Preface
With the discovery of Th17 cells, the past decade has witnessed a major revision of the T helper subset paradigm and significant progress has been made deciphering the molecular mechanisms for T cell lineage commitment and function. In this review, we focus on the recent advances on the transcriptional control of Th17 cell plasticity and stability as well as the effector functions of Th17 cells—highlighting IL-17 signaling mechanisms in mesenchymal and barrier epithelial tissues. We also discuss the emerging clinical data showing anti-IL-17 and anti-IL-23 treatments are remarkably effective for many immune-mediated inflammatory diseases.
Introduction
The discovery of interleukin-23 (IL-23) and elucidation of the biology governed by this cytokine has led to important new insights within immunology1–3. IL-23 has critical roles in the pathogenesis of autoimmunity as it induces a cell population with a unique inflammatory gene signature that includes Il17a, Il17f, Il6, Csf2, Tnf, Ccl20, Ccl22, Il1r1, and Il23r4,5. Based on this distinct gene profile and unique Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway, a novel subset of T helper (TH) cells was coined and designated ThIL-17 or Th176,7. The discovery of the IL-23–Th17 pathway has led to fundamental changes in our understanding of cellular immunity (Figure 1). In 1989, Mossman and Coffman proposed to divide CD4+ Th cells into interferon- γ (IFNγ)-producing Th1 cells (which are dependent on STAT1) and IL-4-producing Th2 cells (which are dependent on STAT6) to explain the tendency of T cells to diverge into either a cellular or humoral immune response8, 9. At that time, it was predicted that additional CD4+ T cell subsets may exist that drive the full spectrum of immune responses. Indeed, the Th1-Th2 hypothesis does not adequately explain the regulation of STAT3-dependent CD4+ T cells during autoimmunity and infections3. In 2003, the recognition that IL-23 is indispensable for promoting autoimmunity led to rethinking of the established paradigm1, 3, 4, 10, 11.
Figure 1.
Discovery of the IL-23/Th17 immune pathway
In addition to Th17 cells, many innate immune cells respond to IL-23 and are important in both resistance to infection and mediating autoimmune pathology. These cells are characterized by expression of the transcription factor retinoic-acid-receptor-related orphan receptor-γ (RORγt, which is encoded by Rorc)12 and include subsets of γδT cells, natural killer T (NKT) cells, ‘natural’ Th17 cells and innate lymphoid cells (ILCs) collectively, and are termed “Type-17” cells13–17 (Box 1). These innate immune cells are located in non-lymphoid tissues where they are poised to respond immediately to tissue injury or pathogenic insults. Stimulation of Th17 cells and Type-17 cells with IL-1β and IL-23 induces local tissue inflammation, which is mainly mediated by Type-17 signature cytokines such as IL-17, IL-22 and granulocyte–macrophage colony-stimulating factor (GM-CSF)4, 18–20.
Box 1.
“Type 17” subsets of cells ubiquitously express RORγt and IL-23R. Their development is Thymic dependent with the exception of Group 3 ILCs. Adaptive CD4+ IL-17-producing cells require IL-6 signaling during initial TCR-mediated activation. All other subsets do not require IL-6 activation and are capable of responding to IL-1 and IL-23 signaling upon emigrating from the thymus. These “innate” immune cells are poised to produce IL-17 upon sensing inflammatory cytokines as well as stress and injury signals. While the adaptive Th17 cells reside primarily in secondary lymphoid organs, the “innate” Type 17 cells are situated in a broad range of peripheral tissues, where they directly survey the interface between the host and the environment.
| Cell Type | TCR usage | Developmental Timing |
Activation requirements |
Primary Tissue location (steady state) |
Refs |
|---|---|---|---|---|---|
| Adaptive Th17 |
Diverse TCR | Thymic, adolescent to adult |
Antigen- specific TCR activation IL-6, TGFb, IL- 1, IL-23 |
Lymphoid organs |
12 |
| Natural Th17 |
Diverse TCR Self reactivity? |
Thymic, adolescent to adult |
IL-1, IL-23 | Skin and mucosal tissues |
17 |
| IL-17- producing gd T cells |
Invariant TCR Vg6, Vg4, Vg1 Variable TCR |
Fetal and neonatal thymic development Adolescent to adult |
Dectins, TLRs, IL-1, IL-23 TCR + IL-1 and IL-23 |
Mucosal and peripheral tissues Lymphoid organs |
13 172 |
| iNKT | Oligoclonal | Thymic, adolescent to adult |
CD1, glycolipids |
Liver | 173 |
| Group 3 ILC | None | Independent of thymic development; throughout life |
Dectins, TLRs, IL-1, IL-23 |
Gut, skin | 174 |
The exploration of Type-17 immunity has led to the identification of previously unrecognized immune cell subsets and new paradigms for disease localization that is dependent on the distribution of cell populations that express the IL-23 receptor (IL-23R) and RORγt+. These concepts are at the core of novel therapeutic strategies based on neutralization of IL-17 or IL-23, which show encouraging results for treatment of psoriasis, multiple sclerosis, Crohn’s disease and ankylosing spondylitis. In this Review, we highlight the roles of cytokines that promote development and stabilization of Th17 cells via complex transcriptional networks. We discuss how IL-17 signaling is unique compared to other receptor families and highlighting recent studies that reveal mechanisms by which IL-17 signaling is constrained. Finally, we discuss disease settings where anti-IL-17 and anti-IL-23 therapeutic agents worked as predicted and where they have shown unexpected effects.
Cytokines that promote Th17 cell development
The cumulative impact of RORγt+ Th17 cells is a function of the effector cytokines they produce and the subsequent impact on target cells and tissues bearing appropriate receptors. RORγt+ Th17 cells can be broadly divided into two groups: host protective cells that express both IL-17 and IL-10 21–23 and a highly inflammatory population that express IL-17, IL-22, IFNγ and GM-CSF4, 18. The ultimate outcome of Th17 cell activity is determined by the balance of these effector functions. Th17 cells activated by transforming growth factor-β (TGFβ) and IL-6 promote mucosal defense, barrier tissue integrity and curtail immunopathogenic responses21–23, whereas IL-23-activated Th17 cells promote chronic tissue inflammation during infection, granuloma formation and autoimmunity24–26.
Initiating cytokines
Studies in 2003 and 2005 showed that IL-23 promotes the development and expansion of a pathogenic T cell population with a unique inflammatory gene signature1, 4, 6, 7. However, IL-23 alone cannot drive differentiation of Th17 cells from naïve CD4+ T cell precursors, indicating that additional factors are required for their lineage fate determination. Shortly thereafter, Stockinger, Weaver, and Kuchroo simultaneously showed that addition of TGFβ and IL-6 during initial T cell recognition of cognate antigen promotes Th17 cell differentation27–29. IL-6 has an essential role in this process by activating signal transducer and STAT330, which directly drives transcription of TH17 lineage-specific genes31 including Rorc, Il17, and Il23r. STAT3 also suppresses TGFβ-induced forkhead box P3 (FOXP3) expression, and thereby inhibit the generation of regulatory T (TReg) cells27. Consequently, Il6−/− mice are unable to generate Th17 cells and are protected from developing experimental autoimmune encephalomyelitis (EAE)32 and collagen-induced arthritis (CIA)33. Although IL-6 was identified as a therapeutic target long before the recognition of Th17 cells, the recent advances have validated IL-6 as a target for the treatment of rheumatoid arthritis and other inflammatory conditions.
IL-1β signaling also has a critical role during the initial stages of Th17 cell differentiation. Il1r1−/− mice fail to develop antigen-specific Th17 cells and are resistant to EAE34. Expression of IL-1 receptor type 1 (IL-1R1) by Th17 cells is induced by IL-6, and signaling through the IL-1R promotes the transcription factor interferon-regulatory factor 4 (IRF4), which reinforces the expression of RORγt35. Mechanistically, IL-1β induces phosphorylation of the mammalian target of rapamycin (mTOR) in Th17 cells and thereby enhances the metabolic fitness of rapidly dividing Th17 cells during inflammation. Indeed, rapamycin treatment or mTOR-deficiency completely abolishes IL-1β-induced Th17 cell proliferation36. These results suggest that IL-6 functions to direct differentiation of Th17 cells, whereas IL-1β functions to enhance expansion of Th17 cells in competition with other T cell subsets in the context of a resource-limited tissue environment.
In contrast to IL-1β and IL-6, the role of TGFβ in Th17 cell differentiation is more complex. Genetic approaches that used a dominant negative TGFβ receptor II, which inhibits the formation of a functional TGFβ signaling complex, and a T cell-specific deletion of TGFβ confirmed that endogenous TGFβ induces Th17 cell development in vivo37–39. However, deletion of TGFβ also led to excessive production of IFNγ and IL-4, which inhibited Th17 cell development and expansion37, 38. Thus, TGFβ can either function as an obligate Th17-inducing factor or could indirectly suppress alternate cell fates. Notably, in the absence of TGFβ, IL-6 alone can promote Th17 development in T cells that lack transcription factors T-bet and STAT640, which suggest that TGFβ represses Th17 differentiation indirectly by repressing T-bet and transcription factor GATA-binding protein 3 (GATA-3). This complexity is further highlighted by human studies; three separate research groups show that TGFβ is required for in vitro Th17 cell development, whereas two other research groups found that a cocktail of IL-1β, IL-6 and IL-23 is sufficient to drive Th17 differentiation41–45. These discrepancies could be due to the source of the human T cells (for example peripheral blood versus umbilical cord blood), which may influence their developmental status and hence, the requirement for TGFβ. However, under in vivo inflammatory conditions, the presence of TGFβ is probably necessary for optimal Th17 cell differentiation.
Although the combination of TGFβ and IL-6 efficiently generates Th17 cells in vitro, the cells generated by this cocktail are only weakly pathogenic, and subsequent exposure to IL-23 is essential to drive pathogenicity21, 46–50. A recent study further supported the notion that TGFβ is not always required for in vitro Th17 differentiation, as Th17 cells could be generated in response to IL-1β, IL-6 and IL-23 in a TGFβ-independent manner50. Interestingly, these TGFβ-independent IL-23R+, RORγt+ Th17 cells are also T-bet+ and co-express IFNγ and GM-CSF4, 19, 20, 50; such “hybrid” T cells with diverse functional plasticity (see below) are frequently detected in lesions from patients with multiple sclerosis (MS) and mice with EAE 51, 52.
The studies mentioned above specifically examined TGFβ1. In a recent report, it was proposed that TGFβ3 promotes pathogenic TH17 cells by upregulating IL-23R expression53. Interestingly, TGFβ3 was induced by IL-23 and acted endogenously in a feed forward loop to enhance IL-23R expression, thereby amplifying the IL-23 signal53. Taken together, these studies indicate that TGFβ together with IL-6 can drive development of IL-17- and IL-10-producing cells that have mucosal defensive functions; however, subsequent exposure to IL-23 is required for development of autoimmune-associated Th17 cells.
Differentiating and stabilizing cytokines
TGFβ- and IL-6-driven Th17 cells have limited inherent pathogenicity, and exposure to IL-23 is essential for the maturation of inflammatory Th17 cells1. This is supported by genetic studies that link IL23R polymorphisms with susceptibility to autoimmune diseases such as psoriasis, psoriatic arthritis, ankylosing spondylitis, multiple sclerosis and Crohn’s disease54–58. Accumulating data clearly indicates that IL-23 promotes pathogenicity of mature Th17 cells by several mechanisms, including maintenance of Th17 signature genes (Rorc and Il17), induction of effector genes (Il22, Csf2 and Ifng), downregulation of repressive factors (IL-2, IL-27 and IL-12) and amplification of its own signal by upregulation of Il23r expression.
By using an approach that restricts IL-23R deficiency to T cells, we have shown that without IL-23 signaling, Th17 cells are arrested at an early activation stage and are unable to mediate encephalitogenicity in EAE 48. Subsequent studies revealed that IL-23R is needed for co-expression of GM-CSF; accordingly, GM-CSF-deficient mice are resistant to EAE and GM-CSF-deficient TH17 cells cannot transfer EAE to naive recipients 19, 20. IL-23 also induces IFNγ expression in Th17 cells, and IFNγ+IL-17+ cells are highly pathogenic52, 59. To gain inflammatory function, Th17 cells must mitigate the impact of inhibitory cytokines such as IL-27 and IL-2 60, 61. Indeed, committed IL-23R+ Th17 cells are highly resistant to IL-27- and IL-2-mediated suppression62. This phenomenon could be due to downregulation of IL-27R in mature Th17 cells, and IL-23R signaling may also induce transcription factors that partner with RORγt to inhibit repressive signals, and thereby stabilize the Th17 lineage. Pathogenic Th17 cells induced by IL-23 in the absence of TGFβ also express reduced levels of aryl hydrocarbon receptor (AHR), C-MAF and IL-10 compared with TGFβ driven Th17 cells53, 59. AHR and C-MAF are induced under TGFβ- and IL-6-driven Th17 differentiation conditions and transactivate IL-10 expression63–65. However, it is unclear whether IL-23 directly suppresses these factors to drive pathogenicity. The ability of IL-23 to drive differentiation and stabilization of pathogenic Th17 cells is upregulation of IL-23R expression, which is STAT3-dependent 10, 59. Taken together, these studies establish that TGFβ, IL-6 and IL-1β are essential elements that initiate Th17 cell development, whereas IL-23 serves as a pivotal factor that drives both the differentiation and inflammatory functions of pathogenic Th17 cells.
Transcriptional control of Th17 cells
CD4+ TH cell populations are considered unique lineages based on their characteristic expression of transcriptional regulators and signature cytokines. RORγt serves as the master regulator for TH17 cells, and Rorc−/− mice lack TH17 cells and exhibit reduced autoimmune disease severity12. In contrast, enhanced expression of RORγt in naïve CD4+ T cells induces key genes essential for TH17 cell differentiation12. RORγt cooperates with a network of transcription factors — most notably, STAT3 IRF4 and basic leucine zipper transcription factor ATF-like (BATF) — to initiate the complete differentiation program. The activation of STAT3 by IL-6 and IL-23 is a key event in Th17 lineage specification. ChIP-Seq analysis has demonstrated that STAT3 directly regulates the Th17 cytokine and cytokine receptor genes Il17, Il17f and Il23r, and it also controls expression of the Th17-specific transcription factors Rorc, Batf, and Irf4 31. In addition, STAT3 binds both the promoter regions of Il17a and Il17f and their intergenic regions with enhancer elements that undergo histone modifications during differentiation31. These findings are also relevant to humans as patients with STAT3 mutations suffer from autosomal dominant hyper-IgE syndrome (AD-HIES; also known as Job’s syndrome), and are susceptible to recurrent pulmonary infections and chronic mucocutaneous candidiasis due to reduced IL-17-producing T cells 66, 67 (see below).
BATF is a member of the activator protein 1 (AP-1) family of transcription factors, which is upregulated in TH cells following T cell receptor (TCR) activation68, 69. A study from 2009 showed that BATF is essential for TH17 differentiation, and mice lacking BATF are resistant to EAE69. Batf−/− mice have normal TGFβ and IL-6 signaling and can upregulate RORγt early on during T cell development, but fail to maintain RORγt expression. However, overexpression of RORγt could only partially rescue Th17 defects in Batf−/− cells and expression of both BATF and RORγt is required to drive IL-17 expression, which suggests that BATF and RORγt cooperatively control Th17 differentiation. Furthermore, BATF forms a dimer with JUNB that binds directly to the promoters of Il17, Il21 and Il22 in Th17 cells. IRF4 was originally reported to be essential for TH2 differentiation70 and has now also been implicated in TH17 differentiation71. IRF4-deficient mice are protected from EAE and exhibit a defect in TH17 differentiation that stems from impaired RORγt expression. However, similar to BATF, overexpression of RORγt in Irf4−/− T cells could only partially restore IL-17 production, which indicates that both RORγt and IRF4 are required to establish the TH17 lineage.
Two recent studies show that BATF and IRF4 are “pioneer factors” that cooperatively bind and govern chromatin accessibility, which enables RORγt recruitment and binding to Th17 signature genes72, 73. A comprehensive ChIP-Seq analysis revealed a striking overlap between BATF and IRF4 promoter occupancy in Th17 cells73. Furthermore, these transcription factors form a complex and regulate each other’s DNA binding activities. Since BATF and IRF4 binding promotes strong association and co-occupancy by Th17-defining transcription factors (STAT3 and RORγt), BATF and IRF4 seem to serve as initiating factors that regulate chromatin accessibility in TH cells. Surprisingly, co-occupancy by these transcription factors was also observed in TCR activated but non-polarized Th0 cells. Thus, BATF and IRF4 are bona fide pioneer factors that remodel chromatin to enable access of lineage-specifying transcription factors induced following environment signals.
Transcription of a gene is regulated by sequence-specific binding of proteins to the promoter region and/or to distant regulatory enhancer sites. Enhancer activity is measured by histone modification marks such as the histone acetyltransferase p300, which are recruited by lineage specifying signals, and the presence of these marks indicates a transcriptionally active regulatory domain. Strikingly, BATF, IRF4 and STAT3 deficiency substantially reduces p300 binding, but a lack of RORγt has almost no effect on the enhancer landscape. Although RORγt is referred to as a master transcription factor, in fact only a few Th17 genes—Il17, Il17f, and Il23r—are exclusively dependent on RORγt 73. Instead, RORγt acts as a modulator that fine-tunes a pre-established Th17 lineage program. RORγt positively regulates the Th17-specific genes while simultaneously repressing alternate lineage fates, by exploiting the altered enhancer landscape initially created by BATF, IRF4 (induced by TCR) and STAT3 (induced by cytokines) (Figure 2). These new insights argue against the notion of a single factor governing lineage specificity, and instead indicates that a transcription factor complex regulates fate commitment 74. Furthermore, these results are consistent with studies that have identified other transcription regulators, such as T-bet (Th1) and Foxp3 (Treg) as factors that take advantage of an enhancer DNA landscape already established by STAT proteins75, 76.
Figure 2.
Transcriptional control of Th17 cells
Another key transcription factor in Th17 fate determination is hypoxia-inducible factor 1α (HIF-1α), which is a key sensor of hypoxia (Figure 2). HIF-1α is induced upon TCR activation, and its expression is further enhanced by hypoxic conditions associated with tissue inflammation as well as IL-6-and STAT3-dependent differentiation signals77. Mechanistically, HIF-1α directly binds and drives transcription of RORγt and forms a complex with RORγt and p300 to drive IL-17 expression. HIF-1α can also form a complex with FOXP3, and targets it for proteasomal degradation. In this way, proinflammatory environmental cues such as hypoxia stabilize the Th17 lineage.
In spite of advances in deciphering the Th17 transcriptional network, we still lack a clear understanding of IL-23-dependent mechanisms that orchestrate the pathogenic Th17 differentiation process. STAT3 activation alone cannot explain the unique requirement for IL-23 in pathogenic TH17 cell commitment since IL-6 is an even more potent activator of STAT3. Additional transcriptional regulators or signaling pathways induced by IL-23 must operate to promote TH17 inflammatory effector cell function. Also, it will be important to assess the Th17 enhancer landscape created by IL-6 compared with IL-23 to determine whether divergence at an epigenetic level impacts functional outcomes.
Th17 cell plasticity versus stability
Plasticity is defined as the ability of Th17 cells to acquire divergent functional capabilities while maintaining the fundamental Th17 program as defined by RORγt and IL-17 expression. Recent published data indicates that Th17 cells retain a high degree of plasticity that enables them to co-express alternate lineage-specifying transcription factors and effector cytokines 23, 50, 52, 53, 78. Such functional heterogeneity endows Th17 cells with an enhanced capacity to traffic to a broad range of anatomical sites and efficiently promote diverse functions including host defense, barrier tissue integrity and wound healing responses.
Factors that define regulation of Th17 plasticity is an area of intensive investigation. Although RORγt is essential for the development of Th17 cells, it is not sufficient to promote all functional capabilities. As mentioned above, IL-23 might be a pivotal factor dictating Th17 functional diversity through induction of alternate lineage specific transcription factors (such as T-bet) and effector genes (such as Il22, Csf2 and Ifng), while still maintaining the core Th17 program (which entails expression of Rorc and Il17)4, 25. We have recently identified that transcription factor B-lymphocyte-induced maturation protein 1 (BLIMP1, which is encoded by Prdm1) is induced by IL-23 and acts both as a gene activator and repressor to induce Th17 maturation (Unpublished data, R. Jain and D. J. Cua). BLIMP1 coordinates with RORγt to enhance expression of IL-23R and GM-CSF while simultaneously repressing IL-2 and alternate fate determining factors (such as Bcl2, Foxp3, Gata3). Interestingly, BLIMP1 directly engages the Il23r, Il17f and csf2 loci in close proximity to RORγt, p300 and STAT3, which suggests that BLIMP1 is part of a complex that regulates Th17 pathogenic function (Unpublished data, R. Jain and D. J. Cua).
Th17 cell plasticity could be explained by epigenetic modifications. Recent advances in genome-wide analysis of histone modifications enables examination of permissive and repressive histone marks, H3K4me3 and H3K27me3, respectively, on lineage specifying genes. Interestingly, Th17 cells display both active and repressive histone marks on Tbx21 and Gata3, whereas Th1 and Th2 cells show only repressive marks on Rorc and Il17 loci. In other words, Th17 cells appear to be highly poised for subsequent activation under appropriate environmental stimuli79. Interestingly, TReg cells are the only other cell type with such dual modifications on master transcription factors, which gives them the unique advantage of functional diversity and plasticity.
The concept of Th17 stability is more than a question of functional malleability. Stability is defined by whether RORγt-expressing Th17 cells can lose their core transcriptional and functional characteristics and become RORγt-”ex-Th17” cells52. The concept of Th17 stability has been explored by employing both in vitro and in vivo approaches. The in vitro polarization approach, in which the inducing signals are constantly present, has been unable to demonstrate that Th17 cells are unstable. However, in vivo systems that use mouse strains with a expressing a Cre recombinase from IL-17A promoter have greatly enhanced our understanding of this important question 22, 52, 80. Crossing the Il17a-Cre allele to Rosa26-enhanced yellow fluorescence protein (YFP) reporter mice generated a fate mapping system in which Il17-expressing cells are permanently marked with YFP. This cell tagging system has demonstrated that “bona fide” Th17 cells can be destabilized and converted to Th1 cells under proper environmental stimuli. Histone modification analysis of cytokine bead captured IL-17+ cells from the inflamed intestine confirmed these results81. However, an epigenomic analysis of ex-Th17 cells at a single cell level is required to gain a deeper molecular understanding of Th17 stability and to avoid any variations caused by heterogeneity of the population. For instance, it would be revealing to analyze epigenetic changes in Th17 cells that acquire Th1 functions. Do they undergo complete epigenetic remodeling of Th17 associated genes, or do they retain epigenetic evidence of their Th17 origins? Are former Th17 cells capable of reacquiring the original Th17 program upon TCR stimulation because they retain a core epigenetic status? Understanding the plasticity of Th17 cells at the epigenetic level may have important advantages. First, it could predict the developmental stages of Th17 cells that would be susceptible to reprogramming towards the Treg cell phenotype. This cellular reprogramming could have added anti-inflammatory benefits, in addition to ‘depleting’ Th17 cells. Second, it is vital to understand the durability of the Th17 effector cell suppression and/or reprogramming. It is critical to recognize whether “ex Th17 cells” can revert back to their original pathogenic phenotype once the effects of drugs have subsided.
IL-17 signal transduction
As outlined above, much effort has been devoted to defining cellular sources of IL-17 and understanding the subsequent regulation of its expression. However, the biological impact of IL-17 is mediated through its receptor in downstream target cells, which is outlined in this section. Cells that respond to IL-17 are mainly, though not exclusively, non-hematopoietic and the effects of IL-17 are most similar to other innate inflammatory effectors, especially those that belong to the IL-1 family of cytokines and TLR ligands82. In particular, IL-17 is a potent regulator of neutrophils, which is mediated by IL-17-induced downstream genes such as those that encode the granulocyte colony-stimulating factor (G-CSF) and CXC chemokines. The IL-17 receptor (IL-17R) has many unique and/or unexpected properties in terms of structure and signaling, and ultimately a detailed understanding of these events may reveal potential novel drug targets or avenues for therapeutic intervention.
Positive activation of signaling
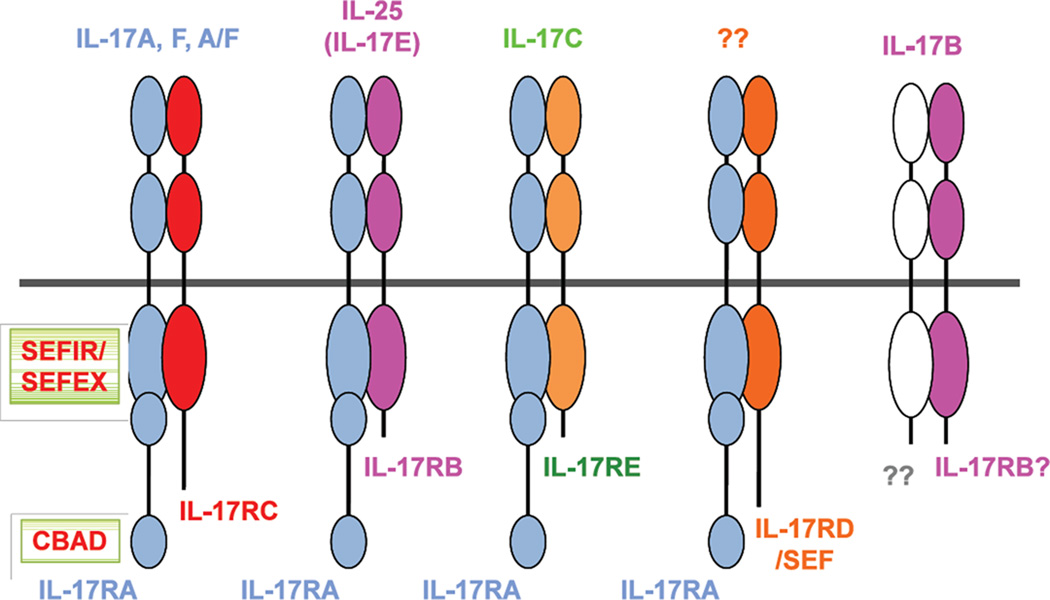
When IL-17R was cloned in 1995 it was noteworthy for its striking lack of similarity to known receptor families83. Subsequently, it became clear that the IL-17R (now known as IL-17RA) was the founding member of a new subclass of cytokine receptors, which comprises five receptor subunits, IL-17RA–IL-17RE, that have homology to one another but not to other receptor families82 (Figure 3). All subunits of the IL-17R family exhibit a broad expression pattern; IL-17RA is most highly expressed in hematopoietic cell types and IL-17RC in non-hematopoietic cells84. Most of the homology within the IL-17R family is located in the conserved cytoplasmic motif SEF/IL-17R (SEFIR), which suggests a common mode of signaling that is probably distinct from other cytokine receptors. A landmark bioinformatics study demonstrated that this domain was related to the Toll/IL-1R (TIR) domain. Recent studies have revealed that the IL-17RA subunit is shared among several members of this family (Figure 3). IL-17RA is essential for signaling through IL-17E (also known as IL-25) when partnered with IL-17RB, and also for signaling through IL-17C when partnered with IL-17RE85, 86. The only other protein identified to have a SEFIR domain is the adaptor ACT1 (also known as CIKS)87; indeed, ACT1 is essential for signal transduction by all analyzed IL-17R family members82, 88, 89.
Figure 3.
IL-17 receptor family members
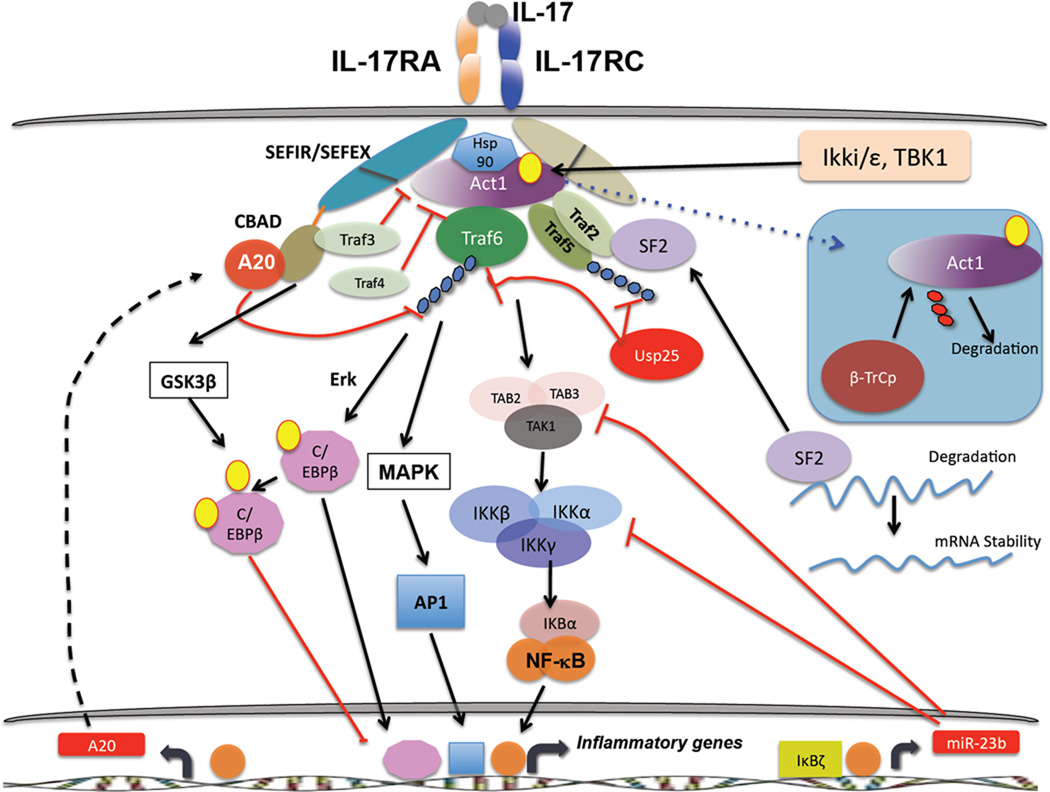
Despite the similarity between IL-17R and TLR/IL-1Rs, studies of IL-17 have revealed several differences in how IL-17R operates. For example, ACT1 is not a positive activator of signaling in any system outside the IL-17R family90. ACT1 serves both as a signaling adaptor, to recruit tumour necrosis factor receptor (TNFR)-associated factor (TRAF) proteins to direct activation of several downstream signaling cascades88, and as an E3 ligase to mediate ubiquitination of TRAF691. Recruitment and ubiquitination of TRAF6 leads to activation of the canonical NF-κB pathway and the mitogen-activated protein kinase (MAPK) pathways, which include extracellular signal-regulated kinase (ERK), p38 and JUN N-terminal kinase (JNK). Consequently, activation of these pathways by IL-17 is impaired in the absence of either ACT1 or TRAF688, 89, 92. Alternatively, ACT1 can recruit TRAF2 and TRAF5, which promote activation of an mRNA stability pathway via the SF2/ASF or HuR complexes93, 94. Regulation of these alternative pathways occurs via inducible ACT1 phosphorylation, which is mediated by Ikki (Ikkε) and TBK1 (Figure 4)95, 96. ACT1 function and stability is in turn regulated by the chaperone protein hsp90. Inhibition of hsp90 results in proteasomal degradation of ACT1, which in turn leads to defective IL-17 signaling97. ACT1 appears to be required for most IL-17-dependent signaling events98 and the majority of phenotypes Act1−/− mice are similar to Il17ra−/− mice. Notably, a single-nucleotide polymorphism (SNP) in ACT1 has been associated with psoriasis99. Thus, in the IL-17R family all known signaling emanates from ACT1, which leads to an activation of pro-inflammatory signaling cascades.
Figure 4.

IL-17 induces signaling mainly in cells of non-hematopoietic origin, including epithelial and mesenchymal cell types. The genes that are induced by IL-17 largely explain its pro-inflammatory activities; for example, IL-17 induces expression of chemokines such as CXC-chemokine ligand 1 (CXCL1), CXCL2 and CXCL5 that promote neutrophil chemotaxis100. IL-17 also induces expression of CC-chemokine ligand 20 (CCL20), which promotes trafficking of mucosal-associated cells expressing CC-chemokine receptor 6 (CCR6); notably, CCR6 is a characteristic receptor on IL-17-expressing cells such as Th17 cells and ILC3s 101. Other cytokines induced by IL-17, such as IL-6 and G-CSF, regulate myeloid lineages and especially neutrophils. Additionally, IL-17 is a strong inducer of antimicrobial peptides (AMPs), particularly β-defensins, which can prevent infection at mucosal surfaces and skin and that may also exert chemotactic activity102. Lipocalin-2 (LCN2; also known as 24p3) is another IL-17-induced antimicrobial protein that controls bacterial growth by restricting access to dietary iron103.
The CCAAT/enhancer-binding protein (C/EBP) family of transcription factors is also activated by IL-17 signal transduction, and is often just as important as NF-κB in the regulation of target genes. For example, the Il6 and Lcn2 promoters require both NF-κB and C/EBP elements, as deletion of either element renders their promoters insensitive to IL-17-mediated signals104, 105. IL-17 induces expression of C/EBPδ mRNA and protein, and to a lesser extent C/EBPβ104, 106. The latter C/EBPδ protein is regulated extensively by posttranscriptional or posttranslational modifications, including alternative translation and phosphorylation107. Phosphorylation of C/EBPβ depends on a GSK3β- and ERK-dependent pathway, which functions to dampen IL-17 dependent signaling (see below)108.
On their own, IL-17 (and IL-17F) are modest activators of signaling, but they function cooperatively with other pro-inflammatory molecules, particularly TNFα but also IFNγ, IL-22, lymphotoxin, IL-1β and lipopolysaccharide109. The molecular basis for this synergy is not completely understood, and probably involves multiple mechanisms. In synovial tissue, IL-17 upregulates TNFRII expression, and thereby enhances responsiveness to TNFα signaling110. For some genes, cooperativity between IL-17 and TNFα occurs at the level of the promoter (for example Il6 and Lcn2) and/or mRNA stability111. IL-17 also upregulates expression of IκBζ — a positive regulator of the NF-κB family — which in turn promotes expression of at least some target genes112. The ability of IL-17 to signal cooperatively with other cytokines is probably an important aspect of its biology, since inflammatory environments contain multiple cytokines that can act in concert.
Negative regulation of IL-17 signaling
Dysregulated IL-17 production and signaling is associated with the development of several autoimmune diseases, thus it is not surprising that there are multiple layers of restriction that control IL-17-mediated inflammatory signaling. As outlined below, a variety of inhibitory mechanisms have been identified (Figure 4).
Deubiquitylating enzymes (DUBs) can temper IL-17-mediated signal transduction by targeting TRAFs; ubiquitin C-terminal hydrolase 25 (USP25) removes K63-linked ubiquitin chains on both TRAF6 and TRAF5, which inhibits transcription of IL-17 target genes as well as IL-17-induced mRNA stability USP25 deficiency is associated with enhanced IL-17-dependent pulmonary inflammation and increased susceptibility to EAE113.
We recently identified A20 (Tnfaip3) as a vital feedback inhibitor of IL-17 signaling114. SNPs in TNFAIP3 are associated with numerous autoimmune and lymphoproliferative diseases, including psoriasis, rheumatoid arthritis (RA) and certain B cell malignancies115. IL-17 induces expression of Tnfaip3 mRNA and A20 protein levels, which triggers deubiquitination of TRAF6 and downregulation of NF-κB and MAPK activation (particularly JNK). Notably, A20 associates directly with IL-17RA through the receptor’s C-terminal “C/EBPβ activation domain” (CBAD)114. The CBAD is also an interaction site for TRAF3, another negative regulator of IL-17 signaling116, and deletion of the CBAD was shown several years ago in a structure-function analysis of IL-17RA to enhance IL-17 signaling117. Intriguingly, A20 appears to inhibit IL-17 in a noncanonical fashion. In other systems, A20 needs to interact with accessory factors, such as TAX1-binding protein 1 (TAX1BP1) and E3 ubiquitin protein ligase ITCH, in order to hydrolyze K63-linked substrates efficiently118 and deletion of TAXBP1 or ITCH results in a phenotype similar to the lack of A20, albeit less severe119, 120. However, knockdown of either TAX1BP1 or ITCH do not inhibit IL-17 signaling, which suggests that A20 might use alternate adaptors. Indeed, we found that A20 associates with anaphase-promoting complex subunit 5 (APC5)121, 122, which is a subunit of a multi-protein E3 ubiquitin ligase complex. Together, these data show that reversal of K63 ubiquitination on TRAF6 via A20 is an important downregulatory strategy for restricting IL-17-mediated inflammation.
K48-linked ubiquitination and proteasomal degradation of adaptor proteins is another means of regulating inflammatory signaling. IL-17R signaling is inhibited by the Ub ligase complex β-TrCP, which induces K48 ubiquitination on phosphorylated forms of ACT1 that in turn leads to its degradation through the proteasome and consequent dampening of IL-17-dependent signaling123. TRAF proteins also play negative roles in regulating inflammatory signaling pathways by IL-17 124. For example, TRAF3 associates with IL-17RA through a consensus TRAF binding site in the CBAD of the receptor. TRAF3 competes with ACT1 for binding to IL-17RA and thereby dampens IL-17-dependent signals116. Similarly, TRAF4 directly binds Act1 upon IL-17 stimulation and competes for its association with TRAF6. Consistently, TRAF3 and TRAF4 limit IL-17-mediated development of EAE116, 126.
Some data implicate microRNAs in regulating both Th17 development and IL-17 signaling. The microRNA MiR-155 expressed in CD4+ T cells positively regulates Th17 differentiation127, which may be due to blockade of suppressor of cytokine signalling 1 (SOCS1), a negative regulator of JAK/STAT signaling128. Consequently, MiR155−/− mice or mice in which this miRNA is silenced are resistant to EAE127, 129. In terms of signaling, MiR-23b keeps IL-17-induced NF-κB activation in check by targeting the degradation of TAB2/3 and IKKα. Consistently, patients with RA and systemic lupus erythematosus (SLE) show reduced expression of miR-23b 130.
As noted above, C/EBPs regulate IL-17-induced gene expression. Interestingly, C/EBPβ is both a positive and negative regulator of the IL-17 pathway. Although C/EBPβ can positively transactivate IL-17 target genes104, C/EBP phosphorylation of C/EBPβ by the kinases Erk and GSK3β correlates with a decrease in IL-17-induced gene expression. Notably, phosphorylation through GSK3β is dependent on the CBAD, another negative regulatory event mediated through the IL-17RA distal domain 108.
Effector cytokines of Th17 cells
Th17 cells in steady state
Naturally, Th17 cells did not evolve to cause autoimmunity, but to provide effective host defense against pathogens. Experiments using IL-17- and IL-17R-knockout mouse models have demonstrated increased susceptibility to a wide variety of infectious organisms, including bacteria, parasites, fungi and viruses. However, examination of families or individuals with specific mutations in the Th17 pathway indicated a surprisingly limited role for Th17 cells and IL-17 in immunity to infections, with susceptibility to commensal fungi (mainly the commensal Candida albicans) by far the dominant disease seen in these settings (Summarized in Table 1). Although the reasons for the dichotomy between mice and humans are still unclear, this is similar to findings in MyD88-deficient humans, who show a relative restricted susceptibility to pyogenic bacteria, in contrast to the broadly susceptible MyD88-knockout mice131.
Table 1.
Human mutations that impact the Th17 pathway
| Gene | Syndrome | Disease susceptibility |
Connection to Th17 pathway |
Refs |
|---|---|---|---|---|
| STAT3 | Autosomal dominant- Hyper-IgE Syndrome (Job’s Syndrome) |
Mucocutaneous candidiasis Staph aureus Cold abcesses Viral susceptibility Etc. |
Reduced Th17 frequency, defects in IL-6, IL- 23, IL22 signal transduction |
67, 175 |
| TYK2 | Autosomal recessive- Hyper-IgE Syndrome (Job’s Syndrome) |
Mucocutaneous candidiasis Staph aureus Cold abcesses Other? |
Loss of IL-23 signaling (presumed) |
176 |
| IL-12p40 | Reduced Th17 (and Th1) frequency |
177 | ||
| IL12RB1 | Reduced Th17 (and Th1) frequency |
178, 179 | ||
| IL17F | Mucocutaneous candidiasis Staph aureus? |
Dominant negative mutation in IL-17F that blocks IL-17F and IL-17A/F signaling |
138 | |
| IL17RA | Mucocutaneous candidiasis Staph aureus |
Loss of function in IL-17RA |
138 | |
| Dectin-1 | Mucocutaneous candidiasis |
Induces Th17 differentiation in response to β- glucans |
175 | |
| CARD9 | Mucocutaneous and disseminated candidiasis |
Induces Th17 differentation in response to fungal PAMPs via CLRs |
133 | |
| STAT1 | Mucocutaneous candidiasis Viral susceptibility? |
Reduced Th17 frequency, increased IL-27 and Type I IFN signaling |
134, 135 | |
| AIRE | APS-1 | Endocrinopathy, mucosal candidiasis |
Neutralizing Abs against Th17 cytokines (IL-17A, IL-17F and/or IL- 22) |
136, 137 |
| ACT1 | Mucocutaneous candidiasis |
139 | ||
| IL17RC | Mucocutaneous candidiasis |
Loss of function in IL- 17RC |
J-L Casanova and A Puel, personal communication |
As noted above, patients with AD-HIES have autosomal dominant-negative mutations in STAT3 that are associated with reduced frequency of Th17 cells and susceptibility to chronic mucocutaneous candidiasis (CMC), Staphylococcus aureus infections and pulmonary infections66, 132. The reduced frequency of Th17 cells in CMC is associated with mutations that impair fungal sensing (CARD9), IL-23 signaling (IL12RB1, IL12B) or enhance negative regulation of Th17 differentiation (STAT1) 133–135. Similarly, CMC but not other infections occurs patients with APS-1/APECED, who have naturally-occurring neutralizing antibodies against Th17 cytokines136, 137. Null mutations in IL17RA or ACT1 and dominant-negative mutations in IL17F also promote CMC and some staphylococcal infections138, 139. Therefore, deficiencies in Th17 and/or IL-17 are strongly linked to defects in fungal immunity and these association studies support data from mouse studies while revealing the apparent resilience of the human immune system in the face of immunodeficiency140.
Th17 cells in disease states
While Th17 cells did not evolve to cause disease, they are programed to provide formidable host protective responses—often associated with rapid recruitment and activation of granulocytes and macrophages capable of producing tissue damaging reactive oxygen species (ROS) 141. During prolonged and dysregulated exposure to IL-1 and IL-23, Th17 cells recruit inflammatory myeloid cells to cause severe local tissue injury35, 142. Experiments using Il17-, Il22-, Il23a- or Il23ra-deficient mice as well as antibody-mediated inhibition have demonstrated a requirement for these cytokines in EAE, CAIA, CIA, IBD, AS, and psoriasis disease models4, 5, 25, 142, 143. Based on these preclinical models and human GWAS studies and pharmacogenomic disease association analyses, psoriasis, IBD, and AS have emerged as the leading disease indications for anti-IL-17 and anti-IL-23 treatments54–58, 144.
Psoriasis is an immune-mediated inflammatory disorder characterized by the activation of both adaptive and innate Type-17 responses145. The pathogenic involvement of IL-23 in psoriasis is supported by the increased expression of IL-23 in psoriatic lesional skin and its ability to induce psoriasiform characteristics in a preclinical model of intradermal IL-23 administration143. Injection of IL-23 promoted dermal acanthosis, neutrophil recruitment, and infiltration of IL-17- and IL-22-producing T cells. In addition, inhibition of IL-23 led to significant reduction of IL-17 and IL-22 in psoriatic lesional skin, which supports that these cytokines are key regulatory factors for this disease. IL-17-producing CD4+ T cells, CD8+ T cells, γδT cells and ILCs can all be found in the psoriatic skin146. Additional skin resident cells such as keratinocytes, fibroblasts and endothelial cells respond to IL-17 and produce autocrine factors that promote rapid cell division and neo-angiogenesis, which strongly contribute to the disease process. Inappropriate secretion of anti-microbial peptides (such as β-defensins, lipocalin, LL-37, S100A7) and chemokines (CXCL 1,2,3,5, IL-18, CCL20) by keratinocytes further recruit and activate mast cells, neutrophils, and inflammatory macrophages147. This interaction between Type-17 lymphoid cells and tissue resident keratinocytes/fibroblasts creates amplification loops that drive epidermal hyperplasia and pro-inflammatory conditions—all of which are hallmarks of psoriasis pathogenesis.
The seronegative spondyloarthropathies (SPA) are a heterogeneous group of immunological diseases that follows specific types of viral and bacterial infections and include ankylosing spondylitis, psoriatic arthritis and the ‘reactive’ arthritis148. In contrast to rheumatoid arthritis where inflammation is accompanied by bony erosion and destruction, spondyloarthropathy is characterized by new bone formation where ligaments attach onto bone—the entheseal organ. We have recently uncovered the presence of RORγt+ IL-23R+CD3+CD4-CD8-Sca1+ entheseal resident cells and that their production of IL-17 and IL-22 are the major cause of STAT3-dependent osteoblast-mediated bone remodeling142. The new bone appears as inappropriate bony outgrowths from spinal vertebrae, which can fuse and immobilize the spine causing the ultimate morbidity for this disease149. This condition is commonly associated with uveitis, aortic valve inflammation, psoriasis and inflammatory bowel disease. Indeed, uveitis accounts for significant comorbidity in ankylosing spondylitis, affecting 40% of patients. In addition, up to 70% of patients with ankylosing spondylitis have microscopically evident bowel inflammation150. These observations suggest that there is a fundamental underlying pathology linking these immune disorders. Recently GWAS have identified multiple SNPs in IL-23R, which are associated with either protection from ankylosing spondylitis, psoriasis, IBD or increased susceptibility to these autoimmune conditions54–58.
The same IL-23R SNPs found in individuals with ankylosing spondylitis and psoriasis were present in patients with Crohn’s disease along with Nod2 / Card15 (sensor for bacterial cell wall)54, 151, 152. This is consistent with the body’s immune system having an elevated Th17 response attacking the intestinal mucosa, possibly against commensal bacterial antigens. Preclinical animal studies have also identified IL-23 as a key inflammatory mediator that promotes chronic tissue injury responses. In Crohn’s disease, increased expression of IL-17A mRNA and intracellular protein in the intestinal mucosa has been reported153. Elevated fecal IL-17A levels were also described in active Crohn’s disease154. Taken together, these observations implicate the IL-23-IL-17 immune axis in the pathogenesis for Crohn’s disease and the related immune disorders such as psoriasis and spondyloarthropathies and provide a rationale for targeted-antibody therapy.
Targeting the Th17 pathway in disease
IL-17 and IL-23 are emerging as essential factors in the disease process of many autoimmune disorders. Currently, a number of pharmaceutical companies are testing antibody drugs targeting IL-17A, IL-17RA, IL-17A/IL-17F (an antibody recognizing common motifs of the 2 related cytokines), IL-17A/TNF (bi-specific antibody with one CDR recognizing IL-17A and the other recognizing TNF), p40 subunit of IL-23 and IL-12, as well as p19 subunit of IL-23—in psoriasis, rheumatoid arthritis, ankylosing spondylitis, psoriatic arthritis, autoimmune uveitis, asthma, and multiple sclerosis (Table 2). To date, the antibody drugs that specifically neutralize IL-23 or IL-17A have shown remarkable effectiveness for the treatment of psoriasis. These agents are also showing initial promising results in AS and MS.
Table 2.
human diseases being treated with anti-p40, anti-p19, anti-IL-17, and anti-IL-17RA
| Company | Agent | Target | Indications | Stage | Clin Trial ID |
|---|---|---|---|---|---|
| Eli Lilly | Ixekizumab (Ly2439821) |
IL-17A | Psoriasis Rheumatoid arthritis |
Phase 3 Ph 2 complete |
NCT01597245 NCT00966875 |
| Novartis | Secukinmab (AIN457) |
IL-17A | Psoriasis Rheumatoid arthritis Ankylosing spondylitis Psoriatic arthritis Asthma Multiple sclerosis Type 1 Diabetes Crohn’s disease |
Phase 3 Ph 3 Ph 3 Phase 3 Ph 2 Ph 2 Ph 2 Ph 2terminated |
NCT01544595 NCT01770379 NCT01358175 NCT01892436 NCT01478360 NCT01874340 NCT02044848 NCT01009281 |
| Amgen/ MedImmun e |
Brodalumab (AMG 827) |
IL-17 Receptor A |
Psoriasis Psoriatic arthritis Asthma Crohn’s disease |
Phase 3 Ph 3 Ph 2 Ph 2suspended |
NCT01708590 NCT02024646 NCT01902290 NCT01150890 |
| Abbott AbbVie |
ABT-122 | IL-17A/ TNFa |
Rheumatoid arthritis | Phase 1 | NCT01853033 |
| Johnson & Johnson Janssen Biotech |
Stelara (Ustekinumab) (CNTO 1275) |
p40 subunit of IL-12 and IL-23 |
Psoriasis Crohn’s disease Ankylosing spondylitis Rheumatoid arthritis Psoriatic arthritis Multiple sclerosis GvHD Atopic dermatitis |
Approved 2009 Phase 3 Phase 2 Phase 2 Phase 2 Phase 2 Phase 2 Phase 2 |
approved NCT01369329 NCT01330901 NCT01645280 NCT01009086 NCT00207727 NCT01713400 NCT01945086 |
| Abbott | Briakinumab ABT-874 |
p40 subunit of IL-12 and IL-23 |
Crohn’s disease Psoriasis Multiple Sclerosis |
Ph 2terminated Phase 3 Phase 2 |
NCT00562887 NCT00626002 NCT00086671 |
| Merck | Tildrakizumab (MK 3222) (SCH 900222) |
IL-23p19 | Psoriasis | Phase 3 | NCT01729754 |
| Johnson & Johnson Janssen Biotech |
Guselkumab CNTO 1959 |
IL-23p19 | Psoriasis Rheumatoid arthritis |
Phase 2 Phase 2 |
NCT01483599 NCT01645280 |
| Amgen/ MedImmun e |
AMG 139 | IL-23p19 | Psoriasis Crohn’s disease |
Phase 1 Phase 1 |
NCT01094093 NCT01258205 |
| Eli Lilly | LY3074828 | IL-23p19 | Psoriasis | Phase 1 | NCT01947933 |
| Boehringer Ingelheim |
BI 655066 | IL-23p19 | Ankylosing spondylitis Crohn’s disease Psoriasis (single rising dose) |
Phase 2 Phase 2 Phase 2 |
NCT02047110 NCT02031276 NCT02054481 |
Currently, four companies are in the final stages of clinical testing of anti-IL-23 and anti-IL-17 for psoriasis (Table 3). The phase 2 proof of concept studies for Ixekizumab (anti-IL-17A), Secukinumab (anti-IL-17A), Brodalumab (anti-IL-17RA), and Tildrakizumab (anti-IL-23p19)155–158 showed that inhibition of IL-17, IL-17RA, or IL-23p19 had similar safety profiles and therapeutic efficacy (Table 3), comparable or better than current standard of care drugs such as ustekinumab (anti-p40 of IL-12 and IL-23) and etanercept (anti-TNF)159. All agents showed impressive efficacy with 70–80% of the patients achieving PASI-75, which is defined as having >75% disease clearance, and nearly half reaching PASI-100, a remarkable 100% disease clearance (Table 3). Currently, all four compounds have advanced to Phase 3 trials. These clinical data support the concept that IL-17 and IL-23 are key “check points” for the psoriatic inflammatory skin conditions.
Table 3.
Primary Psoriasis clinical end point in 4 separate Phase II dose-ranging studies
| Phase II Studies | Dosage | Primary endopoint: PASI 75 (% patients) |
Secondary endopoint: PASI 100 (% patients) |
Refs |
|---|---|---|---|---|
| Ixekizumab (anti-IL-17A) n=142 |
5 × Placebo 5 × 10 mg 5 × 25 mg 5 × 75 mg 5 × 150 mg |
8 29 77 83 82 |
0 0 17 38 39 |
155 |
| Secukinmab (anti-IL-17A) n=404 |
3 × Placebo 3 × 25 mg 3 × 75 mg 3 × 150 mg |
9 18 57 81 |
155 | |
| Brodalumab (anti-IL-17RA) n=198 |
3 × Placebo 6 × 70 mg 6 × 140 mg 6 × 210 mg 3 × 280 mg |
0 33 77 82 67 |
0 10 38 62 29 |
155 |
| Tilrakizumab (anti-IL-23p19) n=350 |
6 × Placebo 6 × 5 mg 6 × 25 mg 6 × 100 mg 6 × 200 mg |
2 33 58 62 74 |
158 |
The primary endpoint of the studies is percent of patients with >75% disease clearance as defined by the Psoriasis Area and Severity Index (PASI-75). All four therapeutic agents showed treatment response rate of 74% to 83% for PASI-75 at doses ranging from 25 to 100 mg/kg. The Placebo groups showed 0% to 9% response rates for PASI-75. Two studies also reported a secondary endpoint of PASI-100 (defined as 100% disease clearance). Patients treated with Ixekizumab (150 mg/kg) and Brodalumab (210 mg/kg) showed 38% and 62% PASI-100, respectively.
Conventional treatment for Crohn’s disease includes immunosuppression with corticosteroids, methotrexate and anti-TNF agents. Many patients became secondary failures due to intolerance or loss of initial response (for example development of anti-drug antibody). Ustekinumab, which targets the p40 unit of both IL-12 and IL-23, was tested in patients with moderate to severe Crohn’s disease160. The study showed that patients with elevated C reactive protein (CRP), colonic inflammation, and primaly anti-TNF ‘failures’ had better overall treatment response rates. These results are consistent with the concept of targeting the IL-23-IL-17 immune axis may reduce disease burden in patients with Crohn’s and suggest that targeting additional check points along this pathway may be effective.
Based on preclinical studies of anti-IL-17 blockade in IBD and the encouraging ustekinumab study, two groups evaluated anti-IL-17 in IBD. Unexpectedly, trials of Brodalumab (anti-IL-17RA) and Secukinumab (anti-IL-17A) for Crohn’s Disease were terminated due to lack of efficacy and/or disease exacerbation161, 162. None of the anti-IL-17RA treatment groups showed improvement in Crohn’s Disease Activity Index response compared to placebo. Moreover, results showed a disproportionate worsening of disease in anti-IL-17RA-treated patients. A higher frequency of fungal infections was observed in patients on Secukinumab (9%) compared with placebo (0%), consistent with the possibility that inhibition of IL-17 may increase susceptibility to CMC (see above). However, it is unclear why this condition appeared in Crohn’s disease but not psoriasis patients. To compound the situation, post hoc analysis revealed that the subgroup of anti-IL-17A-treated patients with disease exacerbation had elevated inflammatory markers including CRP and fecal calprotectin (Reg3g), which are known targets of IL-23 and IL-22. It is intriguing that blockade of IL-17 results in mucosal tissue damage associated with up-regulation of IL-23 and RORγt-dependent inflammatory signals. One plausible explanation is that the basal levels of IL-17 may be gut protective. Indeed, Il17-/-mice have increased gut mucosal permeability, predisposing to higher incidence and severity of IBD 163 perhaps due to impaired tight junction integrity (Tato and Cua, unpublished observation). These findings may explain differences between targeting IL-23 p40/p19—which leaves tonic levels of IL-17 intact—versus complete ablation of IL-17.
Conclusions and perspectives
Since the discovery of the IL-23-Th17 immune pathway a decade ago, immunologists and clinicians have worked diligently to bring this novel therapeutic strategy to the clinic, which is now showing encouraging results for psoriasis, Crohn’s disease, rheumatoid arthritis, psoriatic arthritis, and ankylosing spondylitis160, 164. However, this treatment strategy is complex. It was initially assumed that IL-23 controls the production of pathogenic IL-17 and that these cytokines are ‘duplicate’ targets. Recent clinical results suggest that is not the case at all. We are now beginning to appreciate that anti-IL-23p19 versus anti-IL-17 treatments each has its own beneficial effects as well as unique challenges in different disease settings. For example, anti-IL-17 showed good therapeutic efficacy for the treatment of psoriasis—even surpassing anti-TNF therapy159,165, but failed in Crohn’s disease. The search for better clinical efficacy biomarkers is critically needed to improve patient stratification and disease indication selection. In addition, better understanding of Th17 biology and cellular mechanisms would allow discovery of additional targets for inflammatory diseases.
Progress in deciphering IL-17R-mediated signal transduction cascades has advanced considerably in recent years. Data indicate that, although these signaling pathways in many respects overlap with IL-1R and TLR-mediated pathways, there are also distinct processes that seem to be IL-17 specific. Understanding these events, particularly those that are unique to the IL-17 pathway, may reveal novel treatment strategies. Similarly, new insights into the epigenetics and transcriptional control of Th17 cells have revealed new treatment paradigms. A novel approach could be through destabilizing the Th17 lineage to induce reprogramming or functional suppression that could have immense therapeutic potential. Indeed, suppression of JAK and TYK2-dependent STAT3 activation166, 167 as well as direct inhibition of RORγt—using small molecule pharmacologic agents—have demonstrated impressive efficacy in preclinical disease models168–171. It is only a matter of time before these new therapeutic approaches will begin to make a difference in the clinic. The discovery IL-23-IL-17 immune axis has thus brought about fundamental changes in cellular immunology and more importantly improved the quality of life for many patients.
Acknowledgments
COI- SLG has received a research grant from Novartis; travel reimbursements and honoraria from Novartis, Amgen and Janssen; consults for Novartis. RJ and DJC are employed by Merck and Co.
Glossary terms
ChIP-Seq
- Hyper-IgE syndrome
Inherited immune deficiency usually due to mutations in STAT3 and associated with reduced Th17 cell frequency. Disease is characterized by elevated IgE and eosinophilia, “cold” staphylococcal abscesses, eczema, pulmonary infections and CMC.
- APS-1/APECED
Autoimmune polyendocrine syndrome/autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome. Inherited autoimmune disorder due to mutations in the AIRE gene targeting multiple endocrine tissues. Disease is associated with a high incidence of mucocutaneous candidiasis, thought to arise due to neutralizing auto-antibodies against IL-17A, IL-17F and/or IL-22.
- Antimicrobial peptides
short peptides (typically 12–50 amino acids) with bactericidal and fungicidal activities. Some may also exhibit chemotactic activities.
- A20
Gene product of the Tumor necrosis factor alpha induced protein 3 (TNFAIP3) gene. Zinc finger protein with E3 ligase and deubiquitinase activity that downregulates signaling by multiple inflammatory effectors.
- microRNAs
Single-stranded RNA molecules of approximately 21–23 nucleotides in length that regulate the expression of other genes.
psoriasiform
Crohn's disease
References
- 1. Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. • This study shows IL-23 controls a key check point for induction of autoimmune inflammation.
- 2.Oppmann B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 3.Kastelein RA, Hunter CA, Cua DJ. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annual review of immunology. 2007;25:221–242. doi: 10.1146/annurev.immunol.22.012703.104758. [DOI] [PubMed] [Google Scholar]
- 4. Langrish CL, et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. The Journal of experimental medicine. 2005;201:233–240. doi: 10.1084/jem.20041257. • This is the first study to suggest IL-17-producing cells are critical mediators of autoimmunity, which led to proposal of the Th17 hypothesis.
- 5.Murphy CA, et al. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. The Journal of experimental medicine. 2003;198:1951–1957. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nature immunology. 2005;6:1123–1132. doi: 10.1038/ni1254. • This study coined the term "Th17" cells as a unique lineage that is STAT-3 dependent rather than STAT-4 and STAT-6 independent.
- 7. Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nature immunology. 2005;6:1133–1141. doi: 10.1038/ni1261. • This is one of this first papers suggesting existence of IL-17-producing inflammatory T cells.
- 8. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. • The landmark paper proposing the Th1- Th2 hypothesis.
- 9.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annual review of immunology. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 10.Parham C, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 11.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 Family Cytokines and the Expanding Diversity of Effector T Cell Lineages. Annual review of immunology. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 12. Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. • This study discribes the discovery of a novel transcrptional regulator that controls IL-17A expression and provide the definitive proof that Th17 cells belong to a new lineage of CD4+ T helper cells.
- 13.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nature reviews. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 14.Annunziato F, Cosmi L, Liotta F, Maggi E, Romagnani S. Type 17 T helper cells-origins, features and possible roles in rheumatic disease. Nat Rev Rheumatol. 2009;5:325–331. doi: 10.1038/nrrheum.2009.80. [DOI] [PubMed] [Google Scholar]
- 15.Zuniga LA, Jain R, Haines C, Cua DJ. Th17 cell development: from the cradle to the grave. Immunological reviews. 2013;252:78–88. doi: 10.1111/imr.12036. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, et al. Natural and inducible TH17 cells are regulated differently by Akt and mTOR pathways. Nature immunology. 2013;14:611–618. doi: 10.1038/ni.2607. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Marks BR, et al. Thymic self-reactivity selects natural interleukin 17-producing T cells that can regulate peripheral inflammation. Nature immunology. 2009;10:1125–1132. doi: 10.1038/ni.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 19.El-Behi M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nature immunology. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nature immunology. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 21.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nature immunology. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 22.Hirota K, et al. Plasticity of Th17 cells in Peyer's patches is responsible for the induction of T cell-dependent IgA responses. Nature immunology. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esplugues E, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chackerian AA, et al. Neutralization or absence of the interleukin-23 pathway does not compromise immunity to mycobacterial infection. Infection and immunity. 2006;74:6092–6099. doi: 10.1128/IAI.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, et al. Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest. 2006;116:1317–1326. doi: 10.1172/JCI25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman LA, et al. IL-23 provides a limited mechanism of resistance to acute toxoplasmosis in the absence of IL-12. J Immunol. 2004;173:1887–1893. doi: 10.4049/jimmunol.173.3.1887. [DOI] [PubMed] [Google Scholar]
- 27.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 28.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 29. Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. • The Betelli et al, Mangan et al and Veldhoen et al papers showed the importance of TGFβ plus IL-6 in the lineage specification of Th17 cells.
- 30.Yang XO, et al. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. The Journal of biological chemistry. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 31.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samoilova EB, Horton JL, Hilliard B, Liu TS, Chen Y. IL-6-deficient mice are resistant to experimental autoimmune encephalomyelitis: roles of IL-6 in the activation and differentiation of autoreactive T cells. J Immunol. 1998;161:6480–6486. [PubMed] [Google Scholar]
- 33.Alonzi T, et al. Interleukin 6 is required for the development of collagen-induced arthritis. The Journal of experimental medicine. 1998;187:461–468. doi: 10.1084/jem.187.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutton C, Brereton C, Keogh B, Mills KH, Lavelle EC. A crucial role for interleukin (IL)-1 in the induction of IL-17-producing T cells that mediate autoimmune encephalomyelitis. The Journal of experimental medicine. 2006;203:1685–1691. doi: 10.1084/jem.20060285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung Y, et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–587. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gulen MF, et al. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity. 2010;32:54–66. doi: 10.1016/j.immuni.2009.12.003. • IL-1 is a key factor that provides key competitive advantage for in vivo Th17 cell expansion and survival during inflammatory conditions by enabling catabolic energy pathways.
- 37.Veldhoen M, Hocking RJ, Flavell RA, Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nature immunology. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 38.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Gutcher I, et al. Autocrine transforming growth factor-beta1 promotes in vivo Th17 cell differentiation. Immunity. 34:396–408. doi: 10.1016/j.immuni.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das J, et al. Transforming growth factor beta is dispensable for the molecular orchestration of Th17 cell differentiation. The Journal of experimental medicine. 2009;206:2407–2416. doi: 10.1084/jem.20082286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volpe E, et al. A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nature immunology. 2008;9:650–657. doi: 10.1038/ni.1613. [DOI] [PubMed] [Google Scholar]
- 42.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nature immunology. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nature immunology. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 44.Wilson NJ, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nature immunology. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 45.Chen Z, Tato CM, Muul L, Laurence A, O'Shea JJ. Distinct regulation of interleukin-17 in human T helper lymphocytes. Arthritis and rheumatism. 2007;56:2936–2946. doi: 10.1002/art.22866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, et al. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. The Journal of experimental medicine. 2009;206:1549–1564. doi: 10.1084/jem.20082584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McGeachy MJ, et al. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nature immunology. 2009;10:314–324. doi: 10.1038/ni.1698. • This paper describes the critical roles of IL-23 for the in vivo expansion and function of Th17 cells during inflammation.
- 49.Haines CJ, et al. Autoimmune memory T helper 17 cell function and expansion are dependent on interleukin-23. Cell reports. 2013;3:1378–1388. doi: 10.1016/j.celrep.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 50.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kebir H, et al. Preferential recruitment of interferon-gamma-expressing TH17 cells in multiple sclerosis. Annals of neurology. 2009;66:390–402. doi: 10.1002/ana.21748. [DOI] [PubMed] [Google Scholar]
- 52. Hirota K, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nature immunology. 2011;12:255–263. doi: 10.1038/ni.1993. • This elegant study used Il17a-Cre × Rosa26-flox-stop-eYFP fate mapping strategy to demonstrate existence of ex-Th17 cells driving autoimmune pathology.
- 53.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nature immunology. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science (New York, N.Y. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS genetics. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reveille JD, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nature genetics. 2010;42:123–127. doi: 10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lock C, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nature medicine. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 58.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nature genetics. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Diveu C, et al. IL-27 blocks RORc expression to inhibit lineage commitment of Th17 cells. J Immunol. 2009;182:5748–5756. doi: 10.4049/jimmunol.0801162. • IL-27 is a remarkable inhibitor of the Th17 immune pathway. This work explores the mechanisms underlying IL-27 regulation of inflammation.
- 61.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 62.El-behi M, et al. Differential effect of IL-27 on developing versus committed Th17 cells. J Immunol. 2009;183:4957–4967. doi: 10.4049/jimmunol.0900735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 64.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 65.Apetoh L, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nature immunology. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Milner JD, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Minegishi Y, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 68.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature immunology. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lohoff M, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nature immunology. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 72.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. • This study argues against RORgt as a single factor regulating Th17 lineage specificity.
- 74.Oestreich KJ, Weinmann AS. Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nature reviews. 2012;12:799–804. doi: 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morrison PJ, et al. Th17-cell plasticity in Helicobacter hepaticus-induced intestinal inflammation. Mucosal immunology. 2013;6:1143–1156. doi: 10.1038/mi.2013.11. [DOI] [PubMed] [Google Scholar]
- 82.Iwakura Y, Ishigame H, Saijo S, Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 83.Yao Z, et al. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 84.Ishigame H, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 85.Rickel EA, et al. Identification of functional roles for both IL-17RB and IL-17RA in mediating IL-25-induced activities. J Immunol. 2008;181:4299–4310. doi: 10.4049/jimmunol.181.6.4299. [DOI] [PubMed] [Google Scholar]
- 86.Bordon Y. Cytokines: IL-17C joins the family firm. Nature reviews. 2011;11:805. doi: 10.1038/nri3118. [DOI] [PubMed] [Google Scholar]
- 87.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 88.Qian Y, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nature immunology. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 89.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. The Journal of biological chemistry. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 90.Li X. Act1 modulates autoimmunity through its dual functions in CD40L/BAFF and IL-17 signaling. Cytokine. 2008;41:105–113. doi: 10.1016/j.cyto.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 91.Liu C, et al. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Science signaling. 2009;2 doi: 10.1126/scisignal.2000382. ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwandner R, Yamaguchi K, Cao Z. Requirement of tumor necrosis factor-associated factor (TRAF)6 in interleukin 17 signal transduction. J. Exp. Med. 2000;191:1233–1239. doi: 10.1084/jem.191.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun D, et al. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing regulatory factor SF2 (ASF) Nature immunology. 2011 doi: 10.1038/ni.2081. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herjan T, et al. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J Immunol. 2013;191:640–649. doi: 10.4049/jimmunol.1203315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bulek K, et al. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nature immunology. 2011;12:844–852. doi: 10.1038/ni.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Qu F, et al. TRAF6-Dependent Act1 Phosphorylation by the IkappaB Kinase-Related Kinases Suppresses Interleukin-17-Induced NF-kappaB Activation. Mol Cell Biol. 2012;32:3925–3937. doi: 10.1128/MCB.00268-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang C, et al. The psoriasis-associated D10N variant of the adaptor Act1 with impaired regulation by the molecular chaperone hsp90. Nature immunology. 2013;14:72–81. doi: 10.1038/ni.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sonder SU, et al. IL-17-induced NF-{kappa}B Activation via CIKS/Act1: PHYSIOLOGIC SIGNIFICANCE AND SIGNALING MECHANISMS. The Journal of biological chemistry. 2011;286:12881–12890. doi: 10.1074/jbc.M110.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huffmeier U, et al. Common variants at TRAF3IP2 are associated with susceptibility to psoriatic arthritis and psoriasis. Nature genetics. 2010;42:996–999. doi: 10.1038/ng.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shen F, Ruddy MJ, Plamondon P, Gaffen SL. Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol. 2005;77:388–399. doi: 10.1189/jlb.0904490. [DOI] [PubMed] [Google Scholar]
- 101.Acosta-Rodriguez EV, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nature immunology. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 102.Yang D, et al. b-Defensins: Linking innate immunity and adaptive immunity through dendritic and T cell CCR6. Science (New York, N.Y. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 103.Goetz DH, et al. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Molecular cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 104.Ruddy MJ, et al. Functional cooperation between interleukin-17 and tumor necrosis factor-a is mediated by CCAAT/enhancer binding protein family members. The Journal of biological chemistry. 2004;279:2559–2567. doi: 10.1074/jbc.M308809200. [DOI] [PubMed] [Google Scholar]
- 105.Shen F, Hu Z, Goswami J, Gaffen SL. Identification of common transcriptional regulatory elements in interleukin-17 target genes. The Journal of biological chemistry. 2006;281:24138–24148. doi: 10.1074/jbc.M604597200. [DOI] [PubMed] [Google Scholar]
- 106.Patel DN, et al. Interleukin-17 stimulates C-reactive protein expression in hepatocytes and smooth muscle cells via p38 MAPK and ERK1/2-dependent NF-kappaB and C/EBPbeta activation. The Journal of biological chemistry. 2007;282:27229–27238. doi: 10.1074/jbc.M703250200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen F, et al. IL-17 Receptor Signaling Inhibits C/EBP{beta} by Sequential Phosphorylation of the Regulatory 2 Domain. Science signaling. 2009;2 doi: 10.1126/scisignal.2000066. ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Onishi R, Gaffen SL. IL-17 and its Target Genes: Mechanisms of IL-17 Function in Disease. Immunology. 2010;129:311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zrioual S, et al. Genome-wide comparison between IL-17A- and IL-17F-induced effects in human rheumatoid arthritis synoviocytes. J Immunol. 2009;182:3112–3120. doi: 10.4049/jimmunol.0801967. [DOI] [PubMed] [Google Scholar]
- 111.Shen F, Gaffen SL. Structure-function relationships in the IL-17 receptor: Implications for signal transduction and therapy. Cytokine. 2008;41:92–104. doi: 10.1016/j.cyto.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Karlsen JR, Borregaard N, Cowland JB. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. The Journal of biological chemistry. 2010;285:14088–14100. doi: 10.1074/jbc.M109.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhong B, et al. Negative regulation of IL-17-mediated signalling and inflammation by the ubiquitin-specific protease USP25. Nature immunology. 2012 doi: 10.1038/ni.2427. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Garg AV, Ahmed M, Vallejo AN, Ma A, Gaffen SL. The deubiquitinase A20 mediates feedback inhibition of interleukin-17 receptor signaling. Science signaling. 2013;6 doi: 10.1126/scisignal.2003699. ra44. • Zhong, B. et al. and Garg, A. V. et al. reports show that IL-17R signaling is restrained by multiple deubiquitinating enzymes that target TRAF6
- 115.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nature reviews. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu S, et al. Modulation of experimental autoimmune encephalomyelitis through TRAF3-mediated suppression of interleukin 17 receptor signaling. The Journal of experimental medicine. 2010;207:2647–2662. doi: 10.1084/jem.20100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maitra A, et al. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc. Natl. Acad. Sci, USA. 2007;104:7506–7511. doi: 10.1073/pnas.0611589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shembade N, Harhaj EW. Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cell Mol Immunol. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Iha H, et al. Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-kappaB activation. EMBO J. 2008;27:629–641. doi: 10.1038/emboj.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shembade N, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nature immunology. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 121.Garg A, Gaffen SL. IL-17 Signaling and A20: A Balancing Act. Cell Cycle. 2013 doi: 10.4161/cc.26699. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ho AW, et al. The Anaphase-Promoting Complex Protein 5 (AnapC5) associates with A20 and inhibits IL-17-mediated signal transduction. PLoS One. 2013;8:e70168. doi: 10.1371/journal.pone.0070168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shi P, et al. Persistent stimulation with interleukin-17 desensitizes cells through SCFbeta-TrCP-mediated degradation of Act1. Science signaling. 2011;4 doi: 10.1126/scisignal.2001653. ra73. [DOI] [PubMed] [Google Scholar]
- 124.Xie P. TRAF molecules in cell signaling and in human diseases. J Mol Signal. 2013;8:1–31. doi: 10.1186/1750-2187-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hartupee J, et al. IL-17 signaling for mRNA stabilization does not require TNF receptor-associated factor 6. J Immunol. 2009;182:1660–1666. doi: 10.4049/jimmunol.182.3.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zepp JA, et al. Cutting edge: TNF receptor-associated factor 4 restricts IL-17-mediated pathology and signaling processes. J Immunol. 2012;189:33–37. doi: 10.4049/jimmunol.1200470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.O'Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yao R, et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One. 2012;7:e46082. doi: 10.1371/journal.pone.0046082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–2221. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhu S, et al. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nature medicine. 2012;18:1077–1086. doi: 10.1038/nm.2815. • This is the first identification of a microRNA feedback loop in IL-17R signaling.
- 131.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science (New York, N.Y. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ma CS, et al. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. The Journal of experimental medicine. 2008;205:1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Glocker EO, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. The New England journal of medicine. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu L, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. The Journal of experimental medicine. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.van de Veerdonk FL, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. The New England journal of medicine. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 136.Puel A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. The Journal of experimental medicine. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kisand K, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. The Journal of experimental medicine. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Puel A, et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science (New York, N.Y. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Boisson B, et al. A biallelic ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity. 2013;39:676–686. doi: 10.1016/j.immuni.2013.09.002. • Puel, A. et al. and Boisson, B. et al. link mucosal Candida albicans infections directly with the IL-17R-mediated signaling axis.
- 140.Hernández-Santos N, Gaffen SL. Th17 cells in immunity to Candida albicans. Cell Host Microbe. 2012;11:425–435. doi: 10.1016/j.chom.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stark MA, et al. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–294. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 142.Sherlock JP, et al. IL-23 induces spondyloarthropathy by acting on ROR-gammat+ CD3+CD4-CD8- entheseal resident T cells. Nature medicine. 2012;18:1069–1076. doi: 10.1038/nm.2817. [DOI] [PubMed] [Google Scholar]
- 143.Chan JR, et al. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2-dependent mechanisms with implications for psoriasis pathogenesis. The Journal of experimental medicine. 2006;203:2577–2587. doi: 10.1084/jem.20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tonel G, et al. Cutting edge: A critical functional role for IL-23 in psoriasis. J Immunol. 2010;185:5688–5691. doi: 10.4049/jimmunol.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Perera GK, Di Meglio P, Nestle FO. Psoriasis. Annual review of pathology. 2012;7:385–422. doi: 10.1146/annurev-pathol-011811-132448. [DOI] [PubMed] [Google Scholar]
- 146.Villanova F, et al. Characterization of Innate Lymphoid Cells in Human Skin and Blood Demonstrates Increase of NKp44+ ILC3 in Psoriasis. The Journal of investigative dermatology. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guttman-Yassky E, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol. 2008;181:7420–7427. doi: 10.4049/jimmunol.181.10.7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rudwaleit M, et al. The Assessment of SpondyloArthritis International Society classification criteria for peripheral spondyloarthritis and for spondyloarthritis in general. Annals of the rheumatic diseases. 2011;70:25–31. doi: 10.1136/ard.2010.133645. [DOI] [PubMed] [Google Scholar]
- 149.van Echteld I, et al. Identification of the most common problems by patients with ankylosing spondylitis using the international classification of functioning, disability and health. The Journal of rheumatology. 2006;33:2475–2483. [PubMed] [Google Scholar]
- 150.Mielants H, et al. The evolution of spondyloarthropathies in relation to gut histology. II. Histological aspects. The Journal of rheumatology. 1995;22:2273–2278. [PubMed] [Google Scholar]
- 151.Cotterill L, et al. Replication and meta-analysis of 13,000 cases defines the risk for interleukin-23 receptor and autophagy-related 16-like 1 variants in Crohn's disease. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 24:297–302. doi: 10.1155/2010/480458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lesage S, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. American journal of human genetics. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fujino S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dige A, et al. Increased levels of circulating Th17 cells in quiescent versus active Crohn's disease. Journal of Crohn's & colitis. 2013;7:248–255. doi: 10.1016/j.crohns.2012.06.015. [DOI] [PubMed] [Google Scholar]
- 155.Leonardi C, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. The New England journal of medicine. 2012;366:1190–1199. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 156.Rich P, et al. Secukinumab induction and maintenance therapy in moderate-to-severe plaque psoriasis: a randomized, double-blind, placebo-controlled, phase II regimen-finding study. The British journal of dermatology. 2013;168:402–411. doi: 10.1111/bjd.12112. [DOI] [PubMed] [Google Scholar]
- 157. Papp KA, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. The New England journal of medicine. 2012;366:1181–1189. doi: 10.1056/NEJMoa1109017. • Clinical trials reveal the remarkable efficacy of anti-IL-17/IL-17RA therapy for treatment of psoriasis
- 158.Papp K. e.a. Dose-dependent improvement in chronic plaque psoriasis following treatment with anti-IL-23p19 humanized monoclonal antibody (MK-3222). Presented at: American Academy of Dermatology Annual meeting; March 1–5, 2013; Miami Beach, Florida. [Google Scholar]
- 159.Chiricozzi A, Krueger JG. IL-17 targeted therapies for psoriasis. Expert opinion on investigational drugs. 2013;22:993–1005. doi: 10.1517/13543784.2013.806483. [DOI] [PubMed] [Google Scholar]
- 160.Sandborn WJ, et al. Ustekinumab induction and maintenance therapy in refractory Crohn's disease. The New England journal of medicine. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 161.Hueber W, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. 2012;61:1693–1700. doi: 10.1136/gutjnl-2011-301668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Targan SR. #Mo2083: A Randomized, Double-Blind, Placebo-Controlled Study to Evaluate the Safety, Tolerability, and Efficacy of AMG 827 in Subjects With Moderate to Severe Crohn’s Disease. Presented at: the 2012 Digestive Disease Week Annual Meeting; May 19–22, 2012; San Diego. [Google Scholar]
- 163.Ogawa A, Andoh A, Araki Y, Bamba T, Fujiyama Y. Neutralization of interleukin-17 aggravates dextran sulfate sodium-induced colitis in mice. Clinical immunology (Orlando, Fla. 2004;110:55–62. doi: 10.1016/j.clim.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 164.Patel DD, Lee DM, Kolbinger F, Antoni C. Effect of IL-17A blockade with secukinumab in autoimmune diseases. Annals of the rheumatic diseases. 2013;72(Suppl 2):ii116–ii123. doi: 10.1136/annrheumdis-2012-202371. [DOI] [PubMed] [Google Scholar]
- 165.Garber K. Anti-IL-17 mAbs herald new options in psoriasis. Nature biotechnology. 2012;30:475–477. doi: 10.1038/nbt0612-475. [DOI] [PubMed] [Google Scholar]
- 166.Nakamura R, et al. Tyk2-signaling plays an important role in host defense against Escherichia coli through IL-23-induced IL-17 production by gammadelta T cells. J Immunol. 2008;181:2071–2075. doi: 10.4049/jimmunol.181.3.2071. [DOI] [PubMed] [Google Scholar]
- 167.Ishizaki M, et al. Tyk2 is a therapeutic target for psoriasis-like skin inflammation. International immunology. 2013 doi: 10.1093/intimm/dxt062. [DOI] [PubMed] [Google Scholar]
- 168.Solt LA, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huh JR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xiao S, et al. Small-Molecule RORgammat Antagonists Inhibit T Helper 17 Cell Transcriptional Network by Divergent Mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lee JS, Cua DJ. The Emerging Landscape of RORgammat Biology. Immunity. 2014;40:451–452. doi: 10.1016/j.immuni.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 172.Stritesky GL, Jameson SC, Hogquist KA. Selection of self-reactive T cells in the thymus. Annual review of immunology. 2012;30:95–114. doi: 10.1146/annurev-immunol-020711-075035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annual review of immunology. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 174.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 175.Plantinga TS, et al. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2009;49:724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 176.Minegishi Y, et al. Human tyrosine kinase 2 deficiency reveals its requisite roles in multiple cytokine signals involved in innate and acquired immunity. Immunity. 2006;25:745–755. doi: 10.1016/j.immuni.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 177.Prando C, et al. Inherited IL-12p40 deficiency: genetic, immunologic, and clinical features of 49 patients from 30 kindreds. Medicine. 2013;92:109–122. doi: 10.1097/MD.0b013e31828a01f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.de Beaucoudrey L, et al. Revisiting human IL-12Rbeta1 deficiency: a survey of 141 patients from 30 countries. Medicine. 2010;89:381–402. doi: 10.1097/MD.0b013e3181fdd832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ouederni M, et al. Clinical features of Candidiasis in patients with inherited interleukin 12 receptor beta1 deficiency. Clin Infect Dis. 2014;58:204–213. doi: 10.1093/cid/cit722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Langrish CL, et al. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunological reviews. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]