Summary
Little is known about genetic mechanisms that regulate the ratio of cortical excitatory and inhibitory neurons. We show that NPAS1 and NPAS3 transcription factors (TF) are expressed in progenitor domains of the mouse basal ganglia (subpallium, MGE and CGE). NPAS1−/− mutants had increased proliferation, ERK signaling and expression of Arx in the MGE and CGE. NPAS1−/− mutants also had increased neocortical inhibition (sIPSC and mIPSC), and generated an excess of somatostatin+ (SST) (MGE-derived) and vasoactive intestinal polypeptide+ (VIP) (CGE-derived) neocortical interneurons, but had a normal density of parvalbumin+ (PV) (MGE-derived) interneurons. In contrast, NPAS3−/− mutants showed decreased proliferation and ERK signaling in progenitors of the ganglionic eminences and had fewer SST+ and VIP+ interneurons. NPAS1 repressed activity of an Arx enhancer, and Arx over-expression resulted in increased proliferation of CGE progenitors. These results provide novel insights into genetic regulation of cortical interneuron numbers and cortical inhibitory tone.
Keywords: NPAS1, NPAS3, Cortical Interneuron Development, Excitation/Inhibition
Introduction
While rapid progress has been made in understanding the regulation of neural regional and cell fate specification, much less is known about genetic mechanisms underlying brain size and how the correct ratios of excitatory and inhibitory neurons within local circuits are generated. Understanding these mechanisms is essential for elucidating important facets of brain development that underlie neurodevelopmental disorders that may have an imbalance of cortical excitation/inhibition (E/I) (e.g. epilepsy, autism spectrum disorders, and schizophrenia) (Lewis et al., 2012; Marin, 2012; Rubenstein and Merzenich, 2003; Yizhar et al., 2011), and to understanding why many autistic children have macrocephaly that includes a large cerebral cortex (Courchesne et al., 2007; Hazlett et al., 2011). While unifying genetic mechanisms which control cortex size and E/I balance have not yet been elucidated, progress has been made in understanding each of these processes individually.
Genetic causes of human neonatal macrocephaly are present in individuals with mutations in genes regulating growth factor signaling, particularly in pathways that engage the function of AKT and PTEN (Poduri et al., 2012; Rivière et al., 2012; Striano and Zara, 2012; Zhou and Parada, 2012), and RAS and MAPK (Gripp et al., 2013). For instance, patients who are heterozygotes for loss of function PTEN mutations have macrocephaly, and are at increased risk for autism (Zhou and Parada, 2012).
Likewise, progress has been made in understanding the mechanisms that control the generation of cortical excitatory and inhibitory neurons (Gelman et al., 2012; Kwan et al., 2012; Marin, 2013), and the mechanisms that coordinate their relative activities to create the proper E/I balance (Le Magueresse and Monyer, 2013). In this regard, there is particular interest in the mechanisms that control the development of cortical inhibitory neurons (GABAergic interneurons). In rodents, most cortical inhibitory neurons are generated during embryogenesis in subpallial structures called the caudal and medial ganglionic eminences (CGE and MGE) (Rudy et al., 2011).
The specification, migration and differentiation of cortical interneurons is controlled by cascades of transcription factors. Some TFs regulate the development of either CGE or MGE derived neurons. For instance, Lhx6, Nkx2-1, Olig1 and Sox6 regulate for the specification and differentiation of MGE derived interneurons (Azim et al., 2009; Batista-Brito et al., 2009; Liodis et al., 2007; Silbereis et al., 2014; Sussel et al., 1999; Zhao et al., 2008). Other TF’s, such as the Arx and Dlx genes, control development of both CGE and MGE derived interneurons (Cobos et al., 2005; Colasante et al., 2008; Marsh et al., 2009).
Herein, we present evidence that the mouse NPAS basic helix-loop-helix (bHLH)-PAS TF genes (Erbel-Sieler et al., 2004; Pieper et al., 2005; Zhou et al., 1997) are novel regulators of cortex size and E/I balance. NPAS1 and NPAS3 are expressed in telencephalic progenitor domains of the cortex, and the CGE and MGE, and later in immature and mature cortical interneurons (Batista-Brito et al., 2008; Erbel-Seiler et al., 2004; Zhao et al., 2008). Vertebrate NPAS function in embryonic neural progenitors may be related to the function of Trachealess, the NPAS Drosophila homolog. Trachealess modulates fibroblast growth factor (FGF) signaling by transcriptional regulation of the FGF receptor (Ohshiro and Saigo, 1997). In the adult mouse hippocampus, NPAS3 regulates expression of FGFR1 to control proliferation of hippocampal granule neurons (Pieper et al., 2005). Here, we have found that NPAS1 negatively regulates proliferation and MAPK signaling in CGE and MGE progenitors, not by regulating FGF receptor expression, but through an unexpected mechanism, repression of Arx expression. As a result, NPAS1−/− mutants generate excessive cortical interneurons prenatally, which persisted into adulthood. NPAS1−/− mutants also had increased neocortical inhibition (sIPSC and mIPSC), and generated an excess of SST+ (MGE-derived) and VIP+ (CGE-derived) neocortical interneurons, but had normal numbers of PV+ (MGE-derived) interneurons. In contrast, NPAS3−/− mutants had a complementary phenotype, with reduced proliferation and MAPK signaling in progenitors of the ganglionic eminences, and the postnatal cortex had fewer SST+ and VIP+ interneurons.
We propose that our analysis of NPAS1 and NPAS3 functions in mice provide novel mechanistic insights into human neuropsychiatric disorders, as NPAS3 dysfunction is implicated in schizophrenia (Kamnasaran et al., 2003; Macintyre et al., 2010). Furthermore, we have identified sporadic non-synonymous mutations in NPAS1 and NPAS3 in autistic individuals.
Results
NPAS1 and NPAS3 Expression During Interneuron Development
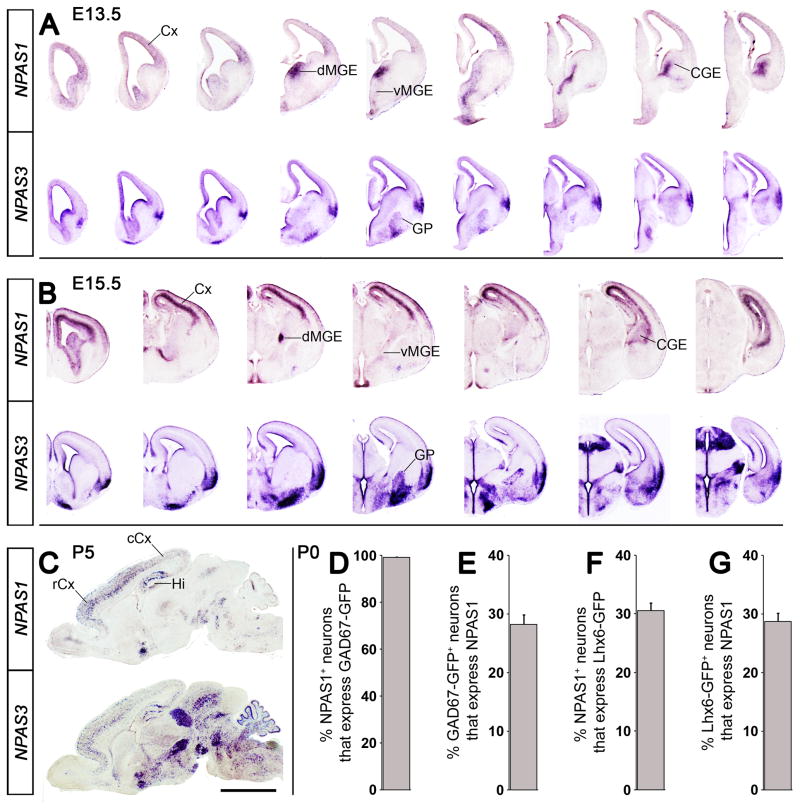
The subpallium generates neocortical interneurons (Flandin et al., 2011; Marin, 2012; Rudy et al., 2011). We examined NPAS1 and NPAS3 RNA expression by in situ hybridization (ISH) at E13.5, E15.5 and P5 (Figure 1), and assessed NPAS1 and NPAS3 expression by immunofluorescence at P0, P5, P15, and P30 (Figure 1 and Figures S1 and S2 and Tables S1 and S2). At E13.5, both had pallial and subpallial ventricular zone (VZ) expression. NPAS1 showed notable expression in the VZ and subventricular zone (SVZ) of the dorsal and ventral MGE, and CGE. By E15.5, NPAS3 was expressed in the MGE mantle zone; NPAS1 expression was prominent in the pallial and subpallial SVZ.
Figure 1. Forebrain expression of mouse NPAS1 and NPAS3 during embryonic and neonatal stages.
(A and B) ISH on rostro-caudal series of coronal hemisections at E13.5 (A) and E15.5 (B). Note strong NPAS1 expression in dMGE and CGE. (C) ISH on neonatal sagittal sections at P5. Note the rostral bias of cortical expression. (D to G) Quantification of percentage of NPAS1+ (immuno-stained) cortical interneurons (GAD67-GFP+ and Lhx6-GFP+) at P0. n = 3 animals for (D) to (G). Abbreviations: CGE: caudal ganglionic eminence, Cx: cortex (rCX: rostral, cCx: caudal), GP: globus pallidus, Hi: hippocampus, MGE: medial ganglionic eminence (dMGE: dorsal, vMGE: ventral). Scale bar, (A) 1.21 mm; (B) 1.74 mm; (C) 3 mm. See also Figure S1 and S2.
Previous studies have described co-expression of NPAS1 or NPAS3 with cortical interneurons using GABA, GAD-67, or calretinin antibodies in the adult mouse brain (Erbel-Sieler et al., 2004). We have extended co-expression analysis of NPAS1 or NPAS3 with various interneuron markers during cortical interneuron development and in the adult (Figure 1 and Figures S1 and S2 and Tables S1 and S2). At P0, NPAS1 was expressed in neocortical interneurons; ~100% of NPAS1+ cells express GAD67-GFP; ~30% had MGE-like properties (Lhx6-GFP+) (Figures 1D and 1F and Figures S1A and S1B). By P5, NPAS1 and NPAS3 were expressed in rostro-caudal gradients in neocortical interneurons; we are unaware of other TFs with this property. Virtually all neocortical NPAS1+ cells (99 ± 0.29%) and the majority of NPAS3+ cells (67 ± 2.94%) expressed GAD67-GFP at P5 (Figures S1C and S1D). By P15, NPAS1 and NPAS3 were expressed by a majority of reelin+ (NPAS1: 68 ± 2.78%; NPAS3: 79 ± 4.79%) and SST+ (NPAS1: 65 ± 1.95%; NPAS3: 75 ± 0.44%) interneurons. Both NPAS1 and NPAS3 were expressed in a small proportion of PV+ cells (NPAS1: 6 ± 0.85%) (NPAS3: 13 ± 1.29%) (Figures S1E, S1F, S1H, S1I, S1J, and S1L). At P30, NPAS1+ cells co-expressed reelin, SST, calretinin (CR), or neuropeptide Y (NPY), but rarely co-expressed PV (reelin: 42 ± 1.94%; SST: 36 ± 3.72%; CR: 28 ± 2.89%; NPY: 12 ± 0.70%; PV: 5 ± 1.24%). On the other hand, NPAS3+ was expressed in a large fraction of all interneuron subtypes assayed, including PV (reelin: 74 ± 2.31%; SST: 75 ± 3.52%; CR: 51 ± 1.23%; PV: 43 ± 1.40%) (Figures S2B–S2E and S2G–S2J and data not shown).
Increased Numbers of Neocortical Interneurons in NPAS1−/− Mutants
We studied the effect of an NPAS1 null allele (NPAS1−/−) (Erbel-Sieler et al., 2004) on neocortical interneuron development using a GAD67-GFP allele to label all of the interneurons (Tamamaki et al., 2003). By E15.5 there was an increased density of GFP+ interneurons through the intermediate zone and then throughout the cortical wall at E17.5 and P0 (26–41%; E15.5: 26 ± 4.67%, p = 0.00099; E17.5: 35 ± 2.08%, p = 9.08E-8; P0: 41 ± 4.46%, p = 2.48E-6) (Figures 2A–2C and 2E–2G). Even though there was ~2-fold increased interneuron cell death at P7 (activated caspase-3, Figures S3A and S3B), P30 mice maintained ~15% (15 ± 5.43%, p = 0.044) more interneurons (Figures 2D and 2H). Surprisingly, while SST+, VIP+, NPY+ and reelin+ interneuron subtypes were increased (28–44%; SST: 32 ± 5.07%, p = 0.001; VIP: 44 ± 7.99%, p = 0.014; NPY: 34 ± 7.42%, p = 0.0019; reelin: 28 ± 3.01%, p = 2.7E-5), PV+ interneuron density was normal (Figures 3A–3E, 3A′–3E′, and 3F–3J). MRI quantification showed that P30 cortical volume was increased 10% (10 ± 1.40%, p = 0.0055) (Figures S3F and S3G and Table S3). Nissl section analysis supported the MRI findings, and showed 11% increased (11 ± 3.13%, p = 0.03) rostral neocortical width (Figures S2A and S2B). Assessment of NeuN+ (neuronal marker) cells in the P21 somatosensory cortex suggests that the enhancement in cortical volume may be the result of increased neuron numbers. In NPAS1−/− mutants, the density of NeuN+ cells was increased by 11% (cortex: 11 ± 2.21%, p = 0.0011; layer I: 22 ± 7.34%, p = 0.030; layer IV: 7 ± 2.09%, p = 0.025; layer V: 16 ± 3.32%, p = 0.0024; layer VI: 13 ± 2.66%, p = 0.0052) (Figures S4A and S4B).
Figure 2. NPAS1−/− mice have increased numbers and density of GAD67-GFP+ cortical interneurons.
(A to D) Increase in GAD67-GFP+ cortical interneurons (somatosensory Cx) beginning by E15.5 shown on coronal cortical sections at E15.5 (A), E17.5 (B), P0 (C) and P30 (D) in NPAS1−/− mice. (E to H) Quantification of GAD67-GFP+ neurons/104 μm2 at E15.5 (E), E17.5 (F), P0 (G), and GAD67-GFP+ neurons/1 mm2 at P30 (H). n = 3 animals per genotype for (E) to (H). Abbreviations: CP: cortical plate, Cx: cortex, IZ: intermediate zone, MZ: marginal zone, SVZ: subventricular zone. *P<0.05. ***P< 0.001. Scale bar, (A) to (C) 200 μm; (D) 400 μm. See also Figure S3.
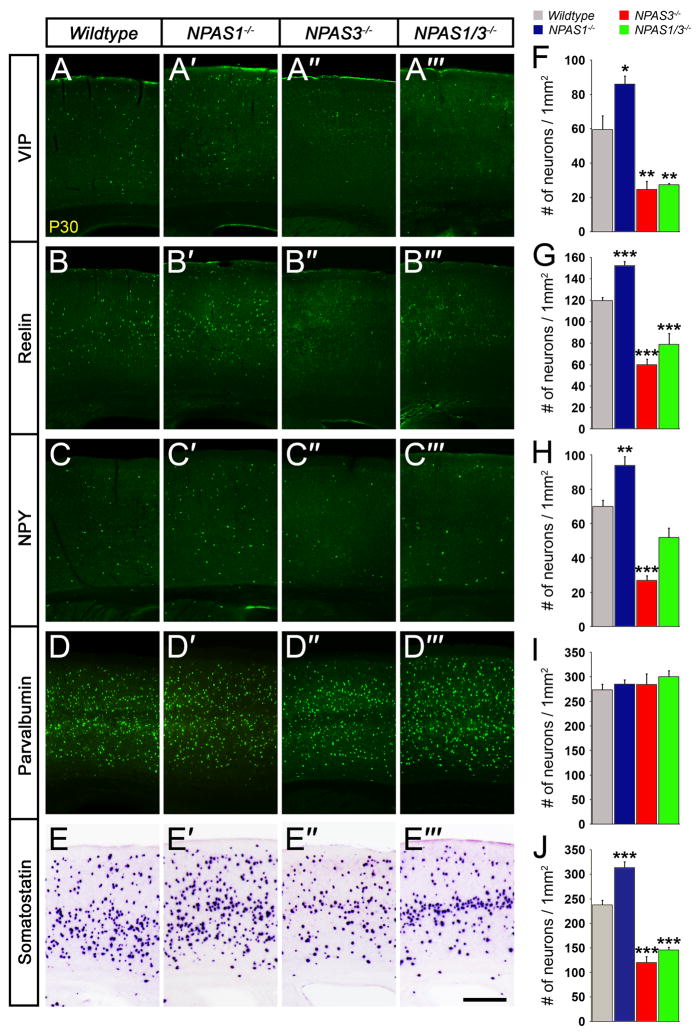
Figure 3. NPAS1−/− mice have increased, whereas NPAS3−/− and NPAS1/3−/− mice have decreased cortical interneurons.
(A to E‴) Interneurons expressing VIP, reelin, NPY and somatostatin are increased in NPAS1−/− mice, but decreased in NPAS3−/− and NPAS1/3−/− mice. All mutants have a normal density of parvalbumin+ interneurons. Immunofluorescence staining of coronal neocortical sections (somatosensory Cx) at P30. (F to J) Quantification of VIP+ (F), reelin+ (G), NPY+ (H), parvalbumin+ (I), and somatostatin+ (J) neurons/1 mm2. n = 3 animals per genotype for (F) to (J). *P<0.05. ***P< 0.001. Scale bar, (A) to (E‴) 400 μm.
Increased Synaptic Inhibition onto Neocortical Pyramidal Neurons in NPAS1−/−
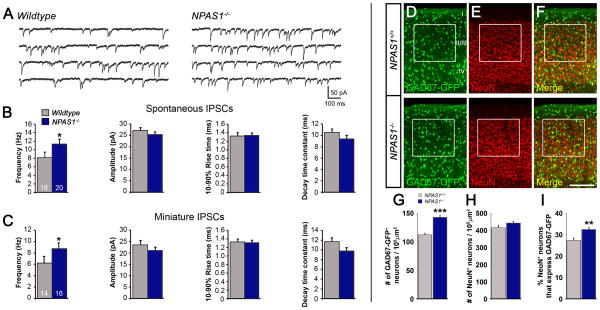
To test whether the increase in cortical interneurons altered the physiology of the NPAS1−/− cortex we performed patch-clamp recordings of layer II/III somatosensory pyramidal neurons in slices from P21-30 NPAS1−/− mice and wildtype (WT) littermates (Figures 4A–4C). Compared to WT littermates, pyramidal neurons from NPAS1−/− mice showed an increase in the frequency of spontaneous IPSCs (sIPSCs) (Wildtype: 8.17 ± 1.29 Hz; NPAS1−/−: 11.18 ± 1.26 Hz; p = 0.042). However, the amplitudes and kinetics of inhibitory events were not significantly changed (Figures 4A and 4B). To further characterize the increase in GABA-mediated synaptic inhibition, tetrodotoxin (TTX) was added to the ACSF to isolate miniature IPSCs (mIPSCs). Similar to spontaneous events, an increase in mIPSC frequencies was observed in NPAS1−/− mice (Wildtype: 6.2 ± 1.24 Hz; NPAS1−/−: 8.74 ± 1.02 Hz; p = 0.036); amplitudes and kinetics of mIPSCs were not significantly changed (Figure 4C).
Figure 4. Synaptic inhibition to neocortical pyramidal cells is increased in NPAS1−/− mice.
(A) Representative traces of sIPSCs recorded from layer II/III pyramidal neurons in somatosensory neocortex of wildtype and NPAS1−/− mice. (B and C) Mean sIPSC (B) and mIPSC (C) frequencies were higher in pyramidal neurons recorded from NPAS1−/− mice than in controls. Numbers of recorded cells are shown in each bar. No significant changes in sIPSC or mIPSC amplitude, 10–90% rise time, or decay time constant were detected. For (B) and (C), n = 7 wildtype and 9 NPAS1−/− animals. (D to F) Increase in GAD67-GFP+ interneurons (D) and percenta bge of neurons (NeuN+) that are inhibitory (GAD67-GFP+) (F) within layer II/III of NPAS1−/− somatosensory cortex (white boxes) shown on coronal sections at P21. The density of NeuN+ cells within layer II/III is not significantly increased in NPAS1−/− mutants (E). (G to I) Quantification of GAD67-GFP+ (G) and NeuN+ (H) neurons/105 μm2 and percentage of NeuN+ cells that express GAD67-GFP within that area (I). n = 3 animals per genotype for (G) to (I). *P<0.05. **P<0.01. ***P< 0.001. Scale bar, (D) to (F) 200 μm. See also Figure S4.
Next, we evaluated the ratio of interneuron numbers to total neuron numbers by quantifying the percentage of NeuN+ cells (total neurons) that express GAD67-GFP in each layer of P21 somatosensory cortex (Figures S4B and S4C). At P21, all NeuN+ cells were GAD67-GFP+ in layer I somatosensory cortex. NPAS1−/− mutant mice displayed a 19% increase (19 ± 5.94%, p = 0.025) in GAD67-GFP+ interneurons within layer I (Figure S4C). In layer II/III, the percentage of NeuN+ cells that express GAD67-GFP was increased by 18% in NPAS1−/− mutants (NPAS1+/+: 27 ± 1.13%; NPAS1−/−: 32 ± 0.96%, p = 0.0024) (Figures 4D–4I). We also estimated the excitatory to inhibitory cell ratio (E/I) in each cortical layer at P21. The E/I ratio, determined as the ratio of NeuN+ and GAD67− cells to GAD67+ cells, was decreased by 23% in layer II/III of the NPAS1−/− somatosensory cortex (NPAS1+/+: 2.74 ± 0.17; NPAS1−/−: 2.12 ± 0.091, p = 0.0040) (Figure S4D). No significant change in the E/I cell ratio was found in deep cortical layers (IV, V, or VI) (Figure S4D). These findings were supported by stereological quantification of NeuN+ and GAD67+ cells within the cerebral cortex. The NPAS1−/− mutant cerebral cortex exhibited a 35% increase (35 ± 5.96%, p = 0.0053) in NeuN+ cells and a 57% increase (57 ± 10.49%, p = 0.0012) in GAD67+ cells at 3 months of age (Figures S4E–S4G). Cortical pyramidal cell density was unchanged in P21 NPAS1−/− mutants as indicated by Cux1+, Ctip2+, and Tbr1+ immunofluorescence staining which labels layer II/III, layer V, and layer VI, respectively (Figures S4H–S4K). Thus, the enhanced level of synaptic inhibition onto layer II/III neocortical pyramidal neurons in the NPAS1−/− mutant was associated with an increased fraction of inhibitory neurons.
NPAS1−/− and NPAS3−/− Had Opposite Effects on Cortical Interneuron Numbers
NPAS3−/− mice display reduced proliferation and size of the adult hippocampal dentate gyrus (Pieper et al., 2005). Given that NPAS1−/− mutants have increased cortical volume (Figures S3F and S3G) and interneuron numbers (Figures 3A–3C, 3E, 3A′–3C′, 3E′, 3F–3H, and 3J), we hypothesized that NPAS1 and NPAS3 may have opposing functions on interneuron development. Therefore, we compared interneuron numbers in NPAS1−/−, NPAS3−/− and NPAS1/3−/− P30 neocortices. While NPAS1−/− mutants had increased density of cortical interneurons (Figures 3A–3C, 3E, 3A′–3C′, 3E′, 3F–3H, and 3J), NPAS3−/− mutants had reduced density of SST+, VIP+, NPY+ and reelin+ interneurons (Figures 3A–3C, 3E, 3A″–C″, 3E″, 3F–3H, and 3J). Like NPAS1−/− mutants, there was not a change in the density of PV+ interneurons (Figures 3D––3D″ and 3I).
MRI quantification revealed that P30 cortical volume was increased in NPAS1−/− mutant mice, as opposed to a 21% decrease (21 ± 1.47%, p = 0.0039) in NPAS3−/− mutants. Also, we found differential changes in P30 NPAS1−/− and NPAS3−/− mutant basal ganglia volumes by MRI. NPAS1−/− mutant mice displayed a 12% increase (12% ± 1.74%, p = 0.042) and NPAS3−/− mutants a 25% decrease (25% ± 2.24%, p = 0.0087) in basal ganglia volume (Figures S3F–S3H and Table S3).
NPAS1/3−/− and NPAS3−/− mutants had similar interneuron phenotypes (Figures 3A–E, 3A″–3E″, 3A‴–3E‴, and 3F–3J). In addition, we observed probable NPAS1−/−, NPAS3−/−, and NPAS1/3−/− mutant basal ganglia phenotypes (N=1), which include subtle cholinergic defects (NPAS1 and NPAS3 are expressed in the pallidum) (Figure 1 and data not shown) (Flandin et al., 2010; Nobrega-Pereira et al., 2010). The distribution of choline acetyltransferase (ChAT)+ cells around the globus pallidus (GP) was abnormal in NPAS1/3−/− and NPAS3−/− mutants. We detected increased ChAT+ cells inside the GP and reduced numbers in the ventral pallidum. While parvalbumin (PV)+ expression in the GP was not grossly abnormal in the mutants, the GP may be enlarged in the NPAS1−/− mutant, and smaller in the NPAS3−/− mutant. PV+ interneuron numbers in the striatum and basolateral nucleus of the amygdala appeared grossly normal in all three mutant genotypes (data not shown).
NPAS1−/− and NPAS3−/− Ganglionic Eminences Had Opposite Proliferation and ERK-Signaling Phenotypes
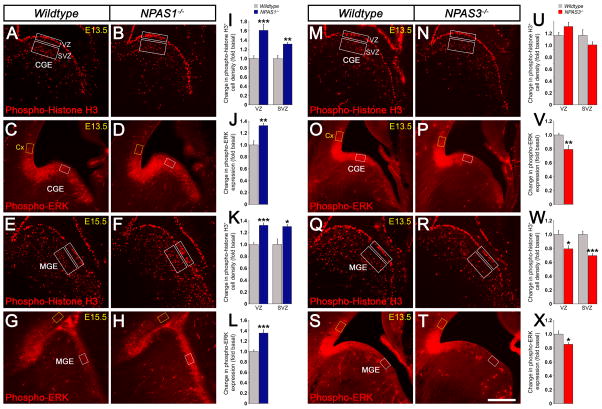
Given that NPAS1 and NPAS3 are expressed in MGE and CGE progenitors (Figures 1A and 1B), we explored whether changes in interneuron numbers in NPAS1−/− and NPAS3−/− mutants were due to changes in proliferation using a marker of M-phase cells (phospho-histone H3, PH3). We found that NPAS1−/− mutants had a 1.6-fold increase (1.61 ± 0.13, p = 0.0007) in PH3+ cells in the VZ, and a 1.3-fold increase (1.31 ± 0.039, p = 0.0033) in the SVZ of the CGE at E13.5; at E15.5 NPAS1−/− mutants had a 1.3-fold increase (VZ: 1.32 ± 0.042, p = 1.09E-5; SVZ: 1.30 ± 0.035, p = 0.022) in PH3+ cells in the VZ and SVZ of the MGE (Figures 5A, 5B, 5E, 5F, 5I, and 5K). In contrast, NPAS3−/− progenitors displayed reduced proliferation in the E13.5 MGE, including a 1.3-fold decrease (1.26 ± 0.043, p = 0.010) in PH3+ cells in the VZ and a 1.4-fold decrease (1.44 ± 0.031, p = 6.54E-5) in the SVZ (Figures 5Q, 5R, and 5W).
Figure 5. NPAS1−/− progenitors have increased proliferation and MAP kinase activity, while NPAS3−/− progenitors exhibit decreased proliferation and MAP kinase activity.
(A to H) Immunofluorescence assays of coronal sections demonstrate increased proliferation (phospho-histone H3+ M-phase cells) and MAP kinase activity (phospho-ERK) in the E13.5 CGE (A to D), and E15.5 MGE (E to H) of NPAS1−/− progenitors. (I to L) Quantification of phospho-histone H3+ (I and K) cells and phospho-ERK (J and L) levels in NPAS1−/− mutants. n = 3 animals per genotype for (I) to (L). (M to T) NPAS3−/− progenitors display reduced proliferation and MAP kinase activity in the E13.5 MGE (Q to T). While MAP kinase activity was decreased in the E13.5 CGE (O to P), proliferation of progenitors was unaltered in the E13.5 CGE (M to N) of NPAS3−/− mutant mice. (U to X) Quantification of phospho-histone H3+ (U and W) cells and phospho-ERK (V and X) levels in NPAS3−/− mutants. Proliferation was measured in the VZ and SVZ (white boxes). Phospho-ERK levels in the CGE/MGE (white box) were normalized by comparison with cortical levels (yellow box). n = 3 animals per genotype for (U) to (X). Abbreviations: CGE: caudal ganglionic eminence, Cx: cortex, MGE: medial ganglionic eminence, SVZ: subventricular zone, VZ: ventricular zone. *P<0.05. **P<0.01. ***P< 0.001. Scale bar, (A) to (H) and (M) to (T) 250 μm. See also Figure S5.
BrdU labeling analysis of E13.5 MGE and CGE progenitors, utilized to detect cells in S-phase, supported the PH3 results. 1 hour BrdU pulse-labeling showed increased proliferation in the SVZ of the NPAS1−/− mutant MGE (total number: 31 ± 1.26%, p = 0.0007; density: 48 ± 2.15%, p = 0.0004) and CGE (total number: 19 ± 3.31%, p = 0.039; density: 34 ± 5.24%, p = 0.022), while NPAS3−/− mutant mice exhibited decreased proliferation in the MGE SVZ (total number: 26 ± 2.05%, p = 0.0018; density: 21 ± 3.08%, p = 0.023) (Figures S5A–S5E). In addition, examination of proliferation in the hippocampal dentate gyrus following intraperitoneal injection of BrdU once daily for 12 days revealed a 31% increase (31 ± 7.89%, p = 0.003) in BrdU+ cells in 5–10 week old NPAS1−/− mice (Figures S5F and S5G).
The NPAS Drosophila homolog Trachealess regulates FGF signaling (Ohshiro and Saigo, 1997); thus we examined activation of the MAP kinase pathway by measuring the level of phospho-ERK immunofluorescence. NPAS1 mutants had a 1.3-fold increase (E13.5 CGE: 1.32 ± 0.043, p = 0.0016; E15.5 MGE: 1.35 ± 0.063, p = 0.00017) in phospho-ERK expression in the E13.5 CGE and E15.5 MGE, whereas NPAS3 mutants exhibited a 1.3-fold decrease (1.26 ± 0.060, p = 0.0079) in phospho-ERK expression in the E13.5 CGE and a 1.2-fold decrease (1.17 ± 0.024, p = 0.023) in the E13.5 MGE (Figures 5C, 5D, 5G, 5H, 5J, 5L, 5O, 5P, 5S, 5T, 5V, and 5X).
NPAS1−/− Ganglionic Eminences Displayed Increased Arx Expression Which Mediated an Increase in Subpallial MAP Kinase Activity and Proliferation
We focused on identifying molecular mechanisms underlying the increased proliferation and phospho-ERK expression in the MGE and CGE of NPAS1 mutants by ISH screening of the MGE and CGE for expression of genes that are implicated in these processes (Arx, COUP-TFI, Cyclin D1, Cyclin D2, Dlx1, ErbB4, FGFR1, FGFR3, Lhx2, Lhx6, NPAS3, Olig1, Sp8, and Sprouty2) (Figure 6 and Figure S5 and data not shown). While we did not find robust changes for most of these, including FGF-signaling components, there were some interesting changes in Arx and Sp8 expression. In accordance with the increase in CGE proliferation, there was a subtle increase in Sp8+ (LGE/CGE marker) migrating interneurons at E13.5 (Figure 6B) (Ma et al., 2012). We also observed a clear increase of Arx TF RNA at E13.5 and E15.5 in regions of the MGE and CGE where NPAS1 is normally expressed (Figures 6A and 6C). Arx function is necessary for proper interneuron migration and maturation (Colombo et al., 2007; Kitamura et al., 2002; Marsh et al., 2009), as well as neocortical neuron progenitor proliferation (Friocourt et al., 2008; Kitamura et al., 2002).
Figure 6. NPAS1−/− GEs have increased Arx expression, whereas NPAS1−/− cortices reveal increased Sp8 expression.
(A) ISH assay on coronal hemisections arrayed in a rostro-caudal series reveal that NPAS1−/− progenitors have increased Arx expression in the VZ/SVZ regions of the E13.5 MGE (arrows) and CGE(arrowheads) [compare white arrow and arrowhead in bottom tier (NPAS1−/−) to middle tier (wildtype)] where NPAS1 is expressed (black arrow and arrowhead in top tier). (B) At E13.5, Sp8 expression appears increased in a pattern consistent with tangentially migrating cortical interneurons (arrows). Data presented in coronal sections at three rostro-caudal planes (left to right). (C) ISH assay on coronal hemisections displayed in a rostro-caudal series show that NPAS1−/− progenitors have increased Arx expression in the VZ/SVZ regions of the E15.5 MGE (arrows) and CGE(arrowheads) [compare white arrow and arrowhead in bottom tier (NPAS1−/−) to middle tier (wildtype)] where NPAS1 is expressed (black arrow and arrowhead in top tier). Abbreviations: CGE: caudal ganglionic eminence, MGE: medial ganglionic eminence. Scale bar, (A) 1 mm; (B) 723 μm; (C) 1.25 mm. See also Figure S6.
To test whether NPAS1 and its co-factor, ARNT, can directly repress Arx expression through a known Arx subpallial enhancer element (Colasante et al., 2008; Visel et al., 2013), we performed luciferase assays in CGE primary cultures (Flandin et al., 2011). We found that this enhancer had two putative NPAS1/ARNT sites, designated A and B (Figure S7D) (Hogenesch et al., 1997). Consistent with the aforementioned phenotype, NPAS1/ARNT induced a ~2-fold repression of the Arx enhancer (0.58 ± 0.035, p = 6.69E-5). Mutation of the Arx enhancer at site A, but not site B, resulted in a rescue of the repression induced by NPAS1/ARNT (p = 0.034) (Figures 7C and 7D).
Figure 7. Increased Arx expression in the CGE mediates increased subpallial proliferation.
(A and B) In utero lentiviral transduction of Arx into the E13.5 CGE results in increased cell proliferation (PH3). Arrowheads in (A) indicate GFP+/PH3+ cells. n = 3 animals per experimental condition for (B). (C) Arx locus on mouse X chr, showing evolutionarily conserved domains, one of which is a subpallial enhancer (UAS3) (Colasante et al., 2008; Visel et al., 2013) that has two predicted NPAS1/ARNT sites (Figure S6). (D) Transcription assay of E12.5 CGE primary culture shows that NPAS1/ARNT expression represses activity of the Arx subpallial enhancer. Mutation of the Arx enhancer at site A, but not site B, resulted in a rescue of the repression induced by NPAS1/ARNT. n = 3 independent experiments, each performed in triplicate for (D). Abbreviations: CGE: caudal ganglionic eminence. *P<0.05. **P<0.01. Scale bar, (A) 70 μm. See also Figure S7.
Next, to investigate whether the increase in Arx expression (Figures 6A and 6C) could alter subpallial development we used ultrasound to guide injection into the E13.5 CGE of a lentivirus encoding either Arx and GFP, or GFP alone. At E15.5, we compared the number of PH3+/GFP+ cells and observed that the Arx-encoding virus increased PH3+/GFP+ cells by ~2-fold (2.18 ± 0.039, p = 0.042) (Figures 7A and 7B and Figure S7A). We also assessed the level of phospho-ERK expression in GFP+ regions of the CGE ventricular zone and found that Arx overexpression increased phospho-ERK expression by 1.4-fold (1.39 ± 0.12, p = 0.046) (Figures S7B and S7C). These findings provide evidence that NPAS1 repression of Arx can regulate CGE proliferation and MAP kinase activity.
Non-Synonymous Mutations in NPAS1 and NPAS3 Are Found in Humans with ASD
NPAS3 mutations are observed in patients with schizophrenia (Kamnasaran et al., 2003; Macintyre et al., 2010). Given that autism spectrum disorders (ASD) and schizophrenia share some genetic risk factors (Murdoch and State, 2013) and, like NPAS1 mutant mice, enlarged brain size is frequent in ASD children (Butler et al., 2005; Hazlett et al., 2011), we screened 947 ASD probands for mutations in NPAS1 and NPAS3 exons, introns, and 5′ and 3′ untranslated regions, and found 10 non-synonymous variants in NPAS1 and seven in NPAS3 (See the Supplemental Text). We assayed 13 of those in 190 control subjects (unscreened for autism) and found that five of seven in NPAS1 and all six in NPAS3 were absent in the controls. Although the data lacks statistical power, and does not unambiguously implicate these variants in genetic susceptibility to ASD, these results support the possibility that NPAS1 and NPAS3 mutations are related to ASD risk.
Discussion
NPAS1−/− mice had increased numbers of neocortical interneurons, whereas NPAS3−/− mice have fewer interneurons (Figures 3A–3C, 3E, 3A′–3C′, 3E′, 3A″–3C″, 3E″, 3F–3H, and 3J). These changes in interneuron numbers were associated with altered ERK signaling and cell proliferation within the MGE and CGE of NPAS1−/− and NPAS3−/− mutants (Figure 5). In addition, the increased numbers of neocortical interneurons in NPAS1−/− mice resulted in enhanced synaptic inhibition onto neocortical pyramidal neurons in layer II/III of the somatosensory cortex (Figures 4A–4C). We focused on how NPAS1 regulates this process in the MGE and CGE, and found that NPAS1−/− mutants had increased Arx expression in these embryonic progenitor zones (Figures 6A and 6C). Using in utero transduction of Arx into the CGE, we showed that increased Arx expression was sufficient to increase subpallial proliferation and MAP kinase activity (Figures 7A and 7B and Figures S7B and S7C). We also demonstrated that NPAS1 could directly repress a bone fide Arx subpallial enhancer (Figures 7C and 7D and Figure S7D) (Colasante et al., 2008).
Our results provide novel insights into mechanisms that control the number of specific subtypes of cortical inhibitory neurons, showing that homologous TF-encoding genes NPAS1 and NPAS3 have opposing functions in regulating the numbers of SST+, and VIP+ interneurons (Figures 3A––3A″, 3E–3E″, 3F, and 3J). Thus, we have provided evidence for a cell autonomous mechanism that controls interneuron numbers, a process proposed to exist based on results from the MGE-transplantation assay (Southwell et al., 2012).
NPAS1 and NPAS3 Regulate the Proliferation of Ganglionic Eminence Progenitors
Proliferation and ERK signaling in progenitor domains of the MGE and CGE were increased and decreased in NPAS1−/− and NPAS3−/− mice, respectively (Figure 5). Vertebrate NPAS function in embryonic neural precursors may be related to the function of Trachealess, the NPAS Drosophila homolog. Trachealess promotes FGF-signaling by increasing expression of the FGF receptor (Ohshiro and Saigo, 1997). This program is evolutionary conserved based on the observation that NPAS3−/− mice have reduced FGFR1 expression in the adult hippocampus (Pieper et al., 2005). However, NPAS3−/− mutant mice did not show this phenotype in the prenatal telencephalon (data not shown). Also, NPAS1−/− mutants did not exhibit decreased FGFR1 expression in the adult hippocampus (Pieper et al., 2005), or in the prenatal telencephalon (Figure S6C). Thus, there is no evidence that changes in phospho-ERK levels, in the subpallium of NPAS1−/− and NPAS3−/− mutants (Figures 5C, 5D, 5G, 5H, 5J, 5L, 5O, 5P, 5S, 5T, 5V, and 5X), is related to altered FGF-signaling.
Our study reveals new insights into NPAS1 regulation of cell proliferation. Here, we have found that the increase in proliferation and MAPK signaling in NPAS1−/− mutant CGE and MGE progenitors was associated with increased Arx expression. Previous studies of Arx mutant mice show that lack of this TF resulted in reduced cortical proliferation (Friocourt et al., 2008; Kitamura et al., 2002). We demonstrated that increased Arx expression was sufficient to enhance subpallial proliferation and MAP kinase activity, using in utero transduction of Arx into the CGE (Figures 7A and 7B and Figures S7B and S7C). Furthermore, we showed that NPAS1 could directly inhibit a bone fide Arx subpallial enhancer (Figures 7C and 7D and Figure S7D) (Colasante et al., 2008).
Given that NPAS1 and NPAS3 have opposite effects on proliferation, an imbalance of either NPAS1 or NPAS3 expression and/or function could potentially be compensated by changes in the expression of the other gene. However, we did not observe changes of NPAS3 expression in the NPAS1−/− E13.5 and P0 forebrain (data not shown).
NPAS1 and NPAS3 Modified the Production of Specific Subtypes of Cortical Interneurons
The increase in Arx and phospho-ERK expression and the increase in proliferation in the MGE and CGE of NPAS1−/− mutants were associated with an increase in subpallially-derived cortical interneurons (Figure 2). As a result, NPAS1 mutants had excessive SST+ and VIP+ interneurons, products of the MGE and CGE, respectively (Figures 3A, 3A′, 3E, 3E′, 3F, and 3J) (Rudy et al., 2011). In contrast, decreased phospho-ERK expression and cell proliferation in the MGE and CGE of NPAS3−/− mutant mice resulted in reduced numbers of SST+ and VIP+ interneurons (Figures 3A, 3A″, 3E, 3E″, 3F, and 3J).
Both NPAS1 and NPAS3 mutants had a normal density of PV+ interneurons (Figures 3D––3D″, and 3I). PV and SST are MGE-derived (Rudy et al., 2011). NPAS1, along with NPAS3, COUP-TFI, Dlx1 and SatB1 (Cobos et al., 2005; Denaxa et al., 2012; Lodato et al., 2011) define a set of TFs that differentially regulate the numbers of interneurons that express PV and SST (Figures 3D––3D″, 3E–3E″, 3I, and 3J). At least one PV+ subtype, the Chandelier neuron, is generated late in gestation (Inan et al., 2012; Taniguchi et al., 2013); perhaps NPAS1 and NPAS3 regulation of MGE proliferation is time-dependent, and thus preferentially affects early-born SST+ interneurons. Based on the embryonic phenotypes of NPAS1−/− and NPAS3−/− mutants, we propose that NPAS1 and NPAS3 control the SST/PV ratio via their functions in MGE progenitor cells. Future studies are needed to examine the roles of NPAS1 and NPAS3 in regulating interneurons via their expression in maturing and mature interneurons (Figure 1C).
The significance of transcriptional repression in the regulation of cortical interneuron number is becoming apparent. A recent study demonstrates that Olig1 inhibits the generation of GABAergic interneurons produced in the MGE by repression of Dlx expression through the Dlx1/2 I12b intergenic enhancer (Silbereis et al., 2014). Likewise, NPAS1 inhibits the generation of cortical interneurons by repression of Arx expression through the subpallial Arx enhancer, which mediates CGE progenitor cell proliferation (Figure 7) (Colasante et al., 2008).
NPAS1 Function in Interneuron Generation Controls Cortical Excitation/Inhibition
NPAS1−/− mutant mice display increased numbers of GAD67-GFP+ interneurons in layer I (Figure S4C). It is likely that an excess of GABAergic neurons in layer I would have an effect on the inhibitory tone of pyramidal cell dendrites within this layer. NPAS1 mutants also exhibit excessive SST+ and VIP+ interneurons. VIP+ interneurons are enriched in layers II/III (Kawaguchi and Kubota, 1997; Miyoshi et al., 2010), and form synapses with pyramidal cell somata and dendrites (Kawaguchi and Kubota, 1996) and other interneurons (Acsády et al., 1996; Dávid et al., 2007; Hajos et al., 1996). Thus, with an increase in both SST+ and VIP+ interneurons, it is difficult to deduce the net physiological effect a priori. We found that NPAS1 mutants had enhanced cortical inhibition in layer II/III of the somatosensory cortex (Figure 4), implying that the increase in interneurons resulted in a net increase in inhibition in superficial cortical layers. The enhanced cortical inhibition in layer II/III somatosensory cortex of NPAS1−/− mutants suggests a decreased E/I ratio in layer II/III somatosensory cortex. This is supported by estimation of the cellular E/I ratio within this cortical region of NPAS1−/− mutant mice (Figure S4D). While increased cortical inhibition is likely to reduce noise in the cortex, and it could be protective for epilepsy, it may be detrimental to cortical function by dampening cortical signals and thereby predispose to neuropsychiatric disorders.
NPAS1 and NPAS3 in Human Neuropsychiatric Disorders
Our examination of NPAS1 and NPAS3 functions in mice provides novel mechanistic insights into human neuropsychiatric disorders. NPAS3−/− mutant mice display behavioral and neuroanatomical deficits associated with human schizophrenia (Erbel-Sieler et al., 2004) and NPAS3 mutations are observed in patients with schizophrenia (Kamnasaran et al., 2003; Macintyre et al., 2010). Furthermore, NPAS1 mutant mice exhibit an increase in brain size (Figures S3D–S3G), similar to the enlargement seen in some children with autism spectrum disorders (ASD) (Butler et al., 2005; Hazlett et al., 2011). Notably, we have found sporadic non-synonymous mutations in NPAS1 and NPAS3 in individuals with ASD, which suggests that NPAS1 and NPAS3 mutations may be associated with ASD risk. However, our analyses lack the statistical power to conclude that these alleles contribute to ASD. Nonetheless, these data, and the experimental analyses of the mouse mutants, strongly suggest that further genetic analyses of NPAS1 and NPAS3 are warranted in human neuropsychiatric disorders.
Experimental Procedures
Mice
NPAS1+/− and NPAS3+/− mice were obtained from Steven McKnight (University of Texas Southwestern Medical Center) and genotyped as described by Erbel-Sieler and colleagues (Erbel-Sieler et al., 2004). NPAS1+/− mice were maintained in a C57BL/6J strain background. NPAS1+/−; NPAS3+/− females mated to NPAS1+/−; NPAS3+/− males also produced C57Bl/6J strain litters. NPAS3−/− mutant mice obtained from this cross were utilized for BrdU analysis of the ganglionic eminences, assessment of cortical layer-specific markers, and in the examination of interneuron markers in the P30 somatosensory cortex. However, due to increased mortality of NPAS3−/− mutants maintained on a C57BL/6J strain background, subsequent experiments were performed with NPAS3−/− mutant mice maintained on a CD-1 strain background. These experiments include assessment of apoptosis within the somatosensory cortex, examination of PH3 and phospho-ERK expression in the ganglionic eminences, and MRI analysis.
The Lhx6-GFP BAC transgenic mouse line was obtained from The Gene Expression Nervous System Atlas Project (GENSAT) at The Rockefeller University (New York, NY). GAD67-GFP mice were genotyped as described by Tamamaki and colleagues (Tamamaki et al., 2003). As a note on nomenclature, experimental samples designated as NPAS1+/+ indicates that the NPAS1 allele is unaltered; however data from these mice are not referred to as wildtype because they have either the Lhx6-GFP or GAD67-GFP alleles. CD-1 wildtype mice were obtained from Charles River Laboratories. Wildtype littermate mice were used as controls for all experiments. For staging of embryos, midday of the vaginal plug was calculated as embryonic day 0.5 (E0.5). Mouse colonies were maintained at the University of California, San Francisco and University of Texas Southwestern Medical Center, in accordance with National Institutes of Health, UCSF, and UT Southwestern guidelines.
Histology
See the Supplemental Experimental Procedures for details on immunohistochemistry, in situ hybridization, BrdU labeling, and Nissl stain analysis.
Magnetic Resonance Imaging (MRI)
P30 4% PFA-fixed brains were washed twice in 20 mL PBS for a total of 24 hours and imaged in Fluorinert FC-40 (Sigma Aldrich) for null background signal. The imaging was done on an 600 MHz NMR spectrometer (Agilent Technologies Inc.) with imaging gradients and the following parameters: 3D gradient echo, TE/TR 15/75 ms, 8 averages, field of view (FOV) 12.8 mm isotropic, resolution of 50 μm × 50 μm × 100 μm, and a total scan time of 5.5 hours. The acquired images were converted on the Varian console to the DICOM format and reformatted into the same orientation as the histology sections using OsiriX, an open source image viewer. Volumetric measurements were made using custom-built software in MATLAB. Cortical and basal ganglia volumes were statistically analyzed in three mice of each genotype. Results are presented as mean ± SEM. Statistical differences between experimental groups were assessed with the Student’s t-test using SPSS 15 software (IBM). See Table S3 in the Supplemental Experimental Procedures for details on cortical and basal ganglia volumes in control, NPAS1−/−, and NPAS3−/− mutants.
Electrophysiology
Coronal brain slices (300 μm) were prepared from P21-30 wildtype and NPAS1−/− mice. Slices were submerged in the recording chamber and continuously perfused with oxygenated ACSF (32–34°C) containing (in mM): 124 NaCl, 3 KCl, 1.25 NaH2PO4-H2O, 2MgSO4-7H2O, 26 NaHCO3, 10 dextrose, and 2 CaCl2 (pH 7.2–7.4, 300–305 mOsm/kg). Whole-cell patch-clamp recordings from layer II/III pyramidal cells in somatosensory neocortex were performed at 40X using an upright, fixed-stage microscope (Olympus BX50WI) equipped with infrared, differential interference contrast (IR-DIC). Patch pipettes (2–4 MΩ) were filled with an internal solution, containing (in mM): 140 CsCl, 1 MgCl2, 10 HEPES, 11 EGTA, 2 NaATP, 0.5 Na2GTP and 1.25 QX-314, pH 7.28. Recordings were obtained with an Axopatch 1D amplifier, filtered at 5 kHz, and recorded to pClamp 10.2 software (Clampfit, Axon Instruments). Spontaneous (s) and miniature (m) IPSCs were examined at a holding potential of −70 mV. GABAergic currents were isolated by adding 1mM kynurenic acid to the ACSF to block glutamate receptors, and tetrodotoxin (TTX, 2μM) was added to isolate mIPSCs. Series resistance was typically <15 MΩ and was monitored throughout the recordings. Data were only used for analysis if the series resistance remained <20 MΩ and changed by ≤20% during the recordings. Data analysis was performed using pClamp 10.2 (Clampfit, Axon Instruments), MiniAnalysis 6.0 (Synaptosoft), Microsoft excel, and Sigmaplot 12.3 programs. Events characterized by a typical fast rising phase and exponential decay phase were manually detected using MiniAnalysis. A 2 min sample recording per cell was used for measuring IPSC frequency, amplitude, 10–90% rise time, and decay time constant. Kinetic analysis of the IPSCs was performed with a single-exponential function. The threshold for event detection was currents with amplitudes greater than three times the root mean square (RMS) noise level. Spontaneous IPSC measurements were recorded from 18 wildtype and 20 NPAS1−/− pyramidal cells. Miniature IPSC measurements were recorded from 14 wildtype and 16 NPAS1−/− pyramidal cells. Seven wildtype and nine NPAS1−/− animals were used for sIPSC and mIPSC measurements. Results are expressed as mean ± SEM. Statistical differences between experimental groups were assessed with the Mann–Whitney U test using SigmaPlot 12.3 software (SYSTAT).
Primary Cell Culture and Luciferase Assays
CGE tissue was dissected from E12.5 WT embryos and mechanically dissociated with a P1000 pipette tip. 200,000 cells were seeded into tissue culture dishes precoated with poly-L-lysine 10 μg/ml, Sigma) then laminin (5 μg/ml, Sigma), and grown in N5 media (DMEM-F-12 with glutamax, N2 supplement, 35 μg/ml bovine pituitary extract, 20 ng/ml bFGF, and 20 ng/ml EGF) as previously described for MGE cells (Flandin et al., 2011).
CGE primary cultures were transfected using Fugene® 6 (Promega) with DNA expression vectors and reporters comprising four conditions: 1) empty firefly-luciferase reporter (PGL4.23) and a control DNA expression vector only expressing GFP (CMV-GFP), 2) empty PGL4.23 and CMV-NPAS1, 3) Arx enhancer (UAS3-PGL4.23) luciferase reporter and CMV-GFP, and 4) UAS3-PGL4.23 and CMV-NPAS1. All conditions also included the same amounts of CMV-ARNT (NPAS co-factor) and Renilla-luciferase (normalization control). Cell lysates were collected at two days post-transfection and procedures were performed according to the manufacturer’s protocol (Promega, dual luciferase assay system). Enhancer activity for each condition was determined from three independent experiments, each performed in triplicate. Results were calculated as mean ± SEM and presented as fold change relative to the basal condition containing CMV-GFP and the PGL4.23 empty luciferase reporter vector. Statistical differences between experimental groups were determined with the Student’s t-test using SPSS 15 software (IBM). See the Supplemental Experimental Procedures for details on expression and luciferase reporter vectors.
Virus Production and In Utero Injection
HEK293T cells grown in DMEM H21 with 10% FBS were transfected with four plasmids to generate lentivirus particles, (pVSV-g, pRSVr, pMDLg-pRRE and the lentiviral vector). Cells were transfected at ~70% confluency and media was completely replaced four hours after transfection, then cultured for four days before harvesting. On day four of culture, all the media (~35 mls) was collected and filtered through a 0.45 low protein binding membrane to remove cells and large debris. The filtered media was pooled and ultracentrifuged at 100,000 × g for 2.5 hours at 4°C. After the ultracentrifuge step, supernatant was removed and the pellet was resuspended overnight in sterile PBS then stored at −80°C until use. For infection, recipient pregnant females (E13.5) were anesthetized with isoflurane, their uterine horns exposed, and mounted under an ultrasound microscope (Vevo 770, VisualSonics). A beveled glass micropipette (~50 μm diameter) was front loaded with CMV-GFP or CMV-GFP-T2a-Arx lentivirus and inserted into the ventricular zone of the CGE under real-time ultrasound guidance imaging. ~1 μl of solution was injected into the ventricular zone of the CGE. Three successfully injected animals were analyzed two days later (E15.5) for each experimental condition. Image analysis of CMV-GFP+/PH3+, CMV-GFP+/phospho-ERK+, CMV-GFP-T2a-Arx+/PH3+ or CMV-GFP-T2a-Arx+/phospho-ERK+ co-expression was performed on images acquired on a confocal microscope (LSM 510 META NLO, Carl Zeiss International) with a 20X objective. The percentage of CMV-GFP+ or CMV-GFP-T2a-Arx+ that express PH3 was calculated in a 10,000 μm2 area of the CGE ventricular zone. Results are presented as mean ± SEM. Statistical differences between experimental groups were assessed with the chi-squared test using SPSS 15 software (IBM). Phospho-ERK expression levels were measured in GFP+ regions of the CGE ventricular zone with the aid of Adobe Photoshop CS4 software. Expression levels were determined as a ratio of the integrated density of phospho-ERK+ CGE ventricular zone cells in a 4,000 μm2 area to the integrated density of phospho-ERK+ cortical ventricular zone cells in a 4,000 μm2 area. Integrated density is equal to the sum of the pixel values in a selected area. Results were calculated as mean ± SEM and presented as fold change relative to control. Statistical differences between experimental groups were determined with the Student’s t-test using SPSS 15 software (IBM). See the Supplemental Experimental Procedures for details on lentiviral vectors.
Human DNA Sequencing
See the Supplemental Experimental Procedures for details on human DNA sequencing.
Supplementary Material
Figure S1, related to Figure 1. Co-localization analysis of interneuron marker and NPAS1 or NPAS3 expression at P0, P5, and P15. NPAS1 or NPAS3 protein co-immunofluorescence with GFP (from GAD67-GFP or Lhx6-GFP alleles) or interneuron markers (i.e., reelin, SST, CR, or PV) in coronal sections of the somatosensory cortex at P0, P5, and P15. (A and B) At P0, virtually all NPAS1+ cells express GAD67-GFP (A), whereas 30% co-express Lhx6-GFP (B). Arrowheads show NPAS1+/Lhx6-GFP− cells. (C and D) Nearly all developing cortical NPAS1+ cells (C) or the majority of NPAS3+ cells (D) express GAD67-GFP at P5. (E to L) At P15, NPAS1 and NPAS3 are expressed by a majority of reelin+ (E and I) and SST+ (F and J) interneurons. NPAS3 is expressed by a small fraction of PV+ cells (L), while NPAS1 expression is nearly absent from PV+ cells (H). n = 3 animals for (A) to (L). Scale bar, (A) and (B) 150 μm; (C) and (D) 350 μm; (E) to (L) 230 μm.
Figure S2, related to Figure 1. Co-localization analysis of interneuron marker and NPAS1 or NPAS3 expression at P30. NPAS1 or NPAS3 protein co-immunofluorescence with GFP (from the GAD67-GFP allele) or interneuron markers (i.e., reelin, SST, CR, or PV) in coronal sections of the somatosensory cortex at P30. (A to J) By P30, most NPAS1+ cells express GAD67-GFP (A), whereas 21% of NPAS3+ cells express GAD67-GFP (F). NPAS1 is expressed predominantly in reelin+ (B), SST+ (C), and CR+ (D) interneurons and rare in PV+ (E) cells. A large proportion of all interneuron subtypes examined express NPAS3 at P30 (G to J). n = 3 animals for (A) to (J). Scale bar, (A) to (J) 220 μm.
Figure S3, related to Figure 2. Interneuron apoptosis and cortical volume is increased in the NPAS1−/− somatosensory cortex, while the NPAS3−/− cortex exhibits a decrease in cortical volume and no change in apoptosis. (A) Increased interneuron (GAD67-GFP) apoptosis (activated caspase-3+ cells) at P7 in the NPAS1−/− somatosensory cortex based on a co-immunofluorescence assay. Arrowheads show GAD67-GFP+/activated caspase-3+ cells. (B and C) Quantification of GAD67-GFP+/activated caspase-3+ or activated caspase-3+ cells at P7 in NPAS1−/− (B) or NPAS3−/− (C) mutants, respectively. n = 3 animals per genotype for (B) and (C). (D and F) Increased cortical (rostral and middle) volume in P30 NPAS1−/− mutants based on measurements of Nissl-stained coronal sections (D) and MRI data (F). Boxed areas in (D) are shown in higher magnification. (F) Illustration of cortical and basal ganglia regions used for volume calculations. (E, G and H) Cortical width (E) and cortical and basal ganglia volume (G and H) measurements. n = 3 animals per genotype for (E), (G), and (H). *P< 0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 300 μm; (D) 2 mm (low magnification images), 740 μm (high magnification images); (F) 2 mm.
Figure S4, related to Figure 4. The neocortex of NPAS1−/− has layer-specific increases in numbers and density of NeuN+ and GAD67-GFP+ neurons. (A) Increase in NeuN+ neurons (somatosensory Cx) at P21 shown on coronal cortical sections in NPAS1−/− mice. (B to D) Quantification of NeuN+ neurons/104 μm2 (B), GAD67-GFP+ neurons/104 μm2 (C), and NeuN+ and GAD67-GFP−/GAD67-GFP+ ratio (D) in somatosensory cortex and individual layers (yellow lines) at P21. n = 3 animals per genotype for (B) to (D). (E) Representative immunohistochemical staining of the cerebral cortex for NeuN and GAD67 in 3 month old mice reveals an increase in the density of all neurons as well as inhibitory interneurons in NPAS1−/− mutants. (F and G) Stereological quantification of NeuN+ neurons/1 mm3 cerebral cortex (F) and GAD67+ neurons/1 mm3 cerebral cortex (G) in NPAS1-deficient mice at 3 months of age. n = 5 wildtype and 7 NPAS1−/− mutant animals for (F) and (G). (H) Density of layer-specific cortical pyramidal cells is unaltered in NPAS1−/− mutants based on Cux1+ (layer II/III), Ctip2+ (layer V), and Tbr1+ (layer VI) immunofluorescence staining of coronal sections from P21 somatosensory cortex. There are no significant changes in the density of Cux1+ or Ctip2+ cells in NPAS3−/− mutant mice. However, NPAS3−/− mutants displayed a 16% (16 ± 3.05%, p = 0.0001) increase in Tbr1+ cell density. (I to K) Quantification of Cux1+ (I), Ctip2+ (J), and Tbr1+ (K) neurons/104 μm2 of somatosensory cortex. n = 3 animals per genotype for (I) to (K). Abbreviations: Cx: cortex. *P<0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 300 μm; (E) 100 μm; (H) 300 μm.
Figure S5, related to Figure 5. NPAS1−/− mutant mice show increased neurogenesis in the ganglionic eminence and hippocampus, while NPAS3−/− mutants exhibit decreased neurogenesis in the ganglionic eminence. (A) BrdU immunofluorescence staining of E13.5 coronal sections following 1 hr BrdU pulse labeling reveals increased neurogenesis in the subventricular zone of the NPAS1−/− mutant MGE and CGE, whereas NPAS3−/− mutant mice display reduced neurogenesis in the MGE subventricular zone. (B to E) Quantification of BrdU+ cell density (C and E) and total cell number (B and D) in ventricular and subventricular zones of the E13.5 MGE (B and C) and CGE (D and E). n = 3 animals per genotype for (B) to (E). (F) Increased BrdU+ cells in the hippocampal dentate gyrus of 5–10 week old NPAS1−/− mice following intraperitoneal injection of BrdU once daily for 12 days. (G) Quantification of BrdU+ cells/1 mm3 hippocampal dentate gyrus. For (G), n = 22 wildtype and 28 NPAS1−/− mutant animals. Abbreviations: CGE: caudal ganglionic eminence, MGE: medial ganglionic eminence, SVZ: subventricular zone, VZ: ventricular zone. *P< 0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 150 μm; (F) 250 μm.
Figure S6, related to Figure 6. ISH assay of regulatory genes in NPAS1−/− mutants. (A to L) RNA expression of most genes that we studied did not change at E13.5 (A to F) and E15.5 (G to L) in the MGE and CGE of NPAS1−/− mutants. Data presented in coronal sections at three rostro-caudal planes (left to right). Scale bar, (A) to (F) 800 μm; (G) to (L) 1 mm.
Figure S7, related to Figure 7. Increased Arx expression in the CGE mediates increased subpallial MAP kinase activity. (A) Arx over-expression in the dorsal LGE/CGE VZ (arrowheads) two days after in utero injection of a CMV-GFP-T2a-Arx lentivirus. Compare the left (non-injected) and right (CMV-GFP-T2a-Arx-injected) hemisphere. This is a section of the dorsal LGE/CGE taken from the same brain series used to illustrate Arx (from CMV-GFP-T2a-Arx) and PH3 co-expression in Figure 7A. (B and C) In utero lentiviral transduction of Arx into the E13.5 CGE results in increased MAP kinase activity (phospho-ERK). Phospho-ERK levels in the CGE/MGE (white box) were normalized by comparison with cortical levels (yellow box). Asterisks in (B) indicate that phospho-ERK immunohistochemistry panels contain a blood vessel staining artifact. n = 3 animals per experimental condition for (C). (D) Arx locus showing evolutionarily conserved domains, one of which is a subpallial enhancer (UAS3) (Colasante et al., 2008; Visel et al., 2013) that has two predicted NPAS1/ARNT sites (Hogenesch et al., 1997). Abbreviations: CGE: caudal ganglionic eminence, Cx: cortex. *P<0.05. Scale bars, (A) 500 μm; (B) 120 μm.
Table S1. Percentage of NPAS1+ or NPAS3+ neurons that express various interneuron markers.
Table S2. Percentage of interneuron marker+ neurons that express NPAS1 or NPAS3.
Table S3. MRI quantification of P30 cortical and basal ganglia volumes in control, NPAS1−/−, and NPAS3−/−.
Table S4. Rare non-synonymous coding variants in NPAS1.
Table S5. Rare non-synonymous coding variants in NPAS3.
Table S6. Synonymous coding and untranslated variants in NPAS1.
Table S7. Synonymous coding and untranslated variants in NPAS3.
Highlights.
NPAS1 represses cortical interneuron generation, regulating excitation/inhibition
NPAS1 represses an enhancer of Arx, which mediates proliferation of CGE progenitors
NPAS1&3 transcription factors have opposite roles in regulating interneuron number
Acknowledgments
We are grateful to S. McKnight and his lab at University of Texas Southwestern Medical Center for the generous gift of NPAS1 and NPAS3 antibodies, the NPAS1+/− and NPAS3+/− mice, the NPAS1 in situ hybridization construct (J. Walker), and the CMV-ARNT and CMV-NPAS1 mammalian expression vectors (L. Wu). We acknowledge the Autism Genetic Resource Exchange (AGRE) and Simons Foundation Autism Research Initiative (SFARI) for ASD genomic DNA samples, and SFARI as well for phenotypic data made available on SFARI Base. We also wish to thank all the families who donated samples and the principal investigators involved in their collection (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, E. Wijsman). Approved researchers can obtain the Simons Simplex Collection (SSC) population dataset by applying at https://base.sfari.org. This work was supported by research grants to JLRR (Simons Foundation, Nina Ireland, Althea Foundation, NIMH R37 MH049428, Weston Havens Foundation), to AS (NIMHT32 MH089920), to SCB (R01NS071785), to RFH (F32NS077747) to DX (R01EB009756), to SPH (Jaffe Family Foundation), and to AAP and Steven L. McKnight (NIMH 5-RO1-MH087986). This work was also funded in part by an unrestricted endowment provided to Steven L. McKnight by an anonymous donor used as support for JW.
Footnotes
Supplemental Information includes seven figures, Supplemental Experimental Procedures, and Supplemental Text.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acsády L, Arabadzisz D, Freund TF. Correlated morphological and neurochemical features identify different subsets of vasoactive intestinal polypeptide-immunoreactive interneurons in rat hippocampus. Neuroscience. 1996;73:299–315. doi: 10.1016/0306-4522(95)00610-9. [DOI] [PubMed] [Google Scholar]
- Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Machold R, Klein C, Fishell G. Gene expression in cortical interneuron precursors is prescient of their mature function. Cereb Cortex. 2008;18:2306–2317. doi: 10.1093/cercor/bhm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Rossignol E, Hjerling-Leffler J, Denaxa M, Wegner M, Lefebvre V, Pachnis V, Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JLR. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JL, Broccoli V. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci. 2008;28:10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Collombat P, Colasante G, Bianchi M, Long J, Mansouri A, Rubenstein JL, Broccoli V. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J Neurosci. 2007;27:4786–4798. doi: 10.1523/JNEUROSCI.0417-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. Mapping early brain development in autism. Neuron. 2007;56:399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Dávid C, Schleicher A, Zuschratter W, Staiger JF. The innervation of parvalbumin-containing interneurons by VIP-immunopositive interneurons in the primary somatosensory cortex of the adult rat. Eur J Neurosci. 2007;25:2329–2340. doi: 10.1111/j.1460-9568.2007.05496.x. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2012;2:1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci U S A. 2004;101:13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JL. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci. 2010;30:2812–2823. doi: 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Zhao Y, Vogt D, Jeong J, Long J, Potter G, Westphal H, Rubenstein JL. Lhx6 and Lhx8 coordinately induce neuronal expression of Shh that controls the generation of interneuron progenitors. Neuron. 2011;70:939–950. doi: 10.1016/j.neuron.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friocourt G, Kanatani S, Tabata H, Yozu M, Takahashi T, Antypa M, Raguenes O, Chelly J, Ferec C, Nakajima K, Parnavelas JG. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J Neurosci. 2008;28:5794–5805. doi: 10.1523/JNEUROSCI.1067-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marin O, Rubenstein JLR. The Generation of Cortical Interneurons. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. Bethesda, MD: National Center for Biotechnology Information; 2012. [PubMed] [Google Scholar]
- Gripp KW, Zand DJ, Demmer L, Anderson CE, Dobyns WB, Zackai EH, Denenberg E, Jenny K, Stabley DL, Sol-Church K. Expanding the SHOC2 mutation associated phenotype of noonan syndrome with loose anagen hair: Structural brain anomalies and myelofibrosis. Am J Med Genet A. 2013:36098. doi: 10.1002/ajmg.a.36098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajos N, Acsady L, Freund TF. Target selectivity and neurochemical characteristics of VIP-immunoreactive interneurons in the rat dentate gyrus. Eur J Neurosci. 1996;8:1415–1431. doi: 10.1111/j.1460-9568.1996.tb01604.x. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Gerig G, Styner M, Chappell C, Smith RG, Vachet C, Piven J. Early brain overgrowth in autism associated with an increase in cortical surface area before age 2 years. Arch Gen Psychiatry. 2011;68:467–476. doi: 10.1001/archgenpsychiatry.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Chan WK, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, Perdew GH, Bradfield CA. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- Inan M, Welagen J, Anderson SA. Spatial and temporal bias in the mitotic origins of somatostatin- and parvalbumin-expressing interneuron subgroups and the chandelier subtype in the medial ganglionic eminence. Cereb Cortex. 2012;22:820–827. doi: 10.1093/cercor/bhr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnasaran D, Muir WJ, Ferguson-Smith MA, Cox DW. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J Med Genet. 2003;40:325–332. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 1996;16:2701–2715. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Magueresse C, Monyer H. GABAergic interneurons shape the functional maturation of the cortex. Neuron. 2013;77:388–405. doi: 10.1016/j.neuron.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Tomassy GS, De Leonibus E, Uzcategui YG, Andolfi G, Armentano M, Touzot A, Gaztelu JM, Arlotta P, Menendez de la Prida L, Studer M. Loss of COUP-TFI alters the balance between caudal ganglionic eminence- and medial ganglionic eminence-derived cortical interneurons and results in resistance to epilepsy. J Neurosci. 2011;31:4650–4662. doi: 10.1523/JNEUROSCI.6580-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Zhang Q, Cai Y, You Y, Rubenstein JL, Yang Z. A subpopulation of dorsal lateral/caudal ganglionic eminence-derived neocortical interneurons expresses the transcription factor Sp8. Cereb Cortex. 2012;22:2120–2130. doi: 10.1093/cercor/bhr296. [DOI] [PubMed] [Google Scholar]
- Macintyre G, Alford T, Xiong L, Rouleau GA, Tibbo PG, Cox DW. Association of NPAS3 exonic variation with schizophrenia. Schizophr Res. 2010;120:143–149. doi: 10.1016/j.schres.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Marin O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- Marin O. Cellular and molecular mechanisms controlling the migration of neocortical interneurons. Eur J Neurosci. 2013;38:2019–2029. doi: 10.1111/ejn.12225. [DOI] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, et al. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain. 2009;132:1563–1576. doi: 10.1093/brain/awp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JD, State MW. Recent developments in the genetics of autism spectrum disorders. Curr Opin Genet Dev. 2013;23:310–315. doi: 10.1016/j.gde.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Nobrega-Pereira S, Gelman D, Bartolini G, Pla R, Pierani A, Marin O. Origin and molecular specification of globus pallidus neurons. J Neurosci. 2010;30:2824–2834. doi: 10.1523/JNEUROSCI.4023-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro T, Saigo K. Transcriptional regulation of breathless FGF receptor gene by binding of TRACHEALESS/dARNT heterodimers to three central midline elements in Drosophila developing trachea. Development. 1997;124:3975–3986. doi: 10.1242/dev.124.20.3975. [DOI] [PubMed] [Google Scholar]
- Pieper AA, Wu X, Han TW, Estill SJ, Dang Q, Wu LC, Reece-Fincanon S, Dudley CA, Richardson JA, Brat DJ, McKnight SL. The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci U S A. 2005;102:14052–14057. doi: 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O’Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2:255–267. doi: 10.1034/j.1601-183x.2003.00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Nobuta H, Tsai HH, Heine VM, McKinsey GL, Meijer DH, Howard MA, Petryniak MA, Potter GB, Alberta JA, et al. Olig1 function is required to repress Dlx1/2 and interneuron production in mammalian brain. Neuron. 2014;81:574–587. doi: 10.1016/j.neuron.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, Alfaro-Cervello C, Tang Y, Garcia-Verdugo JM, Rubenstein JL, et al. Intrinsically determined cell death of developing cortical interneurons. Nature. 2012;491:109–113. doi: 10.1038/nature11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striano P, Zara F. Genetics: mutations in mTOR pathway linked to megalencephaly syndromes. Nat Rev Neurol. 2012;8:542–544. doi: 10.1038/nrneurol.2012.178. [DOI] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Lu J, Huang ZJ. The spatial and temporal origin of chandelier cells in mouse neocortex. Science. 2013;339:70–74. doi: 10.1126/science.1227622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O’Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta M Dela, Westphal H, Rubenstein JLR. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci U S A. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Parada LF. PTEN signaling in autism spectrum disorders. Curr Opin Neurobiol. 2012;22:873–879. doi: 10.1016/j.conb.2012.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1, related to Figure 1. Co-localization analysis of interneuron marker and NPAS1 or NPAS3 expression at P0, P5, and P15. NPAS1 or NPAS3 protein co-immunofluorescence with GFP (from GAD67-GFP or Lhx6-GFP alleles) or interneuron markers (i.e., reelin, SST, CR, or PV) in coronal sections of the somatosensory cortex at P0, P5, and P15. (A and B) At P0, virtually all NPAS1+ cells express GAD67-GFP (A), whereas 30% co-express Lhx6-GFP (B). Arrowheads show NPAS1+/Lhx6-GFP− cells. (C and D) Nearly all developing cortical NPAS1+ cells (C) or the majority of NPAS3+ cells (D) express GAD67-GFP at P5. (E to L) At P15, NPAS1 and NPAS3 are expressed by a majority of reelin+ (E and I) and SST+ (F and J) interneurons. NPAS3 is expressed by a small fraction of PV+ cells (L), while NPAS1 expression is nearly absent from PV+ cells (H). n = 3 animals for (A) to (L). Scale bar, (A) and (B) 150 μm; (C) and (D) 350 μm; (E) to (L) 230 μm.
Figure S2, related to Figure 1. Co-localization analysis of interneuron marker and NPAS1 or NPAS3 expression at P30. NPAS1 or NPAS3 protein co-immunofluorescence with GFP (from the GAD67-GFP allele) or interneuron markers (i.e., reelin, SST, CR, or PV) in coronal sections of the somatosensory cortex at P30. (A to J) By P30, most NPAS1+ cells express GAD67-GFP (A), whereas 21% of NPAS3+ cells express GAD67-GFP (F). NPAS1 is expressed predominantly in reelin+ (B), SST+ (C), and CR+ (D) interneurons and rare in PV+ (E) cells. A large proportion of all interneuron subtypes examined express NPAS3 at P30 (G to J). n = 3 animals for (A) to (J). Scale bar, (A) to (J) 220 μm.
Figure S3, related to Figure 2. Interneuron apoptosis and cortical volume is increased in the NPAS1−/− somatosensory cortex, while the NPAS3−/− cortex exhibits a decrease in cortical volume and no change in apoptosis. (A) Increased interneuron (GAD67-GFP) apoptosis (activated caspase-3+ cells) at P7 in the NPAS1−/− somatosensory cortex based on a co-immunofluorescence assay. Arrowheads show GAD67-GFP+/activated caspase-3+ cells. (B and C) Quantification of GAD67-GFP+/activated caspase-3+ or activated caspase-3+ cells at P7 in NPAS1−/− (B) or NPAS3−/− (C) mutants, respectively. n = 3 animals per genotype for (B) and (C). (D and F) Increased cortical (rostral and middle) volume in P30 NPAS1−/− mutants based on measurements of Nissl-stained coronal sections (D) and MRI data (F). Boxed areas in (D) are shown in higher magnification. (F) Illustration of cortical and basal ganglia regions used for volume calculations. (E, G and H) Cortical width (E) and cortical and basal ganglia volume (G and H) measurements. n = 3 animals per genotype for (E), (G), and (H). *P< 0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 300 μm; (D) 2 mm (low magnification images), 740 μm (high magnification images); (F) 2 mm.
Figure S4, related to Figure 4. The neocortex of NPAS1−/− has layer-specific increases in numbers and density of NeuN+ and GAD67-GFP+ neurons. (A) Increase in NeuN+ neurons (somatosensory Cx) at P21 shown on coronal cortical sections in NPAS1−/− mice. (B to D) Quantification of NeuN+ neurons/104 μm2 (B), GAD67-GFP+ neurons/104 μm2 (C), and NeuN+ and GAD67-GFP−/GAD67-GFP+ ratio (D) in somatosensory cortex and individual layers (yellow lines) at P21. n = 3 animals per genotype for (B) to (D). (E) Representative immunohistochemical staining of the cerebral cortex for NeuN and GAD67 in 3 month old mice reveals an increase in the density of all neurons as well as inhibitory interneurons in NPAS1−/− mutants. (F and G) Stereological quantification of NeuN+ neurons/1 mm3 cerebral cortex (F) and GAD67+ neurons/1 mm3 cerebral cortex (G) in NPAS1-deficient mice at 3 months of age. n = 5 wildtype and 7 NPAS1−/− mutant animals for (F) and (G). (H) Density of layer-specific cortical pyramidal cells is unaltered in NPAS1−/− mutants based on Cux1+ (layer II/III), Ctip2+ (layer V), and Tbr1+ (layer VI) immunofluorescence staining of coronal sections from P21 somatosensory cortex. There are no significant changes in the density of Cux1+ or Ctip2+ cells in NPAS3−/− mutant mice. However, NPAS3−/− mutants displayed a 16% (16 ± 3.05%, p = 0.0001) increase in Tbr1+ cell density. (I to K) Quantification of Cux1+ (I), Ctip2+ (J), and Tbr1+ (K) neurons/104 μm2 of somatosensory cortex. n = 3 animals per genotype for (I) to (K). Abbreviations: Cx: cortex. *P<0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 300 μm; (E) 100 μm; (H) 300 μm.
Figure S5, related to Figure 5. NPAS1−/− mutant mice show increased neurogenesis in the ganglionic eminence and hippocampus, while NPAS3−/− mutants exhibit decreased neurogenesis in the ganglionic eminence. (A) BrdU immunofluorescence staining of E13.5 coronal sections following 1 hr BrdU pulse labeling reveals increased neurogenesis in the subventricular zone of the NPAS1−/− mutant MGE and CGE, whereas NPAS3−/− mutant mice display reduced neurogenesis in the MGE subventricular zone. (B to E) Quantification of BrdU+ cell density (C and E) and total cell number (B and D) in ventricular and subventricular zones of the E13.5 MGE (B and C) and CGE (D and E). n = 3 animals per genotype for (B) to (E). (F) Increased BrdU+ cells in the hippocampal dentate gyrus of 5–10 week old NPAS1−/− mice following intraperitoneal injection of BrdU once daily for 12 days. (G) Quantification of BrdU+ cells/1 mm3 hippocampal dentate gyrus. For (G), n = 22 wildtype and 28 NPAS1−/− mutant animals. Abbreviations: CGE: caudal ganglionic eminence, MGE: medial ganglionic eminence, SVZ: subventricular zone, VZ: ventricular zone. *P< 0.05. **P< 0.01. ***P< 0.001. Scale bars, (A) 150 μm; (F) 250 μm.
Figure S6, related to Figure 6. ISH assay of regulatory genes in NPAS1−/− mutants. (A to L) RNA expression of most genes that we studied did not change at E13.5 (A to F) and E15.5 (G to L) in the MGE and CGE of NPAS1−/− mutants. Data presented in coronal sections at three rostro-caudal planes (left to right). Scale bar, (A) to (F) 800 μm; (G) to (L) 1 mm.
Figure S7, related to Figure 7. Increased Arx expression in the CGE mediates increased subpallial MAP kinase activity. (A) Arx over-expression in the dorsal LGE/CGE VZ (arrowheads) two days after in utero injection of a CMV-GFP-T2a-Arx lentivirus. Compare the left (non-injected) and right (CMV-GFP-T2a-Arx-injected) hemisphere. This is a section of the dorsal LGE/CGE taken from the same brain series used to illustrate Arx (from CMV-GFP-T2a-Arx) and PH3 co-expression in Figure 7A. (B and C) In utero lentiviral transduction of Arx into the E13.5 CGE results in increased MAP kinase activity (phospho-ERK). Phospho-ERK levels in the CGE/MGE (white box) were normalized by comparison with cortical levels (yellow box). Asterisks in (B) indicate that phospho-ERK immunohistochemistry panels contain a blood vessel staining artifact. n = 3 animals per experimental condition for (C). (D) Arx locus showing evolutionarily conserved domains, one of which is a subpallial enhancer (UAS3) (Colasante et al., 2008; Visel et al., 2013) that has two predicted NPAS1/ARNT sites (Hogenesch et al., 1997). Abbreviations: CGE: caudal ganglionic eminence, Cx: cortex. *P<0.05. Scale bars, (A) 500 μm; (B) 120 μm.
Table S1. Percentage of NPAS1+ or NPAS3+ neurons that express various interneuron markers.
Table S2. Percentage of interneuron marker+ neurons that express NPAS1 or NPAS3.
Table S3. MRI quantification of P30 cortical and basal ganglia volumes in control, NPAS1−/−, and NPAS3−/−.
Table S4. Rare non-synonymous coding variants in NPAS1.
Table S5. Rare non-synonymous coding variants in NPAS3.
Table S6. Synonymous coding and untranslated variants in NPAS1.
Table S7. Synonymous coding and untranslated variants in NPAS3.