Abstract
The identification of mutationally activated BRAF in many cancers altered our conception of the role played by the RAF family of protein kinases in oncogenesis. In this review we describe the development of BRAF inhibitors and the results that have emerged from their analysis in both the laboratory and the clinic. We discuss the spectrum of RAF mutations in human cancer and the complex interplay between tissue of origin and response to RAF inhibition. Finally, we enumerate mechanisms of resistance to BRAF inhibition that have been characterized and postulate how strategies of RAF pathway inhibition may be extended in scope to benefit, not only the thousands of patients diagnosed annually with BRAF-mutated metastatic melanoma, but also the larger patient population with malignancies harboring mutationally activated RAF genes that is ineffectively treated with the current generation of BRAF kinase inhibitors.
RAF kinases have been associated with cancer since their discovery in 1983 when Ulf Rapp and colleagues first described v-raf, a murine retroviral oncogene possessing mammalian cell homologs, termed CRAF (also known as RAF1)1. Contemporaneously, Bister and colleagues characterized v-Mil an avian retroviral oncogene orthologous to v-raf2. In 1984 both v-raf and v-mil oncoproteins were demonstrated to possess serine/threonine kinase activity, the first oncoproteins identified to have such activity3. Two genes related to CRAF were subsequently found in mouse and human: ARAF and BRAF4,5. Furthermore, homologs of BRAF were identified in Drosophila melanogaster (D-Raf) and in Caenorhabditis elegans (lin-45), where they act during development downstream of receptor tyrosine kinase (RTK) signaling6,7. Ten years after the identification of CRAF8–10, the dual-specificity protein kinase MEK1 was identified as a physiological substrate of CRAF11. Concurrently, several groups identified a direct interaction between RAF proteins and GTP-bound RAS proteins, implicating RAF proteins as direct effectors of activated RAS12,13. Interaction with RAS-GTP at membranes promotes RAF kinase activation that, in turn, leads to direct RAF-mediated activating phosphorylation of MEK1 and MEK2. MEK1 and MEK2 in turn activate the ERK1 and ERK2 mitogen activated protein (MAP) kinases via phosphorylation. Thus RAF proteins are crucial regulators of the ERK MAP kinase signaling cascade, relaying signaling cues from the extracellular environment throughout the cell thereby directing cell proliferation, differentiation, migration and survival.
In 2002 sequencing efforts identified a high frequency of BRAF point mutations in melanoma and in other human cancers14. The ensuing decade witnessed myriad publications further characterizing the roles of mutant BRAF in numerous solid tumors and hematological malignancies. Further, it has become evident that mutations in CRAF and ARAF also occur in cancer, thus implicating the RAF family protein kinases both as drivers of oncogenesis and also as direct targets for therapeutic intervention. Discovery of the BRAF oncogenes prompted several structure-based drug design campaigns that have yielded several highly potent and selective ATP-competitive small molecule BRAF inhibitors. Two compounds (vemurafenib and dabrafenib) have achieved approval by the Food and Drug Administration (FDA) for the treatment of metastatic and unresectable BRAF-mutated melanomas. Initially, the success of BRAF inhibitors appeared to unequivocally reinforce the paradigm of using predictive markers to molecularly stratify patients in clinical trials testing pathway-targeted therapeutics. However, it has since become apparent that BRAF mutational status alone does not predict therapeutic response in all cancers. Efficacy of BRAF inhibitors is limited to a subset of cancer patients with BRAF-mutated metastatic melanoma, despite the abundance of BRAF-mutated tumors identified in colorectal, thyroid, glioblastoma and non-small cell lung cancers, as well as the minority of ARAF and CRAF mutations observed in lung adenocarcinoma. Furthermore, the durability of responses in BRAF-mutated melanoma is restricted by the onset of drug resistance. Although the era of RAF-targeted therapeutics remains in its infancy, the challenge in the coming years lies in determining how to employ RAF inhibitors across multiple tumor types to achieve the greatest immediate clinical benefit, while simultaneously forestalling the emergence of drug resistant disease.
RAF mutations in cancer
The spectrum of BRAF mutations
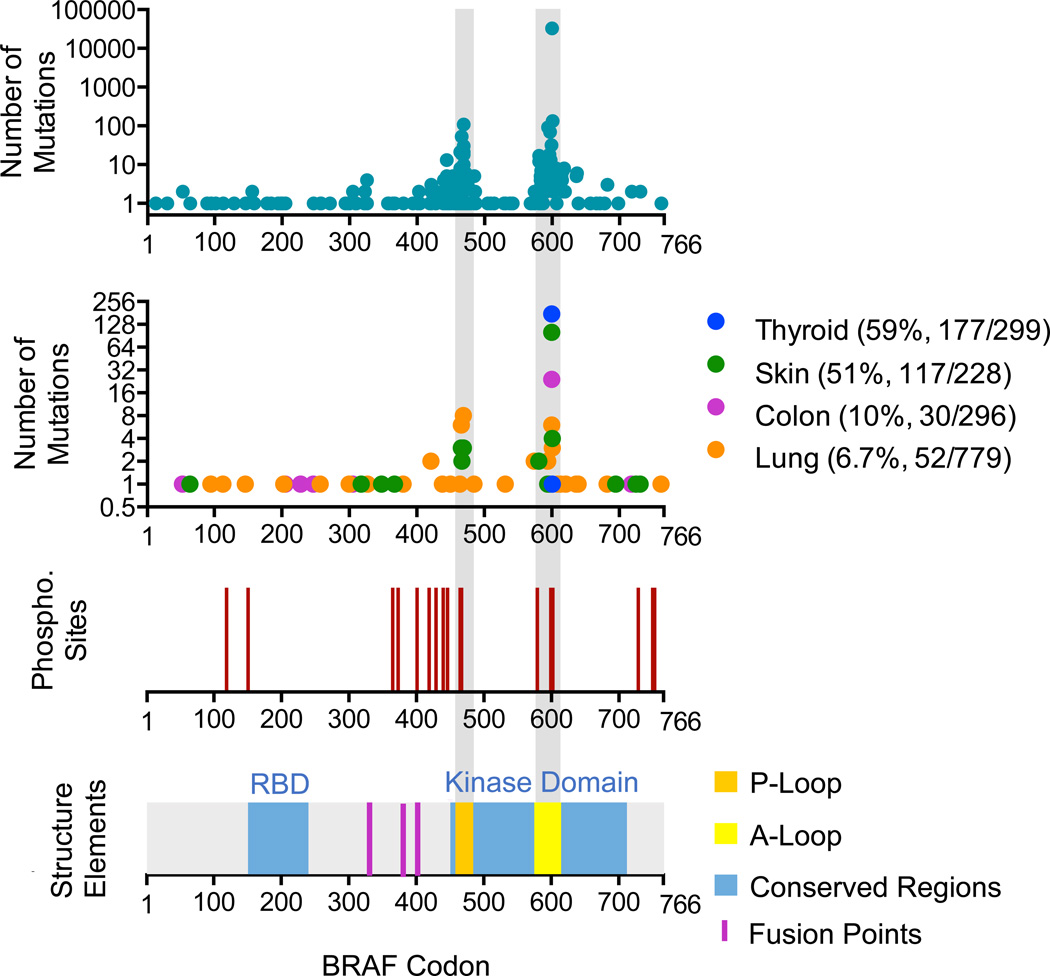
Identification of BRAF mutations in cancer ushered in a new era in the treatment of advanced melanomas. BRAF is mutated in ~8% of all cancers, and roughly half of all melanomas harbor a BRAFT1799A transversion, which encodes the constitutively active BRAF-V600E oncoprotein. In the original description of BRAF mutations in cancer, BRAF-V600E was only one of 14 BRAF alterations identified in cell lines and primary tumor samples14. Since then, nearly 30015 distinct missense mutations have been observed in tumor samples and cancer cell lines (Figure 1). These missense mutations encompass 115 of the 766 BRAF codons, yet the majority of mutations are observed in the activation loop (A-loop) near V600, or in the GSGSFG phosphate binding loop (P-loop) at residues 464–46915,16 (Figure 1). Crystallographic analysis revealed that the inactive conformation of BRAF is stabilized by interactions between the A- and P-loops of the BRAF kinase domain, specifically involving V600 interacting with F46817. Under normal circumstances, reversible phosphorylation of T599 and S602 in the A-loop regulates the A-loop–P-loop interaction allowing BRAF to convert back and forth from its kinase-active to the kinase-inactive state. Consequently, BRAF mutations that lead to amino acid substitutions in either the A-loop or the P-loop mimic T599 and S602 phosphorylation and, by disrupting the A-loop–P-loop interaction, irreversibly shift the equilibrium of BRAF to the kinase-active conformation.
Figure 1. BRAF mutations in cancer.
BRAF codon positions (1 through 766) are depicted on the × axis. Graphs from top to bottom show the number of mutations reported for each codon15 (top panel), the spectrum of BRAF mutations compiled from multiple studies75 in thyroid19, skin138,139, colon cancers140,141 and lung21,40,142 (second panel), the position of putative phosphorylation sites that are reported to have a functional consequence on kinase activity, stability or localization (third panel), and BRAF functional domains: RAS binding domain (RBD) and kinase domain are highlighted in blue, phosphate binding loop (P-loop) highlighted in orange, activation loop (A-loop) highlighted in yellow, fusion points highlighted in magenta (lower graph).
BRAF V600 point mutations are clearly the most common oncogenic driver in melanoma, but BRAF-V600E melanoma represents only a subset of tumors with BRAF alterations. BRAF point mutations also occur in 60% of thyroid, 10% of colorectal carcinomas and in 6% of lung cancers, as well as nearly all papillary craniopharyngioma18, classical hairy cell leukemia19,20, and metanephric kidney adenoma21. Unlike other indications where V600 mutations predominate, BRAF alterations in lung cancer often occur in the P-loop at G466 and G469 (Figure 1). While the frequency of BRAF mutation in colon and lung cancer are considerably lower, the relative morbidity for these indications (50,000 and 158,000 deaths respectively in the US22) may compose an even larger population of patients with BRAF-mutated cancers that are currently ineffectively treated. If 10% of colorectal and 6% of lung cancer patients carry BRAF mutations, that amounts to nearly 16,000 deaths annually due to BRAF-mutated cancers.
The biochemistry of the various altered BRAF proteins varies substantially. While the V600E alteration dramatically elevates kinase activity, several of the less common alterations diminish BRAF kinase activity, yet they promote MEK phosphorylation in a CRAF-dependent manner23. In one genetically engineered mouse model, conditional melanocyte-specific expression of either KRAS-G12D or BRAF D594A (a kinase impaired P-loop mutation) was insufficient to induce nevi or melanomas, yet co-expression of both alleles promoted cellular dimerization of the catalytically inert BRAF-D594A with catalytically competent CRAF and elicited rapidly growing, pigmented tumors24. These data strongly indicate that kinase impaired BRAF mutations are oncogenic drivers, but require RAS and a catalytically competent RAF isoform to activate downstream signaling. Additionally, the P-loop contains an auto-phosphorylation site that dramatically reduces activity of the wild-type enzyme25. At least some of the BRAF mutant proteins appear to bypass this effect, which may explain why even mutations that impair intrinsic kinase activity can constitutively activate the enzyme by preventing auto-inhibition. Therefore, the various BRAF P-loop and A-loop mutations may have different mechanisms of activation, relying partly on constitutive kinase activity or acting entirely as a scaffold for RAS signal transduction through CRAF.
Although the tumor initiating potential of the les common BRAF mutations have yet to be demonstrated in vivo, the oncogenicity of BRAF-V600E has been validated in numerous genetically engineered mouse (GEM) models. To study BRAF-V600E driven tumorigenesis, a mouse carrying a Cre recombinase-activated allele of Braf (BrafCA) was developed, such that normal BRAF is expressed prior to Cre-mediated recombination, and only after exposure to Cre is mutant BRAF-V600E expressed from the endogenous locus.26 Using this model, lungspecific expression of BRAF-V600E caused lung adenomas while concomitant BRAF-V600E expression and homozygous excision of floxed Tp53 alleles caused progression to adenocarcinoma. Expression of BRAF-V600E in melanocyte lineage also cooperated with loss of tumor suppressors (Pten or p16INK4A) to yield melanoma, with metastatic potential in BRAFV600E PTENnull melanocytes27,28. Targeted expression of BRAF-V600E in thyrocytes produced papillary or anaplastic thyroid cancers29–32. Expression of BRAF-V600E in the proliferative cells of the mouse gastrointestinal tract has been shown to act as an early driver mutation in the pathogenesis of serrated colorectal cancer33,34. Finally, recent years have heralded the publication of additional BRAFV600E-driven GEM models of pancreatic ductal adenocarcinoma35, prostate cancer36, pediatric malignant astrocytoma37, soft tissue sarcoma38, and neoplasms of the histiocyte/monocyte lineage39. Together, these models of BRAF-V600E-driven malignancies highlight the oncogenicity of the mutant protein in a wide array of tissue types, and also emphasize the therapeutic potential of targeting BRAF oncogenes.
CRAF and ARAF mutations
In contrast to BRAF, ARAF and CRAF mutations are exceptionally rare in cancer. Recent data indicate that a small subset (~1%) of patients with adenocarcinoma of the lung carry activating ARAF or CRAF mutations. It has not yet been determined if all ARAF and CRAF mutations constitute oncogenic drivers in all cases, but initial cell culture studies confirmed the transforming potential of ARAF S124C, CRAF S257L and CRAF S259A and as well as the sensitivity of these mutants to RAF inhibition40.
Although somatic CRAF point mutations are rare in human cancers, several germ-line CRAF mutations are the cause of Noonan syndrome (NS) (germline mutations in seven other MAP kinase pathway genes are also reported to cause NS; thus the disorder is described as a “RASopathy”)41,42. NS is an autosomal dominant genetic disorder that is characterized by craniofacial deformations, short stature, cardiac anomalies and a propensity for neurocognitive delay43. One-third of NS patients with CRAF mutations also display multiple nevi, lentigines, or café-au-lait spots, suggesting that germ-line CRAF mutations may predispose patients to cutaneous hyperpigmented lesions, similar to the ability of BRAF-V600E to elicit benign nevi27,44. However, the majority of CRAF point mutations identified in NS are not observed in human cancers, and fewer than 2% of human cancer derived cell lines harbor mutated CRAF45. The oncogenic impotence of CRAF, compared to the potent oncogenicity of BRAF, is thought to arise because CRAF possesses low basal kinase activity compared to that of BRAF46. Importantly, one NS mutation, CRAF S259F, has also been identified in lung cancer. Additionally, CRAF S259 mutations have been identified in patients with either colon (S259P) or ovarian cancer (S259A). Phosphorylation of CRAF S259 is associated with reduced activity and promotes direct binding to 14-3-3 proteins, which stabilize the inactive state41,47. Thus, dephosphorylation or mutation of S259 disrupts the stability of the inactive CRAF conformation and facilitates binding to RAS-GTP at the plasma membrane48. The Melan-a cell line, a non-transformed mouse melanocyte-derived cell line that is sensitive to BRAF-V600E induced transformation, is also transformed by expression of CRAF-S259A.49 In contrast to the scarcity of CRAF point mutations identified in cancer, increased CRAF expression has been identified in a number of malignancies, the most notable being bladder cancer50. The oncogenic significance and therapeutic implications of CRAF amplifications remain unclear.
RAF fusion proteins as oncogenic drivers
Analyses of prostate cancer and pediatric astrocytoma or glioma have revealed that point mutation is not the only mechanism that can reveal the oncogenic potential of RAF protein kinases. The presence of the Philadelphia chromosome in chronic myelogenous leukemia is the archetypal example of an oncogenic protein kinase (BCR-ABL) that is generated via a chromosome translocation51. Similarly, RAF fusion transcripts have been identified in melanoma, prostate, gastric, thyroid and breast cancers and gliomas52–57. Early studies demonstrated the constitutive activity and transforming potential of amino-terminally truncated RAF proteins58–60. In every case where chromosomal break points have been identified within the BRAF locus in human cancer, the C-terminal portion of the enzyme is fused in-frame using the start codon of another gene, resulting in a RAF kinase fusion protein lacking the aminoterminal domain. Several 5’ fusion partners have been identified for BRAF, including angiotensin II receptor-associated protein (AGTRAP) and solute carrier family 45, member 3 (SLC45A3) in prostate cancer52; A kinase anchor protein (AKAP9) in thyroid cancer53; and MARVEL domain containing 1 (MALD1), acylglycerol kinase (AGK)61 and heat shock 27kDa associated protein 1 (PASS1) in melanoma62. CRAF fusions with epithelial splicing regulatory protein 1 (ESRP1) have also been described in prostate cancers52, although most RAF kinase fusion events seem to occur in pediatric gliomas. Despite the low frequency of BRAF point mutations in low-grade pediatric astrocytomas (~1%), BRAF gene fusions are observed in the majority (70%) of such cases. To date, the KIAA1549-BRAF fusion is most commonly observed63,64, while FAM131B-BRAF54 and SLIT-ROBO Rho GTPase activating protein 3 (SRGAP3)-CRAF fusions appear less frequently (~2–8% of cases)57.
The biochemistry of RAF fusion proteins remains poorly characterized, but canonical RAF signaling is thought to rely heavily on RAS-mediated dimerization of BRAF and CRAF protomers. Deletion of the amino-terminal RAS binding domain (RBD) has long been known to constitutively activate RAF kinases59,65 and is thought to promote dimerization, suggesting that the amino-terminus of RAF contains an auto-inhibitory domain that prevents dimerization. Dimerization is associated with increased MEK phosphorylation, and disruption of the dimerization interface identified by the crystal structures17,23 nearly abolishes catalytic activity66,67. Indeed, disrupted dimerization also prevents KIAA1549-BRAF-induced oncogenic transformation in vitro68, indicating that loss of the RAF RBD through fusion of the kinase domain to KIAA1549 may constitutively activate RAF proteins through a similar mechanism.
Development of ATP competitive inhibitors of RAF kinase activity
In 2000, sorafenib was the first RAF inhibitor to enter clinical trials; this was prior to the discovery of BRAF, CRAF or ARAF mutations in cancer. Originally developed as a CRAF inhibitor intended to treat RAS-mutated cancers, sorafenib was discovered by screening a chemical library for inhibitors of recombinant, activated CRAF69. Sorafenib competes with ATP for binding directly to the active site of the CRAF kinase domain. Due to the shared structural homology of the ATP-binding pocket several additional kinase targets have been described, including FMS-related tyrosine kinase 3 (FLT3), platelet-derived growth factor receptor-beta (PDGFR-beta), v-Kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (cKIT), and vascular endothelial growth receptor 2 (VEGFR2)70. Although sorafenib is now approved for the treatment of renal cell, hepatocellular and thyroid cancers, it remains unclear if CRAF is the predominant therapeutic target. Efficacy in renal cell carcinomas is likely due to inhibition of VEGFR2, and although responses in hepatocellular carcinomas are correlated with ERK phospohorylation71, responses are not correlated with RAS mutational status.
Melanoma emerged as the ideal test case for BRAF-V600E kinase inhibitors, not only because the tumor genetics of melanoma suggest MAP kinase pathway dependence, but also because patients with metastatic melanoma historically suffered a dearth of efficacious treatment options. Unfortunately, sorafenib and other compounds that were developed to inhibit CRAF lack efficacy in BRAF-mutated melanomas. Therefore, a second-generation of ATP-competitive RAF inhibitors was developed specifically to target BRAF-V600E. Specificity for the mutated oncoprotein was first shown for the vemurafenib analogue PLX-4720, which has a GI50 that is approximately 50-fold more potent for cells containing BRAF-V600E versus cells with wild-type BRAF72. Selectivity for the mutant protein was originally explained by the type I binding mode of vemurafenib (and dabrafenib), which favors the active enzyme conformation imparted by the oncogenic mutation. By contrast, sorafenib and other type II inhibitors preferentially bind the inactive conformation and are thus relatively poor inhibitors of BRAF-V600E. Clinically, this oncoprotein binding selectivity has translated into an unusually high therapeutic index for BRAF-V600E inhibitors, allowing for high exposures of the drug while avoiding acute toxicities associated with RAF inhibition73,74. The high therapeutic index of BRAF-V600E inhibitors has proven critical for treatment of BRAF-mutated melanomas because >80% target inhibition is required for a clinical response75. Recent studies have shown that all ATP-competitive RAF inhibitors—including vemurafenib, dabrafenib and sorafenib—are not only poor inhibitors of wild-type BRAF, but also paradoxically activate the MAP kinase pathway in BRAF wild-type cells24,76,77 (Figure 2). Thus, the high potency of BRAF-V600E inhibitors observed in the treatment of BRAF-V600E mutated melanoma may be attributed to inhibitor binding mode, but the lack of potency in BRAF wild-type cells and the high therapeutic index likely stem from the unique mechanism of action for wild-type RAF kinase, as described below.
Figure 2. Biochemistry of RAF inhibitors in BRAF-V600E and BRAF wild-type cells.
BRAF oncoproteins promote proliferation by constitutively activating the MAP kinase pathway. (a) ATP competitive RAF inhibitors (I) disrupt MEK activation and prevent proliferation of BRAF-V600E mutated melanomas in a dose dependent manner. (b) Wild-type RAF proteins exist largely in an auto-inhibited state. In the context of activated RAS, RAF inhibitors stimulate MEK activation at sub-saturating inhibitor concentrations by relieving auto-phosphorylation (P) of wild-type RAF, promoting RAF dimerization and association with RAS. Saturating inhibitor concentrations prevent MEK phosphorylation by blocking all RAF catalysis.
The RAF inhibitor paradox
Use of RAF inhibitors in RAS-mutated cancers has unveiled the complexity of RAF signal transduction. In 2009 three groups showed that catalytic inhibition of RAF kinases resulted in paradoxical activation of CRAF, increased proliferation of RAS mutated cells in vitro, and in some cases induction of tumorigenesis in vivo24,76,77. While each group proposed a distinct mechanism to explain the effect, several themes overlapped (Figure 2B). RAF inhibitors promoted co-immunoprecipitation of RAF isoforms as well as association with RAS-GTP at cell membranes25,76,78, suggesting that the inhibitors promote CRAF homo- and heterodimerization and RAS-mediated activation. Moreover, site-directed mutagenesis of the putative dimer interface rendered CRAF resistant to paradoxical activation77. The drug-induced association of RAF with RAS-GTP as well as increased RAF dimerization likely both contribute to the activation of ERK signaling, but it remains unclear if either event alone is sufficient to induce paradoxical activation.
Interestingly, paradoxical activation by RAF inhibitors can be recapitulated genetically using catalytically-impaired BRAF24. In this model, BRAF catalytically suppresses CRAF activity in RAS-mutated cells such that disrupting BRAF function, either genetically or with a BRAF inhibitor, alleviates CRAF suppression and activates it in a RAS-dependent manner. This mechanism is further supported by the discovery of a RAF auto-inhibitory phosphorylation site that is required for paradoxical activation25 (Figure 2B), indicating that paradoxical activation may be intrinsic to all catalytic RAF inhibitors.
Clinical use of RAF inhibitors
Prior to the approval of BRAF inhibitors, BRAF-mutated melanoma patients faced a worse prognosis than that of patient’s whose disease expressed wild-type BRAF. However, the recent approval of two BRAF inhibitors (vemurafenib and dabrafenib) and one MEK inhibitor (trametinib) for the treatment of metastatic BRAF-V600E-mutated melanoma has altered the situation; BRAF-mutated melanoma patients treated with BRAF inhibitors now have a longer median survival than that of BRAF wild-type patients79. In Phase I clinical trials, treatment with vemurafenib induced disease stabilization as well as some cases of dramatic regression of BRAF-V600E-mutated melanomas73. The objective response rate (OR) in Phase II increased from 19% in the dacarbazine control arm to 53% in the vemurafenib arm, and the average duration of response increased from 2.7 months to 6.7 months80. In 2011, the Phase III trial reported a 48% OR when patients were treated with vemurafenib compared to a 5% OR when patients were treated with dacarbazine as well as an estimated 5.3 month progression free survival (PFS) in vemurafenib treatment group compared to a 1.6 month median PFS in the dacarbazine treatment group81. Although the overall survival (OS) benefit was limited, approximately 14% of patients experienced durable responses and remained free from relapse after 18 months of vemurafenib treatment82. Moreover, the full measure of the effect of vemurafenib on OS could not be calculated due to the cross-over design of the Phase III trial. A second BRAF inhibitor, dabrafenib also gained FDA approval in 2013, not only for the treatment of BRAF-V600E expressing melanomas, but also for those expressing BRAF-V600K83. Phase III results reported dabrafenib to provide a 50% OR compared to 6% for dacarbazine and 5.1 month PFS with dabrafenib compared to 2.7 for the dacarbazine group84. FDA approval was also granted for trametinib, the first MEK inhibitor marketed for cancer treatment, which increased the median PFS from 1.5 months with dacarbazine to 4.8 months with trametinib and the OR from 8% with dacarbazine to 22% with trametinib85.
The remarkable efficacy of BRAF inhibition in melanoma has been recapitulated in early data emerging from the treatment of classical Hairy Cell Leukemia (HCL-C) patients. In 2011, sequencing of a cohort of HCL-C patient samples revealed the BRAF-T1799A mutation in virtually all HCL-C patients20. Anecdotes from the clinic suggest that HCL-C patients whose disease is refractory to conventional chemotherapy respond well to vemurafenib86. Thus HCL-C may be a neoplasm in which mutated BRAF is a key oncogenic driver, and the paucity of BRAF-independent signaling pathways precludes the development of primary chemoresistance.
Cutaneous lesions induced by BRAF inhibitors
The ability of RAF inhibitors to paradoxically activate ERK MAP kinase signaling carries critical clinical implications. Results from the vemurafenib phase III clinical trial indicate that 18% of patients experienced squamous cell carcinomas (SCC) and/or keratoacanthomas (KAs)81. The list of secondary cutaneous lesions induced by BRAF inhibitors has since expanded to include papillomas, cystic lesions, milia, eruptive nevi, and basal cell carcinomas87. While all drug-induced lesions are BRAF wild-type, multiple other genetic mutations have been identified. Importantly, samples of secondary KAs display a high frequency of HRAS mutations88–90. This suggests that the ability of BRAF inhibitors to elicit paradoxical MAP kinase pathway activation activates a proliferative program in dormant keratinocytes harboring a latent RAS mutation. Indeed, studies using the K5-SOS-F transgenic mouse model, in which mice are predisposed to epidermal tumors through keratinocyte-specific expression of a dominant active, farnesylated Son of Sevenless (SOS-F), have shown that RAF inhibitor treatment promotes RAF dimerization, activates ERK signaling, and suppresses Rho-Associated, Coiled-Coil Containing Protein Kinase 2 (Rokα) to potentiate Ras-driven epidermal tumorigenesis.78 In addition to RAS mutations, oncoviruses have been implicated as initiating factors in a subset of cutaneous lesions. Papilloma virus and Merkel cell polyomavirus DNA were identified in BRAF inhibitor treated patient samples and vemurafenib promoted tumorigenesis in an HPV driven mouse model of SCC91. Thus far the secondary cutaneous malignancies arising in patients receiving BRAF inhibitor therapy have been well-differentiated and amenable to local resection with no reports of metastasis92.
More worrisome than the cutaneous toxicities is the possibility that BRAF inhibitors may induce the proliferation of pre-malignant cells harboring RAS mutations in non-cutaneous tissues, such as the lung, colon, or pancreas. The ability of BRAF inhibitors to promote the growth of pre-malignant precursor lesions along the path to adenocarcinoma is of considerable concern. Indeed, a recent report described a patient with metastatic BRAFV600K melanoma who developed NRAS-mutant chronic myelomonocytic leukemia during the course of vemurafenib treatment93,94. Upon withdrawal of vemurafenib, the patient’s leukemic cell counts dropped, thus necessitating an empirically determined intermittent dosing schedule of vemurafenib and cobimetinib (a MEK inhibitor) to simultaneously control the patient’s metastatic melanoma and also to prevent the outgrowth of leukemia95. A second case report described a patient presenting with BRAF-mutated metastatic melanoma, who had undergone a prior successful surgical resection of a KRAS-G12D colorectal cancer96. While the combination of dabrafenib and trametinib caused a PR of the patient’s melanoma, at 12 weeks the patient presented with a brain metastasis that, upon resection, was characterized as originating from a KRAS-mutated colon cancer. After progressive metastatic colon cancer necessitated cessation of dabrafenib and trametinib combination therapy, the patient received trametinib monotherapy and experienced a brief improvement in performance status. In vitro studies performed using tissue derived from the aforementioned patients suggested that MEK inhibitors suppress the growth of BRAF-V600E melanoma and also prevent expansion of the RAS-mutated malignancies. These case reports not only emphasize the caution that must be exercised when considering the use of RAF inhibitors as an adjuvant therapy, but also demonstrate the importance of determining which node of the MAP kinase pathway is most appropriate to target therapeutically in patients with BRAF-mutated melanoma.
Primary resistance to RAF inhibition
BRAF mutation is not always prognostic of responsiveness to BRAF inhibitor therapy. Although BRAF mutated melanoma is heralded as the malignancy in which BRAF inhibitors prove most beneficial, primary resistance occurs even in BRAF mutated melanoma. In the dabrafenib phase II clinical trial, 16% of BRAF-V600E and 31% of BRAF-V600K melanoma patients experienced progressive disease despite dabrafenib treatment83. In some cases inhibition of the BRAF oncogene may activate signaling through the wild-type RAF alleles by relieving feedback mechanisms and elevating RAS-GTP97. Moreover, results using BRAF inhibitors in non-melanoma malignancies illustrate that the cell of origin of the cancer - and thus the signaling pathways inherently available to the cancer cell - predicts the efficacy of BRAF inhibition. For example, although BRAF-V600E is expressed in ~10% of patients with metastatic colorectal carcinomas (mCRC), a Phase I clinical trial of vemurafenib in BRAF-mutated mCRC patients was associated with only a 3.7 month PFS, about half of the PFS observed in BRAF-mutated advanced melanoma98. Subsequent studies suggested that the diminished efficacy of vemurafenib in BRAF-mutated mCRC can be attributed to increased epidermal growth factor receptor (EGFR) signaling in these cells in response to vemurafenib. At baseline, BRAF-V600E→MEK→ERK signaling activates a negative feedback loop that serves to attenuate RTK EGFR signaling99–101. However, BRAF inhibition relieves this negative feedback, leading to increased signaling through the EGFR (Figure 3); in sum, the on-target activity of BRAF inhibitors activates a rapid, adaptive mechanism of chemoresistance. In contrast to mCRC, basal EGFR levels are low in melanoma, allowing for evasion of this resistance mechanism. Thus the cellular context in which BRAF inhibitors are used is highly relevant to therapeutic efficacy.
Figure 3. Loss of feedback inhibition and activation of the PI’3 kinase pathway mediates primary resistance to BRAF-V600E inhibition.
Resistance to BRAF inhibitors in metastatic colorectal and thyroid cancers is mediated by loss of a feedback loop (EGF→EGFR signaling in colorectal cancers or NRG1→HER2 and HER3 signaling in papillary thyroid cancers) whereby the MAP kinase pathway activity inhibits mitogen dependent signaling (highlighted in blue). The negative feedback loop is disrupted when treated with a RAF inhibitor and proliferation is restored through RTK, RAS and PI3K signaling (highlighted in red).
Papillary thyroid cancer emerges as another malignancy for which BRAF mutational status does not predict response to BRAF-V600E inhibition. By a mechanism conceptually concordant with that observed in mCRC, inhibition of BRAF-V600E in papillary thyroid cancer causes a relief of negative feedback, leading to an induction of human epidermal growth factor receptor 2 (HER2) and HER3 signaling102. The Fagin group showed that BRAF inhibitormediated induction of HER2 and HER3 signaling in thyroid cancer is dependent on the secretion of the HER2 and HER3 ligand neuregulin 1. This ligand is not expressed in melanoma or colorectal cancer cells, thereby allowing the evasion of HER2 and HER3-mediated de novo resistance in these tissues.
The prognostic value of BRAF mutation for pathway-targeted therapy in NSCLC or ovarian cancer remain largely unknown at this time but, at least with NSCLC, the response of BRAF-V600E-initiated lung cancers to pathway-targeted blockade in genetically-engineered mouse models and a case report of the response of an ARAF-mutated lung cancer patient to sorafenib offers grounds for optimism26,40,103,104.
Acquired resistance to RAF inhibition
Although the reported sample sizes remain small, the survival benefit in BRAF-mutated melanoma patients treated with BRAF inhibitors appears to be limited to less than 1 year with the durability of patient responses to RAF inhibitors limited by the onset of drug resistant disease80,81,83. Initial attempts to anticipate resistance mechanisms were informed by experience with acquired resistance to imatinib in BCR-ABL mutated leukemia, where mutation of the “gatekeeper” threonine prevents drug binding without drastically affecting normal kinase activity105,106. Surprisingly, although engineering of mutations at the analogous gatekeeper residue (T529) in BRAF confers vemurafenib resistance in vitro, T529 mutations have never been reported in BRAF inhibitor resistant patient tumor samples106. One possible explanation for the failure to find such an obvious, and highly predicted, mechanism of resistance may be that cells expressing BRAF doubly mutated at codons 529 and 600 are not viable in the absence of drug. Hence, there would be no reservoir of such cells prior to drug treatment and therefore this would not score as a mechanism of drug resistance. Although Marais and colleagues have shown that a myeloid cell line (Ba/F3) remains viable in the absence of BRAF inhibitor when transfected with BRAF doubly mutated at codons 529 and 600106, this observation is likely cell-type specific and may also reflect the outgrowth of cells transduced with lower—and thus non-toxic—levels of doubly mutated BRAF.
Despite the absence of T529 mutations in BRAF, numerous other mechanisms of acquired resistance to BRAF inhibitors that contribute to clinical drug resistance have been described. The majority of resistance mechanisms promote re-activation of the MAP kinase signaling pathway in the presence of BRAF inhibitor (Figure 4). For example, mutational activation of NRAS, MEK1 or MEK2 can reactivate the MAP kinase pathway in the presence of BRAF inhibition107–110 and elevated CRAF protein levels have also been shown to confer resistance to BRAF inhibition in cell culture melanoma models110,111. CRAF protein elevation has yet to be identified in clinical samples of BRAF inhibitor resistance, and it has been shown that in some contexts CRAF negatively regulates BRAF-V600E112, so further analysis is necessary to determine the clinical relevance of CRAF protein elevation as a bona fide BRAF inhibitor resistance mechanism. Through an unbiased screen, the serine/threonine MAP kinase kinase kinase (MAP3K) COT kinase (encoded by MAP3K8) was shown to activate MEK in the presence of BRAF inhibition113. Elevated COT copy number and mRNA expression was identified in biopsy specimens of metastatic melanoma following vemurafenib treatment. Overexpression of the mutant BRAF protein itself has also been reported, further emphasizing the importance of increased expression of the drug target as a relevant mechanism of cancer drug resistance114,115. Additionally, the identification of BRAF-V600E splice-variants, which endow the proteins with the ability to dimerize in a RAS-independent manner and increases the kinase activity of the proteins, represents the only resistance mechanism that involves a structural change to BRAF itself and one of the first examples where altered mRNA splicing can render an oncoprotein resistant to a drug116.
Figure 4. Mechanisms of acquired resistance to BRAF-V600E inhibition leading to reactivation of the MAP kinase pathway.
Multiple mechanisms of acquired resistance (highlighted in blue) to BRAF-V600E inhibitors lead to reactivation of the MEK-ERK pathway (highlighted in red) in the presence of a RAF inhibitor. The mechanisms of resistance include: activating mutations (*) of NRAS, overexpression (↑) of CRAF, overexpression of BRAF-V600E itself, alternative splicing of BRAF that renders the protein immune to inhibitors (p61), overexpression of COT, mutational activation of MEK1 and MEK2, ligand dependent RAS signaling through PDGFRβ or IGF-1R, or stromal cell secretion of HGF to activate through c-MET signaling.
In addition to MAP kinase pathway reactivation in the presence of BRAF inhibition, it has been suggested that about 30% of patients develop MAPK pathway independent mechanisms of resistance117. Initial reports propose that the majority of MAPK pathway independent mechanisms of resistance involve alterations that lead to upregulation of the PTEN-PI3’-K-AKT signaling axis. Elevated expression of either platelet-derived growth factor receptor-β (PDGFRβ) or insulin-like growth factor I receptor (IGF1R) expression was identified in cultured cells and in specimens from patients with vemurafenib-resistant melanomas. It is claimed that PDGFRβ or IGF1R signaling allows for activation of MAP kinase pathway-independent prosurvival signaling pathways, such as the PI3’-K/AKT axis, which render cells resistant to the effects of BRAF inhibition107,118. Further, activation ERBB3 signaling has been identified as an adaptive mechanism of resistance in a subset of melanoma patients. It is thought MAPK pathway inhibition promotes upregulation of the transcription factor FOXD3, which in turn directs increased expression of ERBB3 and allows for enhanced ERBB2/ERBB3 signaling119. Additionally, amplification of the melanocyte lineage specific transcription factor MITF, or its effector BCL2A1, has been shown to confer resistance to BRAF and MEK inhibitors in cell culture models, and MITF upregulation occurs in a minority of BRAF-mutated melanoma patients120–122. Finally, secretion of growth factors by the tumor stroma has been shown to confer resistance to BRAF inhibition. Specifically, stromal cell directed secretion of hepatocyte growth factor has been shown to signal through MET receptor expressed on the surface of melanoma cells, and thereby re-active MAPK and PI3’-K-AKT signaling pathways in the presence of BRAF inhibition123,124.
While numerous on- or off-pathway mechanisms of resistance have been identified through preclinical studies, the challenge lies in characterizing which of these resistance mechanisms are clinically relevant, and the relative frequencies of the resistance mechanisms observed in the clinic. The development of second-generation BRAF inhibitors has definitively demonstrated the potential of pathway-targeted therapeutics in the treatment of BRAF-mutated melanona. However, the emergence of drug resistant disease stands as a formidable obstacle that remains to be addressed to enhance the durability of patient responses.
Improving responses to RAF inhibition with other therapeutic targets
It is currently unclear if survival benefit alone will cause RAF inhibitors to be used as single agents, but preclinical and early clinical data strongly suggest that some combination of RAF and either MEK or conceivably an ERK inhibitor will have the greatest efficacy. FDA approval was recently announced for the combination of dabrafenib and trametinib in advanced melanoma, and detailed results of the phase III trial are forthcoming. Phase I/II results demonstrated that the combination increased PFS from 5.8 months for single agent treatment to 9.4 months in the combination group, and OR increased from 54% to 76%125. Perhaps unexpectedly, tolerability of either agent improved in the combination group. Acneiform dermatitis, the most common dose limiting toxicity associated with trametinib (8%), was not observed in the combination group. Likewise, the frequency of hyperkeratosis, cutaneous SCCs and papillomas, all RAF inhibitor class effects, each decreased when dabrafenib was co-administered with trametinib. Mechanistically, this most likely occurs because the paradoxical activation of the MEK1/2→ERK1/2 MAP kinase pathway induced by RAF inhibitors is suppressed by MEK inhibition.
Tumors that have acquired on-pathway mechanisms of resistance to BRAF inhibitors compose an important group in which to test the efficacy of second-line MEK or ERK inhibitors. Unfortunately, early clinical data suggest limited efficacy of MEK inhibitor monotherapy for BRAF-mutated melanoma patients with drug resistant disease. Within a cohort of 40 patients who had progressed on either vemurafenib or dabrafenib monotherapy, subsequent treatment with trametinib offered only a 1.8 month median PFS and a 5.8 month median OS126. Only two patients within the cohort had a complete or partial response and both of these patients had discontinued BRAF inhibitor therapy due to adverse events rather than the onset of drugresistant disease. Thus second-line MEK inhibitor monotherapy to treat BRAF inhibitor resistant disease seems limited. These results may be partially explained by a recent report demonstrating that oncogenic BRAF over-expression or alternative splicing – previously known to confer resistance to BRAF inhibitor monotherapy – also emerges as a resistance mechanism among patients who receive first-line BRAF and MEK combination therapy127. Another tactic to target tumors that have acquired MAP kinase pathway-dependent resistance mechanisms is the use of ERK inhibitors. While ERK inhibitors have only recently entered clinical trials, preclinical data suggest that cells that have developed resistance to RAF and MEK inhibitors remain sensitive to a selective ERK1 and ERK2 inhibitor128. The acquisition of MAP kinase pathway-independent mechanisms of resistance also may sensitize tumors to other pathway-targeted therapies. For example, elevated PDGFRβ or IGF1R signaling might be combated with receptor tyrosine kinase or PI3’-K inhibitors118.
BRAF-V600E oncogene overdose and intermittent drug dosing
Although BRAF-V600E constitutively activates the MEK→ERK pathway, many cancer cells remain sensitive to the quantity of MAP kinase pathway signaling (as measured by ERK phosphorylation or transcription of target genes). More than ten years ago, it was demonstrated that mouse or human fibroblasts display a peak proliferative response with an intermediate amount of RAF→MEK→ERK pathway activation129,130. Indeed, in nonimmortalized human IMR-90 fibroblasts, sustained RAF activation induced an irreversible cell cycle arrest with features of cellular senescence131. This further substantiated the hypothesis that RAF oncogene induced transformation is determined not only by activation of the MAPK pathway, but the quantity of pathway activation, as reviewed by Marshall132.
The notion that malignant cells might remain sensitive to the strength of MAP kinase pathway signaling was recently substantiated using a patient-derived melanoma xenograft model. Upon transplantation into immunocompromised mice, a chemonaïve, BRAF mutated melanoma displayed striking regression in response to vemurafenib treatment130. However, sustained drug treatment led to the emergence of lethal drug-resistant disease due to elevated BRAF-V600E expression. Remarkably, the drug resistant tumors and cell cultures were not merely drug-resistant but also drug-dependent for their peak proliferation (Figure 5A). Moreover, when vemurafenib was removed from drug resistant cells or tumors, striking antiproliferative effects were observed (Figure 5B). These data indicate that BRAF-V600E ‘addicted’ melanoma cells can remain sensitive to the magnitude of BRAFV600E→MEK→ERK signal pathway activation such that too much of the oncoprotein to which they are addicted can lead to “oncogene overdose”. The demonstration that vemurafenibresistant melanoma cells can have a fitness deficit in the absence of drug has led to the design of clinical trials to test the efficacy of intermittent dosing to forestall the onset of drug resistant disease and therefore enhance the durability of patient responses.
Figure 5. Intermittent BRAF inhibitor therapy to forestall the onset of drug resistant melanoma.
Prior to the initiation of BRAF inhibitor therapy to BRAF mutated melanomas, the bulk of the cells have optimal BRAF-V600E→MEK→ERK activity (A) and are drug sensitive (blue) with only rare variant cells with intrinsic drug resistance (black). The addition of BRAF inhibitor leads to insufficient BRAF-V600E→MEK→ERK activity in the bulk of the tumor (A) leading to tumor regression (B, response 1). However, BRAF inhibitor therapy immediately starts to select for expansion of melanoma cells with elevated BRAF-V600E expression and therefore intrinsic drug resistance, because they have an optimal level of BRAF-V600E→MEK→ERK activity—therefore a fitness benefit—in the presence of drug. Upon cessation of drug administration, drug resistant melanoma cells with elevated BRAF-V600E expression experience an excess of BRAF-V600E→MEK→ERK activity (A) that confers a fitness deficit on the cells (B, response 2). However, cessation of drug administration also allows residual drug sensitive cells to restore their optimal level of BRAF-V600E→MEK→ERK activity (A) and therefore re-initiate their proliferation (B, response 2).
Conclusions
Development of BRAF inhibitors has changed preclinical understanding and clinical treatment of late-stage melanoma, and offers a more hopeful option to patients with a historically devastating diagnosis. Despite the short duration of responses, BRAF inhibitor treated patients have a high overall response rate and frequently experience dramatic tumor regression. Several complete responses have been observed (by Response Evaluation Criteria in Solid Tumors)73,81. Drug resistant disease progression is observed in the majority of cases, often occurring within a few months after initial response. Current efforts to co-target BRAF and MEK125 may prevent some of the identified mechanisms of resistance, though it is not clear to what extent. Identifying and understanding common resistance mechanisms in BRAF and MEK inhibitor resistant melanomas will inform additional rational pathway-targeted therapeutic combinations, such as co-targeting PI3’-kinase signaling, as well as dosing strategies to prevent or delay drug resistance and achieve long-term survival benefit.
A more thorough understanding of the mechanism(s) of oncogenesis has led to potent and selective RAF inhibitors that exploit the unique biology of BRAF mutated melanomas72. However, BRAF-mutated melanoma only represents a small subset of RAF-mutated cancers. Several RAF fusion proteins appear insensitive to the current generation of BRAF inhibitors and instead exhibit “paradoxical activation” similar to wild-type RAF alleles (Table 1). Additionally, CRC and NSCLC comprise a large cohort of BRAF-mutated cancers that are currently ineffectively treated. Just as rational structure-based drug design has yielded selective and targeted treatments for BRAF-mutated melanomas, understanding the mechanisms of other BRAF oncogenes and CRAF-dependent tumors may lead to effective treatments in these malignancies. Similarly, while our understanding of rapid adaptive resistance in BRAF-mutated CRC has led to hypotheses for rational combinations using receptor tyrosine kinase or PI3’-K pathway inhibitors99,100,102, understanding BRAF inhibitor resistance in papillary thyroid and ovarian cancers will likely yield additional rational therapeutic approaches and predictive biomarkers for these indications.
Table 1.
MAP kinase pathway oncogenes and effects of RAF or MEK inhibitors
| Cancer Mutation | Biochemical effect of RAF inhibition |
Clinical effect of RAF inhibition |
Biochemical effect of MEK inhibition |
Clinical Effect of MEK inhibition |
|---|---|---|---|---|
| BRAF V600E/K | Inhibits MEK phosphorylation and inhibits growth of melanoma cells72. | Tumor regression and improved survival in late stage melanoma80–84. | Inhibits ERK phosphorylation and inhibits growth of melanoma cells143. | Tumor regression and improved survival in late stage melanoma85, 126. |
| BRAF point mutations other than V600E/K | Inhibits MEK phosphorylation in some settings25, no effect or enhanced MEK phosphorylation in others136. | Unknown. | Unknown. | Unknown. |
| BRAF fusion proteins | Paradoxical activation of pediatric astrocytoma fusion proteins54, sorafenib but not vemurafenib inhibits MEK phosphorylation in patient derived cell line with AGK-BRAF fusion62. | Unknown. | Inhibits ERK phosphorylation in PASS1-BRAF fusion expressing cells72. | Unknown. |
| CRAF fusion proteins | Unknown. | Unknown. | Unknown. | Unknown. |
| CRAF point mutations | Sorafenib inhibits MEK phosphorylation and inhibits growth of CRAF S257L, S259A transformed cells40. | Unknown. | Inhibits ERK phosphorylation and inhibits growth of S257L and S259A transformed cells40. | Unknown. |
| ARAF point mutations | ARAF-S124C transformed cells are sensitive to RAF inhibition40. | 5 year tumor remission treated with sorafenib40. | Unknown. | Unknown. |
| RAS mutations | Paradoxical activation24,76,77. | Ineffective. | Inhibits ERK phosphorylation and inhibits growth in some settings143. | Largely ineffective, though early reports indicate responses in some NRAS mutated melanomas137. |
While BRAF-V600E and BRAF-V600K are bona fide drug targets in melanoma, it remains unclear if ARAF and CRAF alterations constitute driver mutations, but there is reason to suspect that they are therapeutic targets. A recent case study indicates that ARAF mutations could be treated with currently available RAF inhibitors. An NSCLC patient with an ARAF-S124C mutation, and a lack of alterations in any other known oncogenes or tumor suppressor genes, achieved a sustained tumor remission (~5 years) on sorafenib treatment40, giving some promise of treatment for cancer patients with these rare mutations. Interest has also been renewed in wild-type CRAF as a therapeutic target for KRAS-mutated lung cancer after demonstrations of CRAF dependence for the onset of Kras-mutated NSCLC in GEM models133–135. Interestingly, BRAF was dispensable, yet concurrent loss of MEK1 and MEK2 or ERK1 and ERK2 also prevented tumor initiation, suggesting that CRAF, but not BRAF, is essential for tumor formation in Kras-driven oncogenesis. CRAF but not BRAF dependence was also demonstrated in a SOS-F-induced skin cancer model135 further supporting CRAF as a therapeutic target in RAS dependent cancers. However, the utility of catalytic CRAF inhibitors remains uncertain due to the paradoxical activation of ERK MAP kinase signaling by RAF kinase domain inhibitors. Alternatively, the mechanisms that regulate RAF activation may reveal novel modes of therapeutic intervention. Indeed, efforts to develop “dimer-blockers”136 and RAF inhibitors that do not exhibit paradoxical activation90 have begun. Additionally, preliminary data suggest that MEK inhibitors may be effective in some NRAS mutated melanomas137. A deeper understanding of RAS→RAF→MEK→ERK biochemistry, in conjunction with the identification of new biomarkers to predict response, will be essential for the future success of drug development programs in RAF-driven cancers.
References
- 1.Rapp UR, et al. Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proceedings of the National Academy of Sciences of the United States of America. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen HW, Ruckert B, Lurz R, Bister K. Two unrelated cell-derived sequences in the genome of avian leukemia and carcinoma inducing retrovirus MH2. The EMBO journal. 1983;2:1969–1975. doi: 10.1002/j.1460-2075.1983.tb01686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moelling K, Heimann B, Beimling P, Rapp UR, Sander T. Serine- and threonine-specific protein kinase activities of purified gag-mil and gag-raf proteins. Nature. 1984;312:558–561. doi: 10.1038/312558a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonner TI, et al. Structure and biological activity of human homologs of the raf/mil oncogene. Molecular and cellular biology. 1985;5:1400–1407. doi: 10.1128/mcb.5.6.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonner T, et al. The human homologs of the raf (mil) oncogene are located on human chromosomes 3 and 4. Science. 1984;223:71–74. doi: 10.1126/science.6691137. [DOI] [PubMed] [Google Scholar]
- 6.Mark GE, MacIntyre RJ, Digan ME, Ambrosio L, Perrimon N. Drosophila melanogaster homologs of the raf oncogene. Molecular and cellular biology. 1987;7:2134–2140. doi: 10.1128/mcb.7.6.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han M, Golden A, Han Y, Sternberg PWC. elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature. 1993;363:133–140. doi: 10.1038/363133a0. [DOI] [PubMed] [Google Scholar]
- 8.Kozak C, Gunnell MA, Rapp UR. A new oncogene, c-raf, is located on mouse chromosome 6. Journal of virology. 1984;49:297–299. doi: 10.1128/jvi.49.1.297-299.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen HW, Trachmann C, Bister K. Structural relationship between the chicken protooncogene c-mil and the retroviral oncogene v-mil. Virology. 1984;137:217–224. doi: 10.1016/0042-6822(84)90028-x. [DOI] [PubMed] [Google Scholar]
- 10.Jansen HW, et al. Homologous cell-derived oncogenes in avian carcinoma virus MH2 and murine sarcoma virus 3611. Nature. 1984;307:281–284. doi: 10.1038/307281a0. [DOI] [PubMed] [Google Scholar]
- 11.Kyriakis JM, et al. Raf-1 activates MAP kinase-kinase. Nature. 1992;358:417–421. doi: 10.1038/358417a0. [DOI] [PubMed] [Google Scholar]
- 12.Moodie SA, Willumsen BM, Weber MJ, Wolfman A. Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 13.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 15.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer cell. 2004;6:313–319. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 18.Brastianos PK, et al. Exome sequencing identifies BRAF mutations in papillary craniopharyngiomas. Nature genetics. 2014;46:161–165. doi: 10.1038/ng.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer discovery. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiacci E, et al. BRAF mutations in hairy-cell leukemia. The New England journal of medicine. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.U.S. Cancer Statistics Working Group. United States Cancer Statistics: 1999–2010 Incidence and Mortality Web-based Report. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2013. [Google Scholar]
- 23.Garnett MJ, Rana S, Paterson H, Barford D, Marais R. Wild-type and mutant BRAF activate C-RAF through distinct mechanisms involving heterodimerization. Molecular cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holderfield M, et al. RAF inhibitors activate the MAPK pathway by relieving inhibitory autophosphorylation. Cancer cell. 2013;23:594–602. doi: 10.1016/j.ccr.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Dankort D, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes & development. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhomen N, et al. Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer cell. 2009;15:294–303. doi: 10.1016/j.ccr.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 29.McFadden DG, et al. p53 constrains progression to anaplastic thyroid carcinoma in a Braf-mutant mouse model of papillary thyroid cancer. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1600–E1609. doi: 10.1073/pnas.1404357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knauf JA, et al. Targeted expression of BRAFV600E in thyroid cells of transgenic mice results in papillary thyroid cancers that undergo dedifferentiation. Cancer Res. 2005;65:4238–4245. doi: 10.1158/0008-5472.CAN-05-0047. [DOI] [PubMed] [Google Scholar]
- 31.Charles RP, Iezza G, Amendola E, Dankort D, McMahon M. Mutationally activated BRAF(V600E) elicits papillary thyroid cancer in the adult mouse. Cancer Res. 2011;71:3863–3871. doi: 10.1158/0008-5472.CAN-10-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charles RP, Silva J, Iezza G, Phillips WA, McMahon M. Activating BRAF and PIK3CA Mutations Cooperate to Promote Anaplastic Thyroid Carcinogenesis. Molecular cancer research : MCR. 2014 doi: 10.1158/1541-7786.MCR-14-0158-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carragher LA, et al. V600EBraf induces gastrointestinal crypt senescence and promotes tumour progression through enhanced CpG methylation of p16INK4a. EMBO molecular medicine. 2010;2:458–471. doi: 10.1002/emmm.201000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rad R, et al. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer cell. 2013;24:15–29. doi: 10.1016/j.ccr.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collisson EA, et al. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer discovery. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72:4765–4776. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huillard E, et al. Cooperative interactions of BRAFV600E kinase and CDKN2A locus deficiency in pediatric malignant astrocytoma as a basis for rational therapy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8710–8715. doi: 10.1073/pnas.1117255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mito JK, et al. Oncogene-dependent control of miRNA biogenesis and metastatic progression in a model of undifferentiated pleomorphic sarcoma. The Journal of pathology. 2013;229:132–140. doi: 10.1002/path.4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamata T, et al. Hematopoietic expression of oncogenic BRAF promotes aberrant growth of monocyte-lineage cells resistant to PLX4720. Molecular cancer research : MCR. 2013;11:1530–1541. doi: 10.1158/1541-7786.MCR-13-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Imielinski M, et al. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. The Journal of clinical investigation. 2014 doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandit B, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nature genetics. 2007;39:1007–1012. doi: 10.1038/ng2073. [DOI] [PubMed] [Google Scholar]
- 42.Razzaque MA, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nature genetics. 2007;39:1013–1017. doi: 10.1038/ng2078. [DOI] [PubMed] [Google Scholar]
- 43.Roberts AE, Allanson JE, Tartaglia M, Gelb BD. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pollock PM, et al. High frequency of BRAF mutations in nevi. Nature genetics. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 45.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emuss V, Garnett M, Mason C, Marais R. Mutations of C-RAF are rare in human cancer because C-RAF has a low basal kinase activity compared with B-RAF. Cancer research. 2005;65:9719–9726. doi: 10.1158/0008-5472.CAN-05-1683. [DOI] [PubMed] [Google Scholar]
- 47.Light Y, Paterson H, Marais R. 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Molecular and cellular biology. 2002;22:4984–4996. doi: 10.1128/MCB.22.14.4984-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubicek M, et al. Dephosphorylation of Ser-259 regulates Raf-1 membrane association. The Journal of biological chemistry. 2002;277:7913–7919. doi: 10.1074/jbc.M108733200. [DOI] [PubMed] [Google Scholar]
- 49.Dumaz N, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 50.Simon R, et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer research. 2001;61:4514–4519. [PubMed] [Google Scholar]
- 51.Nowell PCHDA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;142 [Google Scholar]
- 52.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nature medicine. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciampi R, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. The Journal of clinical investigation. 2005;115:94–101. doi: 10.1172/JCI23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cin H, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta neuropathologica. 2011;121:763–774. doi: 10.1007/s00401-011-0817-z. [DOI] [PubMed] [Google Scholar]
- 55.Badiali M, et al. KIAA1549-BRAF fusions and IDH mutations can coexist in diffuse gliomas of adults. Brain pathology (Zurich, Switzerland) 2012;22:841–847. doi: 10.1111/j.1750-3639.2012.00603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones DT, et al. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nature genetics. 2013;45:927–932. doi: 10.1038/ng.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones DT, et al. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene. 2009;28:2119–2123. doi: 10.1038/onc.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ikawa S, et al. B-raf, a new member of the raf family, is activated by DNA rearrangement. Molecular and cellular biology. 1988;8:2651–2654. doi: 10.1128/mcb.8.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stanton VP, Jr, Nichols DW, Laudano AP, Cooper GM. Definition of the human raf amino-terminal regulatory region by deletion mutagenesis. Molecular and cellular biology. 1989;9:639–647. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kerkhoff E, Rapp UR. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Molecular and cellular biology. 1997;17:2576–2586. doi: 10.1128/mcb.17.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botton T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment cell & melanoma research. 2013;26:845–851. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hutchinson KE, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6696–6702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones DT, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfister S, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. The Journal of clinical investigation. 2008;118:1739–1749. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heidecker G, et al. Mutational activation of c-raf-1 and definition of the minimal transforming sequence. Molecular and cellular biology. 1990;10:2503–2512. doi: 10.1128/mcb.10.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rushworth LK, Hindley AD, O'Neill E, Kolch W. Regulation and role of Raf-1/BRaf heterodimerization. Molecular and cellular biology. 2006;26:2262–2272. doi: 10.1128/MCB.26.6.2262-2272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 68.Sievert AJ, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyons JF, Wilhelm S, Hibner B, Bollag G. Discovery of a novel Raf kinase inhibitor. Endocrine-related cancer. 2001;8:219–225. doi: 10.1677/erc.0.0080219. [DOI] [PubMed] [Google Scholar]
- 70.Wilhelm SM, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 71.Abou-Alfa GK, et al. Phase II study of sorafenib in patients with advanced hepatocellular carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4293–4300. doi: 10.1200/JCO.2005.01.3441. [DOI] [PubMed] [Google Scholar]
- 72.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joseph EW, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bollag G, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 76.Hatzivassiliou G, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 77.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doma E, et al. Skin tumorigenesis stimulated by Raf inhibitors relies upon Raf functions that are dependent and independent of ERK. Cancer Res. 2013;73:6926–6937. doi: 10.1158/0008-5472.CAN-13-0748. [DOI] [PubMed] [Google Scholar]
- 79.Long GV, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 80.Sosman JA, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McArthur GA, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. The lancet oncology. 2014;15:323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ascierto PA, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3205–3211. doi: 10.1200/JCO.2013.49.8691. [DOI] [PubMed] [Google Scholar]
- 84.Hauschild A, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 85.Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. The New England journal of medicine. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 86.Dietrich S, et al. Continued response off treatment after BRAF inhibition in refractory hairy cell leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:e300–e303. doi: 10.1200/JCO.2012.45.9495. [DOI] [PubMed] [Google Scholar]
- 87.Anforth R, Fernandez-Penas P, Long GV. Cutaneous toxicities of RAF inhibitors. The lancet oncology. 2013;14:e11–e18. doi: 10.1016/S1470-2045(12)70413-8. [DOI] [PubMed] [Google Scholar]
- 88.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. The New England journal of medicine. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oberholzer PA, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lacouture ME, et al. Analysis of dermatologic events in vemurafenib-treated patients with melanoma. The oncologist. 2013;18:314–322. doi: 10.1634/theoncologist.2012-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holderfield M, et al. Vemurafenib cooperates with HPV to promote initiation of cutaneous tumors. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-13-1065-T. [DOI] [PubMed] [Google Scholar]
- 92.Gibney GT, Messina JL, Fedorenko IV, Sondak VK, Smalley KS. Paradoxical oncogenesis--the long-term effects of BRAF inhibition in melanoma. Nature reviews. Clinical oncology. 2013;10:390–399. doi: 10.1038/nrclinonc.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Abdel-Wahab O, et al. Efficacy of Intermittent Combined RAF and MEK Inhibition in a Patient with Concurrent BRAF- and NRAS-Mutant Malignancies. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-13-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Callahan MK, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. The New England journal of medicine. 2012;367:2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdel-Wahab O, et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF and NRAS mutant malignancies. Cancer discovery. 2014 doi: 10.1158/2159-8290.CD-13-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Andrews MC, et al. BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:e448–e451. doi: 10.1200/JCO.2013.50.4118. [DOI] [PubMed] [Google Scholar]
- 97.Lito P, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kopetz SDJ, Chan E, Hecht JR, O’Dwyer PJ, Lee RJ, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28 Abstract nr 3534. [Google Scholar]
- 99.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 101.Samuels ML, McMahon M. Inhibition of platelet-derived growth factor- and epidermal growth factor-mediated mitogenesis and signaling in 3T3 cells expressing delta Raf-1:ER, an estradiol-regulated form of Raf-1. Molecular and cellular biology. 1994;14:7855–7866. doi: 10.1128/mcb.14.12.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Montero-Conde C, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer discovery. 2013;3:520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trejo CL, Juan J, Vicent S, Sweet-Cordero A, McMahon M. MEK1/2 inhibition elicits regression of autochthonous lung tumors induced by KRASG12D or BRAFV600E. Cancer Res. 2012;72:3048–3059. doi: 10.1158/0008-5472.CAN-11-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Trejo CL, et al. Mutationally activated PIK3CA(H1047R) cooperates with BRAF(V600E) to promote lung cancer progression. Cancer Res. 2013;73:6448–6461. doi: 10.1158/0008-5472.CAN-13-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nature structural & molecular biology. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whittaker S, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Science translational medicine. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 107.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Emery CM, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clara Montagut SVS, Shioda Toshi, McDermott Ultan, Ulman Matthew, Ulkus Lindsey E, Dias-Santagata Dora, Stubbs Hannah, Lee Diana Y, Singh Anurag, Drew Lisa, Haber Daniel A, Settleman Jeffrey. Elevated CRAF as a Potential Mechanism of Acquired Resistance to BRAF Inhibition in Melanoma. Cancer Research. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Su F, et al. Resistance to selective BRAF inhibition can be mediated by modest upstream pathway activation. Cancer Res. 2012;72:969–978. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 112.Karreth FA, DeNicola GM, Winter SP, Tuveson DA. C-Raf inhibits MAPK activation and transformation by B-Raf(V600E) Molecular cell. 2009;36:477–486. doi: 10.1016/j.molcel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 113.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schimke RT, Alt FW, Kellems RE, Kaufman RJ, Bertino JR. Amplification of dihydrofolate reductase genes in methotrexate-resistant cultured mouse cells. Cold Spring Harbor symposia on quantitative biology. 1978;42(Pt 2):649–657. doi: 10.1101/sqb.1978.042.01.067. [DOI] [PubMed] [Google Scholar]
- 115.Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer discovery. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abel EV, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. The Journal of clinical investigation. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haq R, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4321–4326. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smith MP, et al. Effect of SMURF2 targeting on susceptibility to MEK inhibitors in melanoma. Journal of the National Cancer Institute. 2013;105:33–46. doi: 10.1093/jnci/djs471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wilson TR, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim KB, et al. Phase II study of the MEK1/MEK2 inhibitor Trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:482–489. doi: 10.1200/JCO.2012.43.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wagle N, et al. MAP Kinase Pathway Alterations in BRAF-Mutant Melanoma Patients with Acquired Resistance to Combined RAF/MEK Inhibition. Cancer discovery. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 129.Woods D, et al. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Molecular and cellular biology. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Das Thakur M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes & development. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 133.Blasco RB, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. C-Raf is required for the initiation of lung cancer by K-Ras(G12D) Cancer discovery. 2011;1:128–136. doi: 10.1158/2159-8290.CD-10-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ehrenreiter K, et al. Raf-1 addiction in Ras-induced skin carcinogenesis. Cancer cell. 2009;16:149–160. doi: 10.1016/j.ccr.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 136.Freeman AK, Ritt DA, Morrison DK. Effects of Raf dimerization and its inhibition on normal and disease-associated Raf signaling. Molecular cell. 2013;49:751–758. doi: 10.1016/j.molcel.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ascierto PA, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. The lancet oncology. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 138.Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nature genetics. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seshagiri S, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]