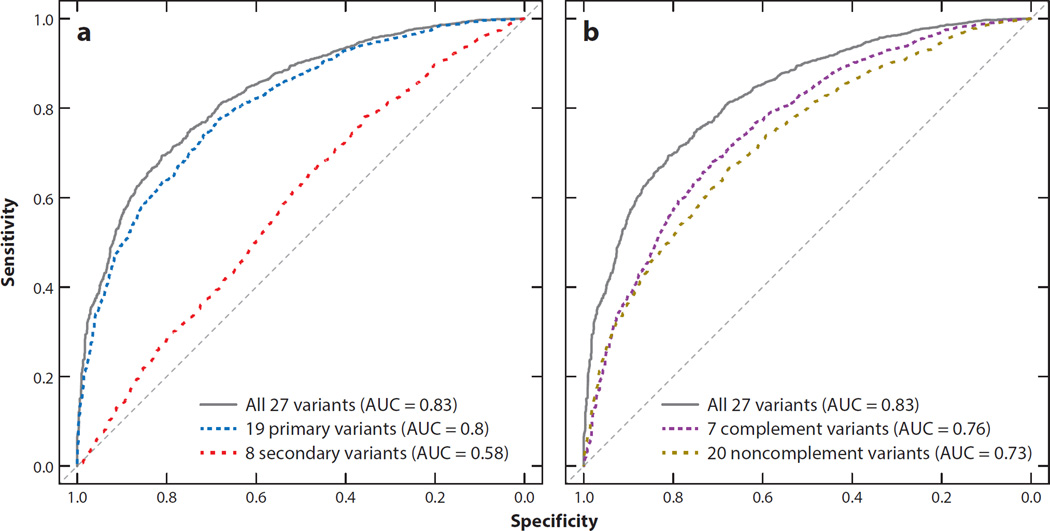
The common risk variants have been utilized recently to predict AMD risk. A risk score analysis of the 19 common risk variants (minor allele frequency ≥1%) can distinguish cases and controls rather well [area under the curve (AUC) = 0.734], at least when compared with the performance of genetic risk scores for other complex diseases (33, 46). Adding the observed secondary signals to the risk model can further improve the ability to differentiate late-AMD cases from controls, as shown in Figure 4, which illustrates data from the Michigan, Mayo, AREDS, Pennsylvania (MMAP) AMD case–control study of 1,628 late-AMD cases and 1,150 controls of European ancestry (AUC = 0.80 and AUC = 0.83, respectively) (7). Recent data from the Three Continent AMD Consortium revealed that nongenetic risk factors (age, sex, AMD baseline grade, smoking, and body mass index) have similar predictive power (AUC = 0.78) (5). A combination of genetic and nongenetic factors reached an AUC of 0.88 in three well-characterized population-based AMD studies.
Figure 4.
Risk scores for individuals in the large Michigan, Mayo, AREDS, Pennsylvania (MMAP) AMD case–control study (7), defined as the product of the number of common risk alleles at each locus and the associated effect size for each allele (measured on the log-odds scale, with each variant conditioned on the other variants). The plot summarizes the ability of these overall genetic risk scores to distinguish late-AMD cases (geographic atrophy and choroidal neovascularization) and controls. Secondary variants—i.e., variants reported to be independently associated within the reported 19 AMD loci—improved discriminatory power (panel a), whereas variants within/near complement genes as well as noncomplement genes contributed to the overall risk score (panel b). Abbreviation: AUC, area under the curve.
To a casual observer, predictive testing for AMD might appear to be within reach, especially for the most extreme end of the risk scale. The distribution of the genetic risk factors in the MMAP AMD case–control study (7) demonstrated that approximately 95% of individuals in the two top risk deciles are AMD patients, whereas almost 90% of the lowest-risk decile represents control individuals (Figure 5a). However, rescaling the AMD prevalence of 65% in the MMAP case–control study (which is highly enriched for AMD patients) to a more realistic disease value of 2.8% [corresponding to the estimated prevalence of late AMD in individuals at age 75 years (82)] suggests that in a population of 75-year-olds,<15% of the individuals within the highest-risk decile will present late AMD (Figure 5b).
Figure 5.
Multilocus genotypes and disease risk. A summary of the proportion of affected individuals in each risk decile is shown, with the highest-risk decile on the left. (a) Michigan, Mayo, AREDS, Pennsylvania (MMAP) AMD case–control samples segregated according to the risk of disease predicted by a simple logistic regression model. (b,c) Equivalent predictions at the population level, after assigning different weights to cases and controls and taking into account that the sample is enriched for cases and that the disease prevalence might actually be 2.8% (panel b) or 10.9% (panel c) (method described in Reference 7). (d) Top 10 risk percentiles of a population with 10.9% disease prevalence.
The proportion of affected individuals among those with a high genetic risk score increases with age. For example, among 85-year-olds, the prevalence of late AMD in populations of European ancestry was estimated at 10.9% (82). We estimate that 46% of 85-year-olds in the top 10% of genetic risk would be affected by late AMD (Figure 5c) and that >80% of 85-year-olds in the top 1% of genetic risk would develop AMD (Figure 5d).
A major limitation of genetic risk scores for AMD is that no prophylactic treatments are available for individuals identified with high disease risk. As shown by the AMD Gene Consortium, predictive power did not increase much when 7 newly identified AMD risk loci were combined with the 12 previously reported loci (33). Although larger studies may uncover additional common variant risk loci, we expect that their effect sizes will be smaller than those of currently known common AMD risk variants (see Figure 3) and result in only minor improvements in AUC. Rare variants with strong risk effects might help predict disease progression in their carriers, but each variant is expected to influence only a small number of individuals. For example, the rare, highly penetrant CFH:p.Arg1210Cys variant (75) was found in only 2 of 4,300 European individuals studied by the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project, whereas three other low-frequency risk variants (CFI:p.Gly119Arg, C3:p.Lys155Gln, and C9:p.Pro167Ser) were observed in 11–66 individuals (ESP6500SI-V2; http://evs.gs.washington.edu).
One approach for evaluating progress in AMD genetics is to compare the variance between disease risk explained by the genetic variants and disease risk estimated through twin and family studies. However, these comparisons are quite sensitive to disease prevalence (33). Depending on the prevalence of AMD, known loci might explain 28–43% of the variance in disease risk, with the lower estimates corresponding to a prevalence of 2.8% and the higher value to a prevalence of 10.9% (Figure 6). Assuming that the genetic factors for late AMD account for 71% of the variation in disease risk among individuals (88), the current set of known risk variants explains 40–60% of this total.
Figure 6.
Estimated disease variance explained by 27 common risk genomic variants reported in 19 AMD loci. Relative risks were estimated using the independent risk effects of each variant conditioned on all other variants, assuming disease prevalences of 2.8%, 5.6%, and 10.9%, which correspond to the predicted prevalences of late AMD in European populations at ages 75, 80, and 85 years, respectively (82). Reference 93 describes the methods applied to measure the variance of the disease. Abbreviation: SNP, single-nucleotide polymorphism. favors combinatorial and synergistic mechanisms involving gene or pathway interactions leading to AMD pathogenesis.
Notably, common risk factors that are near the complement genes CFH, C2/CFB, C3, and CFI account for approximately 57% of the contribution of known variants to disease risk, emphasizing the major role of the complement system in AMD pathogenesis (Figure 6). Thus, although complement-pathway defects play a critical role in AMD pathogenesis, other mechanisms are clearly involved. Additional pathways are also implicated by the failure to replicate a full range of AMD-like retinal pathology in a mouse with laser-induced CNV with complement deficiency (80).
Various groups have reported a preferential association of CFH and ARMS2/HTRA1 risk variants with different forms of advanced AMD. Specifically, CFH risk variants appear to slightly favor progression toward GA, and the ARMS2 risk variant favors progression toward CNV (7, 33, 94). Although this might indicate a divergent impact of the two genes, variants at the two loci significantly increase risk in both forms of late AMD, suggesting their involvement in biological processes active before the onset of advanced disease. Future studies with more accurate phenotyping of patient cohorts using high-resolution imaging techniques might allow the identification of specific risk variants associated with subphenotypes that influence progression toward one of the late-AMD forms, but using genetic risk to make specific predictions about the type of advanced AMD is not currently feasible.
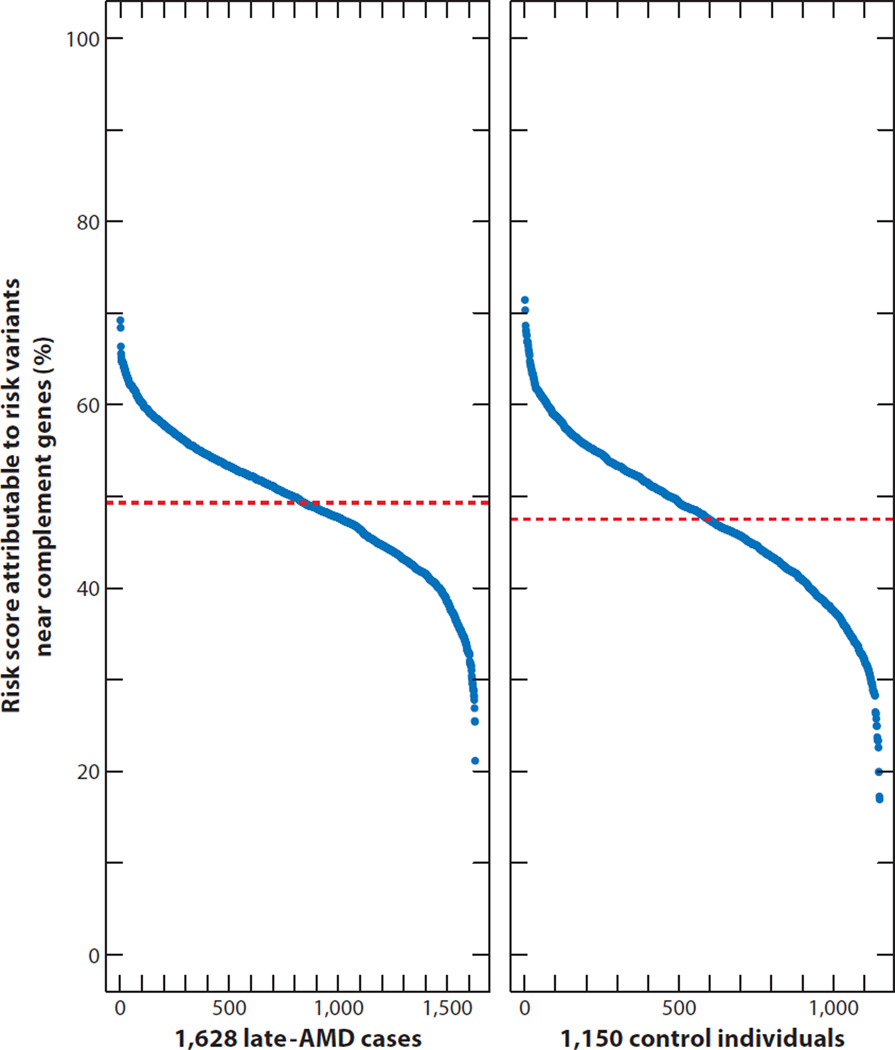
We have also assessed the impact of complement risk gene variants on AMD risk scores. We did identify individuals with differences in the contribution of risk variant groups (ranging from 20% to 70%), but even in the large MMAP AMD case–control study, no affected individual had a risk profile that was based solely on complement or noncomplement risk variants (Figure 7). It is not surprising that no single pathway can explain advanced disease. The current evidence strongly favors combinatorial and synergistic mechanisms involving gene or pathway interactions leading to AMD pathogenesis.
Figure 7.
Attributable contributions of variants within or near complement genes to overall risk scores (19 primary and 8 secondary signals) (33) (see also Figure 3) in 1,628 late-AMD cases and 1,150 control individuals from the large Michigan, Mayo, AREDS, Pennsylvania (MMAP) AMD case–control study (7). The dashed red lines indicate the average contributions in cases (49.3%) and controls (47.5%). Individuals were sorted by increasing percentage of their risk score attributable to risk variants near complement genes. Ticks on the x axis represent 100 individuals.
POPULATION DIFFERENCES
AMD prevalence differs among racial and ethnic groups (72). In the Multi-Ethnic Study of Atherosclerosis (MESA), the frequency of early manifestations of AMD was 4.2% in Hispanics, 4.6% in Chinese Americans, 5.4% in whites, and 2.4% in blacks (54). Observations in the National Health and Nutritional Examination Survey were similar, with a prevalence of 5.1% in Mexican Americans, 7.3% in whites, and 2.4% in blacks (53). These differences could be due to either environmental or genetic factors. A recent reanalysis of the MESA data for common factors such as smoking, body mass index, inflammatory factors, diabetes, and alcohol was unable to explain the significant difference in risk between whites and blacks. In addition, genetic analysis of MESA data with the risk variant CFH:p.Tyr402His did not explain the higher frequency of early AMD in whites compared with blacks (55).
To obtain a better genetic understanding in different populations, we compared the effects of the allele frequencies of the known AMD risk variants between ancestry groups of the 1000 Genomes Project, including 379 European, 286 East Asian, and 244 West African samples (see Figure 3). This comparison confirms reported population differences. For example, the ARMS2/HTRA1 locus (rs10490924) plays a larger role in Asian populations (where it has a risk allele frequency of approximately 40%) than it does in European populations (where it has a risk allele frequency of approximately 20%) (Figure 3). The CFH variant rs10737680 was observed with similar effect allele frequencies in all three ancestry groups (Figure 3); however, the independently associated CFH:p.Tyr402His risk variant (rs1061170) was reported to have markedly lower frequencies in East Asian populations than in European populations (5% and 35%, respectively) (57, 107). Similarly, the nonsynonymous C3 variant rs2230199 is common in Europeans but rare in Asians and Africans (Figure 3). In connection with the observation that the ARMS2 risk variant seems to predispose toward progression to the neovascular form of advanced disease, although CFH variants might be stronger risk factors for GA, one might speculate that the different genetic risk profile in Asians would favor the neovascular disease, which indeed seems to be more common in Asian than in European populations (49). However, differences in the prevalence of GA and CNV between Europeans and Asians are much larger than can be explained by the frequency distribution of CFH and ARMS2 risk alleles.
THE GENETICS OF DISEASE PROGRESSION
Substantial variation exists in the progression from early to late AMD(109).Although current nongenetic and genetic risk factors are suggested to predict this progression (89), our understanding of AMD development is limited because most of the published genome-wide association studies have been conducted on samples from either exclusively or predominantly late-AMD cases. This reflects the use of clinic-based recruitment of cases and controls, where asymptomatic patients are less likely to be seen. The single published genome-wide association study of early AMD (including 4,089 early-AMD cases and more than 20,000 control individuals) reported no significant associations beyond CFH and ARMS2, perhaps hinting at an undiscovered genetic and phenotypic heterogeneity that will require larger sample sizes and more focused investigations (42). Modern imaging tools (such as optical coherence tomography) and next-generation DNA sequencing may be needed to unlock the hidden complexities of early AMD. A better understanding of the genetic and phenotypic architecture of early-stage disease will be helpful in identifying new mechanistic models and effective therapies for slowing progression from early to late stages.
Most of the functional variants at previously reported AMD loci will likely be uncovered by targeted genomic resequencing. These functional variants will include rare nonsense and loss-of-function variants with a high impact on disease risk, which are expected to accelerate the progression from genetic association to biological understanding and eventual new therapies. In particular, the study of individuals with rare but highly penetrant variants, such as naturally occurring “human hypomorphs or knockouts” (22), might reveal new avenues for functional assays and treatment design. The most interesting knockout mutations, if they exist, would be variants that prevent the disease irrespective of other prevailing risk factors. Mimicking such extreme genetic effects by pharmaceutical intervention (for example, through local RNA interference or antibody treatment) may provide an opportunity to postpone or prevent the development of late-stage AMD.
BIOLOGICAL PERSPECTIVES FROM AMD GENETICS: A MULTI-HIT THRESHOLD MODEL
Extensive clinical characterization of patients, in vitro studies of RPE and other cultured cells, research on animal models, and examinations of human donor eyes have provided a wealth of knowledge; however, the field is still struggling to generate a cohesive, integrated, and mechanistic framework that more accurately reflects the complex degenerative processes underlying AMD. Like other complex neurodegenerations, many molecular and cellular pathways are involved in the AMD phenotype, particularly because of the involvement of both neurons (photoreceptors) and their support system (the RPE and choroid). Over the past decade, the picture that has emerged includes pathological changes through complement activation and inflammatory processes surrounding the RPE and BrM, altered lipid transport and accumulation of oxidized lipids beneath the RPE, alteration in the maintenance of BrM, and (in the case of CNV) the activation of proangiogenic processes. Initial genetic findings on CFH strongly corroborate the inflammatory hypothesis of AMD (3, 36); however, complement activation and systemic immune response are not sufficient to produce the complex AMD phenotypes in model organisms.
In an amazing spirit of collaboration and cooperation, numerous groups have joined resources to identify the genetic architecture of AMD. So far, 20 chromosomal regions have been identified that harbor genetic variants associated with AMD. Based on the information in public databases, we can broadly cluster almost all of the genes at the 20 AMD loci into five biologically relevant pathways that are also suggested by biochemical and pathological data: (a) the complement pathway and immune response (CFH, C2/CFB, CFI, C3, and C9), (b) lipid transport (APOE, LIPC, CETP, and BAIAP2L2), (c) ECM remodeling (COL8A1, COL10A1, TIMP3, ADAMTS9, TGFBR1, HTRA1, and B3GALTL), (d) angiogenesis (VEGFA, TGFBR1, and ADAMTS9), and (e) cell survival, including DNA repair, apoptosis, and stress response (ARMS2, RAD51B, and TNFRSF10A).
Although functional validation of most of these genes is still required, we can begin to integrate the genetic information with components of the photoreceptor support system. In any comprehensive attempt to explain AMD disease, we must incorporate the facts that (a) the genetic risk variants are inherited, (b) a single variant or gene alone is unlikely (or even insufficient) to cause the complex disease phenotype, and (c) the disease phenotypes are manifested late in life. Although most patients have multiple risk alleles, many individuals with risk alleles never exhibit the disease phenotype in their lifetimes. The only common associated risk factor in all patients is advanced age, suggesting that disease onset results when physiological damage reaches a certain threshold. We expect that the rate of physiological damage may be modified by a set of risk alleles and environmental factors unique to each individual and that the trigger for disease initiation might be provided by life events, such as chronic inflammation or immune response to infection.
To reconcile existing genetic and biological data into a testable hypothesis, we propose that AMD phenotypes result from a breakdown in complex functional relationships between the photoreceptors and their support system (RPE-choroid complex), which are critical for a stringent control of the two-way flow of nutrients and waste material (Figure 8). All of the above-mentioned biological functions/pathways can be developed into a “threshold” model (117) and linked to discrete aspects of AMD pathogenesis, as discussed below.
Figure 8.
A multi-hit “threshold” model of AMD. Advanced age and environmental factors have an impact on all components (in green) associated with the photoreceptor support system. The assignment of genes to distinct groups is based on published literature and not on functional studies. Each gene group can be associated with a specific function/pathway that may influence one or more components— e.g., alterations in extracellular matrix (ECM)–associated genes can affect both the retinal pigment epithelium (RPE) and Bruch’s membrane. Some of the genetic variants (e.g., those in the complement pathway) are expected to have a stronger impact on specific clinical findings. The cellular changes (in blue) are shown next to (or near) components (in green) that are likely impacted. Some of the changes in one or more components can have a domino effect, leading to pathology phenotypes (red) in AMD. The appearance of late-stage disease [geographic atrophy (GA) or choroidal neovascularization (CNV)] would depend on the acquisition of threshold levels that can be reached by the cumulative effect of multiple hits, including genetic susceptibility, aging-associated changes, and environmental factors.
Advanced age and environmental factors impact all components of the photoreceptor support system concurrently. Oxidative stress is believed to be a major mediator of the effect of age because mitochondrial oxidation is impaired with aging and oxidative damage is widely observed (60, 95, 100). In addition, aging is associated with increased DNA damage and deficiency of repair mechanisms (41). Oxidative damage has also been implicated in AMD (115). Thus, the age of onset and the severity of AMD phenotype could result from combinations of aging, environment, and protective or risk variants. Genetic variants might alter the balance in key biological pathways, leading to AMD pathology. Alternatively, advanced age and environmental insults might lower the threshold for AMD, allowing risk-associated alleles to initiate a process of degeneration in a biological system whose function is compromised. For example, the formation of sub-RPE drusen can be compared with extracellular lipid deposition in a vessel wall of the systemic circulation in atherosclerotic cardiovascular disease (14, 15) and might be promoted by certain variants of lipid transporters. RPE-secreted lipoproteins are a major source of peroxidizable lipids under the RPE, and reactive oxygen species from basolaterally localized RPE mitochondria can lead to oxidized lipids (and proteins). The ECM is implicated in the retention of lipoproteins in BrM. Oxidized cholesterol can also induce inflammation (79).
As illustrated in Figure 8, the development of AMD lesions can be partially explained by the AMD susceptibility genes identified so far. LIPC and CETP are expressed in the subretinal space and may participate in rapid cholesterol transfer from the RPE to the neural retina (111, 112). ApoE and CFH are detected in subretinal and sub-RPE lesions and in BrM. TIMP3 is present in drusen and BrM, where it plays a crucial role in ECM maintenance and remodeling (26, 47). AMD-associated variants in ECM proteins can alter the structure and permeability of BrM, making it more susceptible to aging and environmental abuse.
The presence of inflammatory modulators in AMD lesions suggests that local processes driven by complement dysregulation play a crucial role in the development and progression of AMD (3, 44). The identification of multiple complement genes at AMD susceptibility loci validates a major role of the immune response in AMD. In addition, several lines of evidence implicate inflammation and the recruitment of microglial cells as key mediators of AMD pathology (2, 63,64, 91, 106). RPE and choroidal cells express many immune-modulatory proteins at significant levels (3, 102). Local expression and dysregulation of inflammatory modulators may therefore explain why AMD is not tightly correlated with systemic or diffuse inflammatory processes. If AMD risk alleles were exclusively associated with complement activation or immune response, then modeling the process of degeneration leading to photoreceptor cell death would be less challenging. However, the identification of risk alleles associated with lipid transport and metabolism, ECM deposition and remodeling, cell survival, and angiogenesis increases the challenge of creating a comprehensive model for AMD pathogenesis.
How do we integrate a diverse set of risk factors into an accurate model of pathogenesis that captures the biology of degeneration while leaving room for variability in disease progression and severity? By treating risk alleles and pathways to which they belong as discrete entities operating in isolation, we run the real risk of building models that are not physiologically relevant. Given the extraordinary interdependence of photoreceptors, RPE, BrM, and choroidal capillaries, it may be more fruitful and biologically relevant to envision a scenario where perturbation in one (or more) of the relevant pathways compromises the viability of neighboring and interdependent cellular partners within the support complex. As the viability of one of the components is compromised, so too are the metabolic processes they support. The process of cascading dysfunction, originating at a diverse set of loci but converging through a handful of interdependent physiological pathways, provides a flexible model to accommodate a diverse set of risk alleles with aging and environmental factors as well as an array of shared pathophysiological features.
We emphasize that this is just one way to incorporate genes from many pathways into a pathogenic sequence and that there may be other sequences that are equally plausible, both for the extracellular lesions and for other specific pathologies, such as RPE or photoreceptor death and neovascularization. Nevertheless, it is reassuring that the genetics is now converging with the biology and pathology of AMD.
THE EMERGING PICTURE, GENETIC LOAD, AND OPEN QUESTIONS
We are left with two unresolved dilemmas in AMD genetics. First, the most significantly associated common variants do not have a known functional impact either on the expression or processing of the nearby gene transcripts or on the structural and functional integrity of the encoded proteins. For example, there is strong evidence that the alternative complement pathway plays a role in causing AMD, but this evidence cannot be based on a demonstration of the role of a variant in a single gene. Rather, the influence of complement proteins is generally dependent on the overall impact of risk and protective genetic variants in all components of the complement pathway. Second, we know very little about the tissue-specific triggers for AMD pathogenesis, and discovering these may also require identifying the pathways leading to specific early pathologies, such as RPE atrophy and drusen. Like other complex diseases, AMD is not caused by a single factor, and multiple pathogenic mechanisms that need not be mutually exclusive are almost certainly involved. Therapeutic strategies based on a single pathway are expected to provide only partial treatment. The cure for AMD may come by intervening at an early stage or perhaps even before the disease process is initiated.
We must realize that complex human traits and common clinical phenotypes arise from an individual’s unique set of variants under the influence of nongenetic factors. The currently known risk variants and AMD genes represent a rather atypical situation in the spectrum of complex diseases, in that they include a few strong susceptibility variants (in CFH and ARMS2/HTRA1) and many others with smaller effects (in ECM genes). As a consequence, their combined predictive power is unparalleled in the field of complex diseases. Yet it is evident that nongenetic risk factors, such as age and environment (specifically smoking), still have a major influence on the outcome of the disease. Presymptomatic genetic testing is especially challenged by the lack of preventive treatments and cure. It therefore remains unwise to make population-wide predictions based on limited genetic data, as suggested by some reports (4).
Future genetic studies with larger sample sizes of earlier AMD stages (defined by high-resolution imaging methods) and comprehensive analyses of the genome are likely to add to the overall picture of underlying AMD risk factors. Extensive targeted genomic sequencing, whole-exome sequencing of AMD families where available, and whole-genome sequencing of AMD cases would be valuable for explaining missing heritability and—together with deeper RNA, microRNA, and epigenome profiling—would help in clarifying the biological mechanisms that underlie disease. In addition, the time has come to focus on combining genetic insights with biologically relevant model systems to assess pathogenic mechanisms and initiate knowledge-based interventions for this debilitating disease.
ACKNOWLEDGMENTS
We are grateful to Drs. Emily Chew, Tiziana Cogliati, and Rinki Ratnapriya for assistance and discussions. Our research is supported by intramural research program (including National Eye Institute Computational Medicine initiative) and extramural funds from the National Eye Institute (EY023164, EY022005, and EY06109), the Beckman Initiative for Macular Research, and Research to Prevent Blindness.
DISCLOSURE STATEMENT
L.G.F. has received royalties from patents held by the University of Regensburg, and G.R.A. and A.S. have received royalties from patents held by the University of Michigan. The authors are not aware of any other affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Lars G. Fritsche, Email: larsf@umich.edu.
Robert N. Fariss, Email: farissr@nei.nih.gov.
Dwight Stambolian, Email: stamboli@mail.med.upenn.edu.
Gonçalo R. Abecasis, Email: goncalo@umich.edu.
Christine A. Curcio, Email: curcio@uab.edu.
Anand Swaroop, Email: swaroopa@nei.nih.gov.
LITERATURE CITED
- 1.1000 Genomes Proj. Consort. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog. Retin. Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awh CC, Lane AM, Hawken S, Zanke B, Kim IK. CFH and ARMS2 genetic polymorphisms predict response to antioxidants and zinc in patients with age-related macular degeneration. Ophthalmology. 2013;120:2317–2323. doi: 10.1016/j.ophtha.2013.07.039. [DOI] [PubMed] [Google Scholar]
- 5.Buitendijk GH, Rochtchina E, Myers C, van Duijn CM, Lee KE, et al. Prediction of age-related macular degeneration in the general population: the Three Continent AMD Consortium. Ophthalmology. 2013;120:2644–2655. doi: 10.1016/j.ophtha.2013.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canter JA, Olson LM, Spencer K, Schnetz-Boutaud N, Anderson B, et al. Mitochondrial DNA polymorphism A4917G is independently associated with age-related macular degeneration. PLoS ONE. 2008;3:e2091. doi: 10.1371/journal.pone.0002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chew EY, Clemons TE, Agron E, Sperduto RD, Sangiovanni JP, et al. Long-term effects of vitamins C and E, beta-carotene, and zinc on age-related macular degeneration: AREDS report no. 35. Ophthalmology. 2013;120:1604–1611. doi: 10.1016/j.ophtha.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong NH, Keonin J, Luthert PJ, Frennesson CI, Weingeist DM, et al. Decreased thickness and integrity of the macular elastic layer of Bruch’s membrane correspond to the distribution of lesions associated with age-related macular degeneration. Am. J. Pathol. 2005;166:241–251. doi: 10.1016/S0002-9440(10)62248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins FS. Faces of the genome. Science. 2011;331:546. doi: 10.1126/science.1202894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conley YP, Jakobsdottir J, Mah T, Weeks DE, Klein R, et al. CFH, ELOVL4, PLEKHA1 and LOC387715 genes and susceptibility to age-related maculopathy: AREDS and CHS cohorts and meta-analyses. Hum. Mol. Genet. 2006;15:3206–3218. doi: 10.1093/hmg/ddl396. [DOI] [PubMed] [Google Scholar]
- 12.Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, et al. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curcio CA, Johnson M. Structure, function, and pathology of Bruch’s membrane. In: Ryan SJ, Schachat AP, Wilkinson CP, Hinton DR, Sadda S, Wiedemann P, editors. Retina, Vol. 1, Part 2: Basic Science and Translation to Therapy. 5th ed. London: Elsevier; 2013. pp. 466–481. [Google Scholar]
- 14.Curcio CA, Johnson M, Huang JD, Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog. Retin. Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curcio CA, Johnson M, Rudolf M, Huang JD. The oil spill in ageing Bruch membrane. Br. J. Ophthalmol. 2011;95:1638–1645. doi: 10.1136/bjophthalmol-2011-300344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curcio CA, Messinger JD, Sloan KR, McGwin G, Jr, Medeiros NE, Spaide RF. Subretinal drusenoid deposits in non-neovascular age-related macular degeneration: morphology, prevalence, topography, and biogenesis model. Retina. 2013;33:265–276. doi: 10.1097/IAE.0b013e31827e25e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curcio CA, Millican CL, Bailey T, Kruth HS. Accumulation of cholesterol with age in human Bruch’s membrane. Investig. Ophthalmol. Vis. Sci. 2001;42:265–274. [PubMed] [Google Scholar]
- 18.Curcio CA, Owsley C, Jackson GR. Spare the rods, save the cones in aging and age-related maculopathy. Investig. Ophthalmol. Vis. Sci. 2000;41:2015–2018. [PubMed] [Google Scholar]
- 19.Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J. Comp. Neurol. 1990;292:497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- 20.Day S, Acquah K, Lee PP, Mruthyunjaya P, Sloan FA. Medicare costs for neovascular age-related macular degeneration, 1994–2007. Am. J. Ophthalmol. 2011;152:1014–1020. doi: 10.1016/j.ajo.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delori FC, Goger DG, Dorey CK. Age-related accumulation and spatial distribution of lipofuscin in RPE of normal subjects. Investig. Ophthalmol. Vis. Sci. 2001;42:1855–1866. [PubMed] [Google Scholar]
- 22.Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum. Mol. Genet. 2012;21:R1–R9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards AO, Lee SJ, Fridley BL, Tosakulwong N. Density of common complex ocular traits in the aging eye: analysis of secondary traits in genome-wide association studies. PLoS ONE. 2008;3:e2510. doi: 10.1371/journal.pone.0002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 25.Eldred GE, Miller GV, Stark WS, Feeney-Burns L. Lipofuscin: resolution of discrepant fluorescence data. Science. 1982;216:757–759. doi: 10.1126/science.7079738. [DOI] [PubMed] [Google Scholar]
- 26.Fariss RN, Apte SS, Olsen BR, Iwata K, Milam AH. Tissue inhibitor of metalloproteinases-3 is a component of Bruch’s membrane of the eye. Am. J. Pathol. 1997;150:323–328. [PMC free article] [PubMed] [Google Scholar]
- 27.Feeney-Burns L, Ellersieck MR. Age-related changes in the ultrastructure of Bruch’s membrane. Am. J. Ophthalmol. 1985;100:686–697. doi: 10.1016/0002-9394(85)90625-7. [DOI] [PubMed] [Google Scholar]
- 28.Feero WG, Guttmacher AE, Collins FS. Genomic medicine—an updated primer. N. Engl. J. Med. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- 29.Ferris FL, III, Davis MD, Clemons TE, Lee LY, Chew EY, et al. A simplified severity scale for age-related macular degeneration: AREDS report no. 18. Arch. Ophthalmol. 2005;123:1570–1574. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferris FL, III, Wilkinson CP, Bird A, Chakravarthy U, Chew E, et al. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;120:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher SA, Abecasis GR, Yashar BM, Zareparsi S, Swaroop A, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum. Mol. Genet. 2005;14:2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 32.Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, et al. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 33.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, et al. Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013;45:433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog. Retin. Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 37.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 38.Hammond CJ, Webster AR, Snieder H, Bird AC, Gilbert CE, Spector TD. Genetic influence on early age-related maculopathy: a twin study. Ophthalmology. 2002;109:730–736. doi: 10.1016/s0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 39.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch. Neurol. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 40.Helgason H, Sulem P, Duvvari MR, Luo H, Thorleifsson G, et al. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat. Genet. 2013;45:1371–1374. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 41.Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 42.Holliday EG, Smith AV, Cornes BK, Buitendijk GH, Jensen RA, et al. Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS ONE. 2013;8:e53830. doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, et al., editors. Surveill. Epidemiol. End Results Program. Bethesda, MD: Natl. Cancer Inst.; 2012. SEER cancer statistics review, 1975–2009 (vintage 2009 populations) updated Aug. 20, http://seer.cancer.gov/csr/1975_2009_pops09. [Google Scholar]
- 44.Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. A potential role for immune complex pathogenesis in drusen formation. Exp. EyeRes. 2000;70:441–449. doi: 10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- 45.Jones MM, Manwaring N, Wang JJ, Rochtchina E, Mitchell P, Sue CM. Mitochondrial DNA haplogroups and age-related maculopathy. Arch. Ophthalmol. 2007;125:1235–1240. doi: 10.1001/archopht.125.9.1235. [DOI] [PubMed] [Google Scholar]
- 46.Kalf RR, Mihaescu R, Kundu S, de Knijff P, Green RC, Janssens AC. Variations in predicted risks in personal genome testing for common complex diseases. Genet. Med. 2013;16:85–91. doi: 10.1038/gim.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamei M, Hollyfield JG. TIMP-3 in Bruch’s membrane: changes during aging and in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 1999;40:2367–2375. [PubMed] [Google Scholar]
- 48.Kanda A, Chen W, Othman M, Branham KE, Brooks M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawasaki R, Yasuda M, Song SJ, Chen SJ, Jonas JB, et al. The prevalence of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology. 2010;117:921–927. doi: 10.1016/j.ophtha.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Klein BE, Klein R, Lee KE, Moore EL, Danforth L. Risk of incident age-related eye diseases in people with an affected sibling: the Beaver Dam Eye Study. Am. J. Epidemiol. 2001;154:207–211. doi: 10.1093/aje/154.3.207. [DOI] [PubMed] [Google Scholar]
- 51.Klein ML, Mauldin WM, Stoumbos VD. Heredity and age-related macular degeneration: observations in monozygotic twins. Arch. Ophthalmol. 1994;112:932–937. doi: 10.1001/archopht.1994.01090190080025. [DOI] [PubMed] [Google Scholar]
- 52.Klein R, Cruickshanks KJ, Nash SD, Krantz EM, Nieto FJ, et al. The prevalence of age-related macular degeneration and associated risk factors. Arch. Ophthalmol. 2010;128:750–758. doi: 10.1001/archophthalmol.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein R, Klein BE, Jensen SC, Mares-Perlman JA, Cruickshanks KJ, Palta M. Age-related maculopathy in a multiracial United States population: the National Health and Nutrition Examination Survey III. Ophthalmology. 1999;106:1056–1065. doi: 10.1016/S0161-6420(99)90255-5. [DOI] [PubMed] [Google Scholar]
- 54.Klein R, Klein BE, Knudtson MD, Wong TY, Cotch MF, et al. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the Multi-Ethnic Study of Atherosclerosis. Ophthalmology. 2006;113:373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 55.Klein R, Li X, Kuo JZ, Klein BE, Cotch MF, et al. Associations of candidate genes to age-related macular degeneration among racial/ethnic groups in the Multi-Ethnic Study of Atherosclerosis. Am. J. Ophthalmol. 2013;156:1010–1020. doi: 10.1016/j.ajo.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondo N, Bessho H, Honda S, Negi A. Complement factor H Y402H variant and risk of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology. 2011;118:339–344. doi: 10.1016/j.ophtha.2010.06.040. [DOI] [PubMed] [Google Scholar]
- 58.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 59.LaVail MM. Outer segment disc shedding and phagocytosis in the outer retina. Trans. Ophthalmol. Soc. UK. 1983;103:397–404. [PubMed] [Google Scholar]
- 60.Lesnefsky EJ, Hoppel CL. Oxidative phosphorylation and aging. Ageing Res. Rev. 2006;5:402–433. doi: 10.1016/j.arr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 61.Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat. Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo DG, Xue T, Yau KW. How vision begins: an odyssey. Proc. Natl. Acad. Sci. USA. 2008;105:9855–9862. doi: 10.1073/pnas.0708405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma W, Zhao L, Wong WT. Microglia in the outer retina and their relevance to pathogenesis of age-related macular degeneration. Adv. Exp. Med. Biol. 2012;723:37–42. doi: 10.1007/978-1-4614-0631-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marneros AG. NLRP3 inflammasome blockade inhibits VEGF-A-induced age-related macular degeneration. Cell Rep. 2013;4:945–958. doi: 10.1016/j.celrep.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKibbin M, Ali M, Bansal S, Baxter PD, West K, et al. CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br. J. Ophthalmol. 2012;96:208–212. doi: 10.1136/bjo.2010.193680. [DOI] [PubMed] [Google Scholar]
- 66.McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, Lutty GA. Relationship between RPE and choriocapillaris in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2009;50:4982–4991. doi: 10.1167/iovs.09-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meleth AD, Wong WT, Chew EY. Treatment for atrophic macular degeneration. Curr. Opin. Ophthalmol. 2011;22:190–193. doi: 10.1097/ICU.0b013e32834594b0. [DOI] [PubMed] [Google Scholar]
- 68.Meyers SM, Greene T, Gutman FA. A twin study of age-related macular degeneration. Am. J. Ophthalmol. 1995;120:757–766. doi: 10.1016/s0002-9394(14)72729-1. [DOI] [PubMed] [Google Scholar]
- 69.Miller JW. Age-related macular degeneration revisited—piecing the puzzle: the LXIX Edward Jackson memorial lecture. Am. J. Ophthalmol. 2013;155:1.e13–35.e13. doi: 10.1016/j.ajo.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 70.Moreira EF, Larrayoz IM, Lee JW, Rodriguez IR. 7-Ketocholesterol is present in lipid deposits in the primate retina: potential implication in the induction of VEGF and CNV formation. Investig. Ophthalmol. Vis. Sci. 2009;50:523–532. doi: 10.1167/iovs.08-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 72.Natl. Eye Inst. Age-related macular degeneration (AMD) Bethesda, MD: Natl. Eye Inst.; 2014. http://www.nei.nih.gov/eyedata/amd.asp. [Google Scholar]
- 73.Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc. Natl. Acad. Sci. USA. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Priya RR, Chew EY, Swaroop A. Genetic studies of age-related macular degeneration: lessons, challenges, and opportunities for disease management. Ophthalmology. 2012;119:2526–2536. doi: 10.1016/j.ophtha.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat. Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, et al. Global data on visual impairment in the year 2002. Bull. World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 77.Ridge KD, Abdulaev NG, Sousa M, Palczewski K. Phototransduction: crystal clear. Trends Biochem. Sci. 2003;28:479–487. doi: 10.1016/S0968-0004(03)00172-5. [DOI] [PubMed] [Google Scholar]
- 78.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez IR, Larrayoz IM. Cholesterol oxidation in the retina: implications of 7KCh formation in chronic inflammation and age-related macular degeneration. J. Lipid Res. 2010;51:2847–2862. doi: 10.1194/jlr.R004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rohrer B, Coughlin B, Kunchithapautham K, Long Q, Tomlinson S, et al. The alternative pathway is required, but not alone sufficient, for retinal pathology in mouse laser-induced choroidal neovascularization. Mol. Immunol. 2011;48:e1–e8. doi: 10.1016/j.molimm.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosenfeld PJ, Shapiro H, Tuomi L, Webster M, Elledge J, et al. Characteristics of patients losing vision after 2 years of monthly dosing in the phase III ranibizumab clinical trials. Ophthalmology. 2011;118:523–530. doi: 10.1016/j.ophtha.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 82.Rudnicka AR, Jarrar Z, Wormald R, Cook DG, Fletcher A, Owen CG. Age and gender variations in age-related macular degeneration prevalence in populations of European ancestry: a meta-analysis. Ophthalmology. 2012;119:571–580. doi: 10.1016/j.ophtha.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 83.Rudolf M, Clark ME, Chimento MF, Li CM, Medeiros NE, Curcio CA. Prevalence and morphology of druse types in the macula and periphery of eyes with age-related maculopathy. Investig. Ophthalmol. Vis. Sci. 2008;49:1200–1209. doi: 10.1167/iovs.07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.SanGiovanni JP, Arking DE, Iyengar SK, Elashoff M, Clemons TE, et al. Mitochondrial DNA variants of respiratory complex I that uniquely characterize haplogroup T2 are associated with increased risk of age-related macular degeneration. PLoS ONE. 2009;4:e5508. doi: 10.1371/journal.pone.0005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarks SH, Arnold JJ, Killingsworth MC, Sarks JP. Early drusen formation in the normal and aging eye and their relation to age related maculopathy: a clinicopathological study. Br. J. Ophthalmol. 1999;83:358–368. doi: 10.1136/bjo.83.3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schleicher M, Weikel K, Garber C, Taylor A. Diminishing risk for age-related macular degeneration with nutrition: a current view. Nutrients. 2013;5:2405–2456. doi: 10.3390/nu5072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am. J. Ophthalmol. 1997;123:199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 88.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch. Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 89.Seddon JM, Reynolds R, Yu Y, Rosner B. Validation of a prediction algorithm for progression to advanced macular degeneration subtypes. JAMA Ophthalmol. 2013;131:448–455. doi: 10.1001/jamaophthalmol.2013.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seddon JM, Yu Y, Miller EC, Reynolds R, Tan PL, et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat. Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sene A, Khan AA, Cox D, Nakamura RE, Santeford A, et al. Impaired cholesterol efflux in senescent macrophages promotes age-related macular degeneration. Cell Metab. 2013;17:549–561. doi: 10.1016/j.cmet.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith W, Assink J, Klein R, Mitchell P, Klaver CC, et al. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 93.So HC, Li M, Sham PC. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet. Epidemiol. 2011;35:447–456. doi: 10.1002/gepi.20593. [DOI] [PubMed] [Google Scholar]
- 94.Sobrin L, Reynolds R, Yu Y, Fagerness J, Leveziel N, et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am. J. Ophthalmol. 2011;151:345.e3–352.e3. doi: 10.1016/j.ajo.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sohal RS, Weindruch R. Oxidative stress, caloric restriction, and aging. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp. Eye Res. 2005;80:595–606. doi: 10.1016/j.exer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 97.Sparrow JR, Fishkin N, Zhou J, Cai B, Jang YP, et al. A2E, a byproduct of the visual cycle. Vis. Res. 2003;43:2983–2990. doi: 10.1016/s0042-6989(03)00475-9. [DOI] [PubMed] [Google Scholar]
- 98.Spraul CW, Grossniklaus HE. Characteristics of drusen and Bruch’s membrane in postmortem eyes with age-related macular degeneration. Arch. Ophthalmol. 1997;115:267–273. doi: 10.1001/archopht.1997.01100150269022. [DOI] [PubMed] [Google Scholar]
- 99.Spraul CW, Lang GE, Grossniklaus HE. Morphometric analysis of the choroid, Bruch’s membrane, and retinal pigment epithelium in eyes with age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 1996;37:2724–2735. [PubMed] [Google Scholar]
- 100.Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 101.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 102.Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Swaroop A, Branham KE, Chen W, Abecasis G. Genetic susceptibility to age-related macular degeneration: a paradigm for dissecting complex disease traits. Hum. Mol. Genet. 2007;16:R174–R182. doi: 10.1093/hmg/ddm212. [DOI] [PubMed] [Google Scholar]
- 104.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu. Rev. Genomics Hum. Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swaroop A, Sieving PA. The golden era of ocular disease gene discovery: race to the finish. Clin. Genet. 2013;84:99–101. doi: 10.1111/cge.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thakkinstian A, Han P, McEvoy M, Smith W, Hoh J, et al. Systematic review and meta-analysis of the association between complement factor H Y402H polymorphisms and age-related macular degeneration. Hum. Mol. Genet. 2006;15:2784–2790. doi: 10.1093/hmg/ddl220. [DOI] [PubMed] [Google Scholar]
- 108.Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, Kelly SP. Smoking and age-related macular degeneration: a review of association. Eye. 2005;19:935–944. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- 109.Tikellis G, Robman LD, Dimitrov P, Nicolas C, McCarty CA, Guymer RH. Characteristics of progression of early age-related macular degeneration: the cardiovascular health and age-related maculopathy study. Eye. 2007;21:169–176. doi: 10.1038/sj.eye.6702151. [DOI] [PubMed] [Google Scholar]
- 110.Tilleul J, Richard F, Puche N, Zerbib J, Leveziel N, et al. Genetic association study of mitochondrial polymorphisms in neovascular age-related macular degeneration. Mol. Vis. 2013;19:1132–1140. [PMC free article] [PubMed] [Google Scholar]
- 111.Tserentsoodol N, Gordiyenko NV, Pascual I, Lee JW, Fliesler SJ, Rodriguez IR. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol. Vis. 2006;12:1319–1333. [PubMed] [Google Scholar]
- 112.Tserentsoodol N, Sztein J, Campos M, Gordiyenko NV, Fariss RN, et al. Uptake of cholesterol by the retina occurs primarily via a low density lipoprotein receptor-mediated process. Mol. Vis. 2006;12:1306–1318. [PubMed] [Google Scholar]
- 113.van de Ven JP, Nilsson SC, Tan PL, Buitendijk GH, Ristau T, et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat. Genet. 2013;45:813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 114.Wangsa-Wirawan ND, Linsenmeier RA. Retinal oxygen: fundamental and clinical aspects. Arch. Ophthalmol. 2003;121:547–557. doi: 10.1001/archopht.121.4.547. [DOI] [PubMed] [Google Scholar]
- 115.Winkler BS, Boulton ME, Gottsch JD, Sternberg P. Oxidative damage and age-related macular degeneration. Mol. Vis. 1999;5:32. [PMC free article] [PubMed] [Google Scholar]
- 116.Wong-Riley MT. Energy metabolism of the visual system. Eye Brain. 2010;2:99–116. doi: 10.2147/EB.S9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wright S. An analysis of variability in number of digits in an inbred strain of guinea pigs. Genetics. 1934;19:506–536. doi: 10.1093/genetics/19.6.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, et al. Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 119.Young RW. The organization of vertebrate photoreceptor cells. UCLA Forum Med. Sci. 1969;8:177–210. [PubMed] [Google Scholar]
- 120.Zareparsi S, Branham KE, Li M, Shah S, Klein RJ, et al. Strong association of the Y402H variant in complement factor H at 1q32 with susceptibility to age-related macular degeneration. Am. J. Hum. Genet. 2005;77:149–153. doi: 10.1086/431426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhan X, Larson DE, Wang C, Koboldt DC, Sergeev YV, et al. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat. Genet. 2013;45:1375–1379. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zweifel SA, Imamura Y, Spaide TC, Fujiwara T, Spaide RF. Prevalence and significance of sub-retinal drusenoid deposits (reticular pseudodrusen) in age-related macular degeneration. Ophthalmology. 2010;117:1775–1781. doi: 10.1016/j.ophtha.2010.01.027. [DOI] [PubMed] [Google Scholar]