Abstract
The objective of this analysis was to examine the genetic architecture of diverse cognitive abilities in children and adolescents, including the magnitude of common genetic effects and patterns of shared and unique genetic influences. Subjects included 3,689 members of the Philadelphia Neurodevelopmental Cohort, a general population sample of ages 8-21 years who completed an extensive battery of cognitive tests. We used genome-wide complex trait analysis (GCTA) to estimate the SNP-based heritability of each domain, as well as the genetic correlation between all domains that showed significant genetic influence. Several of the individual domains suggested strong influence of common genetic variants (e.g. reading ability, h2g=0.43, p=4e-06; emotion identification, h2g=0.36, p=1e-05; verbal memory, h2g=0.24, p=0.005). The genetic correlations highlighted trait domains that are candidates for joint interrogation in future genetic studies (e.g. language reasoning and spatial reasoning, r(g)=0.72, p=0.007). These results can be used to structure future genetic and neuropsychiatric investigations of diverse cognitive abilities.
Keywords: Genetics, Cognition, GCTA, Heritability
Introduction
Twin and family studies have consistently evidenced strong genetic influence on ‘g,’ or general intelligence, as well as several specific cognitive domains (1-3). Recent studies using population samples have estimated that common SNPs alone explain 22-46% of variance in intelligence in childhood and 40-51% of the same differences in adulthood (4-7).
There have been few molecular genetic investigations of the unique cognitive domains that may link brain systems to intelligence and behavior—for example: memory, social cognition, executive function, and reasoning. The purpose of these analyses was to examine the genetic architecture of diverse cognitive abilities in children, including the magnitude of common genetic effects and patterns of shared and unique genetic influences.
First, we use genome-wide complex trait analysis(8) (GCTA) to estimate the extent to which the traits are influenced by all common SNPs (chip heritability). The heritability of several of these traits has been previously estimated using twin and family studies (1, 2, 9-11), but a consistent molecular genetic approach has not been used across domains in a single pediatric sample. In doing so, this study aims to highlight cognitive traits, and measurement methods, that are amenable to genetic study in pediatric samples. In addition to nominating phenotypes for GWAS and sequencing analysis, traits with evidence of genetic influence may be strong candidates for endophenotypic research in neuropsychiatric disease. Schizophrenia and autism, for example, have been linked to familial deficits in several neuropsychiatric trait domains (12, 13). As knowledge of the genetic factors relevant to those disorders continues to improve (14), heritable traits measured on samples from the general population can be examined for evidence of shared genetic influence with neuropsychiatric disease and pursued for indicators of common underlying biology.
Second, we estimate genetic correlations between the cognitive traits that show evidence of common variant influence. While it has been shown that some genetic factors influence multiple cognitive abilities (i.e. “generalist genes”) (15-17), weak within-individual correlations between many domains suggest that other genetic effects are likely to be domain-specific or domain-preferential. A map of the genetic correlations between the traits could inform several types of neuropsychiatric research. For neuroscientists, the genetic correlation matrix, in conjunction with the phenotypic correlation matrix, may provide insight into ability domains most likely to share biological underpinnings. For example, there is a strong genetic correlation between schizophrenia and bipolar disorder, but no genetic correlation between either of those disorders and Crohn's disease (18). The difference between the correlations is hypothesized to reflect greater shared biology between schizophrenia and bipolar disorder, brain diseases associated with aberrant behavior, as compared to Crohn's disease, a disease of the gastrointestinal tract and autoimmune system. A similar gradient in biological proximity may be predicted by genetic correlations among cognitive phenotypes. For researchers conducting GWAS and sequencing analysis, traits with strong genetic correlations can be jointly interrogated (19), thereby increasing the probability of collaborative analysis across cohorts with different phenotypic and measurement approaches. As very large sample sizes are required for association research in cognitive and neuropsychiatric disease (5), cross-cohort collaborations will be needed to identify informative genetic signals.
Subjects and Method
The Philadelphia Neurodevelopmental Cohort
The Philadelphia Neurodevelopmental Cohort (PNC) is a population-based sample from the greater Philadelphia area, including over 9000 individuals ages 8-21 years who received medical care within the Children's Hospital of Philadelphia (CHOP) network. The participants presented for a diverse set of medical needs, ranging from a general health checkup and minor problems (e.g. sports related bruise, rash) to chronic condition management (e.g. asthma, type 1 diabetes) to potentially life threatening health problems (e.g. cancer). They were initially enrolled in the genetic study at the Center for Applied Genomics (CAG) at CHOP. Upon assent/consent, the participants were genotyped during the time of their clinical visit and provided written permission to be recontacted for studies of complex pediatric disorders. The PNC participants were selected at random after stratification by sex, age and ethnicity. The overall inclusion criteria included: 1) ability to provide signed informed consent (parental consent was required for participants under age 18), 2) English language proficiency, and 3) physical and cognitive ability to participate in computerized cognitive testing.
All PNC participants completed the Computerized Neurocognitive Battery (CNB) and were assessed psychiatrically with a structured interview. The CNB consists of tests that have been used in functional neuroimaging to probe specific brain systems and is administered with a web browser. It assesses performance on a range of cognitive tasks. The CNB was designed to capture variation in four ability domains, and includes three specific tasks within each domain: 1) executive control (abstraction and mental flexibility, attention, working memory); 2) episodic memory (verbal, facial, spatial); 3) reasoning (verbal, nonverbal, spatial); and 4) social cognition (emotion identification, emotion differentiation, age differentiation). The specific measurement strategy employed for each of the 12 tasks has been described elsewhere (20), but a summary of the measures and their psychometric properties in the PNC is included in the supplemental material (Table S1). The battery also included the reading items from the Wide Range Achievement Test (WRAT) (21).
Cleaning and imputation of genotype data
This study employed genome wide complex trait analysis (GCTA) to estimate the fractional contribution of common SNPs to phenotypic variation in cognitive ability in the general population. One can reduce bias in values estimated through GCTA by minimizing ancestral heterogeneity in the sample (8, 22). As the PNC cohort was drawn from a diverse United States urban population, these analyses were limited at the outset to the subset of participants who identified themselves as white non-hispanic (WNH; n=5,141). All samples were genotyped on one of three Illumina arrays: the HumanHap550, HumanHap610, or OmniExpress v2. Within the self-described WNH group, population outliers were further excluded based on directly genotyped SNP data, prior to imputation. Data were cleaned using a standard approach (23)(Supplement), which reduced the sample by 584 individuals. Over half (62%) of these individuals were excluded for excess relatedness (the PNC included siblings). We conducted a principal components analysis in PLINK (24) (Figures S1a-S1d) which identified 527 individuals with outlying ethnicity, who were subsequently removed. An additional 341 individuals were removed in further phenotypic and genotypic exclusions, described below, resulting in a final analytic sample of 3,689 individuals.
The genotype data were imputed in a separate phase of the study at CHOP. Unobserved genotypes from each chip set were imputed using the IMPUTE2 package and the reference haplotypes in Phase I of the 1000 genomes data (June 2011 release) that included approximately 37,138,905 variants from 1,094 individuals from Africa, Asia, Europe and the Americas. Methodological details regarding the imputation are provided in the Supplement. The imputed genotype data were used in the GCTA analyses.
Phenotypic analyses
We created an index of the overall severity of participant medical conditions based on information obtained from parent or adult proband (ages 18-21) interviews and electronic medical records (EMR). The index included the following levels: 1) None or Minor (43.4% of analytic sample)- no ongoing medical conditions requiring sustained intervention (e.g. well child visit or sprained ankle); 2) Mild (33.5% of analytic sample)- conditions requiring pediatric visits and at times medications but mild in severity (e.g. asthma, allergies); 3) Moderate (22.8% of analytic sample)-medical conditions requiring standing medications and monitoring (e.g. diabetes; lupus); 4) Severe- medical conditions requiring multiple procedures and monitoring that can be life threatening. Individuals with medical rating 4 (n=274) were excluded from the analysis to avoid conflation of their cognitive ability with the physical symptoms of their medical phenotype, which may influence test-taking performance.
Age of the participants was distributed uniformly between 8 and 21 years (mean=13.7 years). Both age and sex were associated with mean differences in CNB performance (20), and were regressed out of all CNB variables, as well as the WRAT, prior to analysis. Individuals with scores more than four standard deviations from the mean of any CNB variable were designated missing for that variable. Using the age- and sex-regressed scores, we conducted common factor and principal components analyses to examine their correlation structure. Common factor analysis (CFA) analyzes the structure of the shared variance of a variable set; principal components analysis (PCA) examines all observed variance. We generated common factor and principal components scores—continuous variables that indicate individual rank within the standardized distribution of the factor or principal component—for use in the genetic analyses. Both the CFA and PCA analyses used promax rotation, allowing the factors to be correlated. Phenotypic analyses were conducted in SAS version 9.3 (25). An unrotated principal components analysis was conducted for purposes of comparison and yielded similar results (Table S2). Given the strong correlation between the variables, only the rotated factor and principal components scores were included in the genetic analyses. There were 46 individuals missing the cognitive battery who were not included in the analysis.
GCTA
The approach underlying GCTA has been described previously (5, 8, 22). In brief, one uses common SNPs to create a genomic relatedness matrix (GRM) that includes pairwise relatedness estimates for all individuals in a sample. The GRM can be estimated using all typed SNPs (across the genome) or using a particular subset of SNPs, for example those on a specific chromosome. Using the GCTA software tool (8), one compares phenotypic similarity with genotypic similarity, which provides an estimate of the fraction of phenotypic variation attributable to the additive effects of common SNPs (SNP-based heritability). For example, a univariate GCTA estimate of 0.35 suggests that 35% of variation in a trait is attributable to the SNPs used to make the GRM, as well as additional variants with which the typed markers are in linkage disequilibrium (LD; correlated variants). If the GRM was estimated using all genotyped SNPs, that fraction reflects the contribution of all common SNPs; if the GRM was estimated using just the SNPs on a single chromosome, that fraction reflects the specific contribution of common SNPs on that chromosome.
We constructed the primary GRM from all imputed, autosomal SNPs with minor allele frequency greater than 1% and Impute2 info score, an imputation quality metric, greater than 0.6 (n=7,635,695 SNPs) (26). This GRM was used in the primary GCTA analyses. After constructing the GRM, we removed the remaining individuals with relatedness above 0.05 (n=20) (26), which yielded the final analytic sample of 3689 individuals. In the univariate analyses, we first estimated the fraction of phenotypic variation in each cognitive trait explained by all common SNPs. Secondly, we estimated genetic correlations between each of the traits with nominally significant (p<0.05) univariate values. Genetic correlations provide an estimate of the degree to which the common variant influences on two traits are shared. A correlation of 1 suggests that the traits share all of their common genetic influences; a correlation of 0 suggests their influences are independent.
The second set of analyses examined the genomic distribution of genetic influence on the accuracy factor scores. We segmented the 7.6 million SNPs used in the primary GRM into: a) 22 chromosome-specific GRMs and b) genic or intergenic space using three genic window definitions(22). For each gene, all genic footprints were defined using the RefSeq transcript with the longest spliced protein-coding DNA sequence (27). All genome files were adjusted for incomplete LD between tagged and causal SNPs, as described by Yang et al. (2011). All analyses controlled for the first ten principal components of ancestry, which collectively explained less than 2% of phenotypic variation in the common factor score.
Results
Phenotypic Analyses
The common factor analysis suggested that the measures have a single factor solution; only one factor had an Eigenvalue exceeding one (Figures S2a). In PCA, accuracy had a dominant first component but two additional interpretable components with Eigenvalues greater than one. The first principal component consisted of the reasoning and executive function items, along with the WRAT. The second principal component included the social cognition items; the third principal component included the memory items (Figure S2b). Factor loadings for both the full sample and age-specific subgroups, as well as the phenotypic correlations, are included in the Supplement (Table S3a-b).
Univariate GCTA Analysis
Table 1 presents the univariate GCTA results. The individual trait scores suggested a wide range of common variant influence, ranging from 0% (emotion differentiation) to 43% (WRAT) of phenotypic variation explained. The reasoning traits had the strongest SNP-based heritabilities, with estimates between 0.30 (s.e.= 0.10; p=2e-04) and 0.41 (s.e.= 0.10; p=4e-06). The heritability estimates of the factor and principal component scores reflected the results of their component traits—the first principal component score (reasoning/executive function) was the most genetically influenced of the summary measures (0.46; s.e.= 0.11; p=8e-06). Strong common variant influence was also suggested for the common factor score (0.36; s.e.=0.11; p=5e-04). As indicated in Table 1, GCTA estimates for 7 of the outcome variables remained significantly different from 0 following a Bonferroni correction for 17 independent tests, which is conservative given that the outcome variables are correlated.
Table 1. Contribution of common genetic variation to population differences in the Computerized Neurocognitive Battery.
| Variable | N | V(G)/Vp | SE | p |
|---|---|---|---|---|
| Domain Scores | ||||
| Abstraction and Mental Flexibility | 3657 | 0.064 | 0.096 | 0.3 |
| Attention | 3618 | 0.148 | 0.097 | 0.07 |
| Working Memory | 3577 | 0.108 | 0.096 | 0.1 |
| Facial Memory | 3652 | 0.064 | 0.093 | 0.2 |
| Spatial Memory | 3643 | 0.028 | 0.090 | 0.4 |
| Verbal Memory | 3638 | 0.244 | 0.097 | 0.005 |
| Language Reasoning | 3651 | 0.302 | 0.098 | 0.0008* |
| Nonverbal Reasoning | 3654 | 0.406 | 0.096 | 4e-06* |
| Spatial Reasoning | 3581 | 0.357 | 0.101 | 0.0002* |
| Age Differentiation | 3637 | 0.039 | 0.098 | 0.4 |
| Emotion Differentiation | 3650 | 0.000 | 0.092 | >0.5 |
| Emotion Identification | 3661 | 0.357 | 0.093 | 1e-05* |
| Wide Range Achievement Test (Reading) | 3680 | 0.433 | 0.098 | 4e-06* |
| Factor and Principal Component Scores | ||||
| Common Factor Score | 3380 | 0.360 | .108 | 0.0005* |
| PC1 (Reasoning & Executive Function) | 3380 | 0.460 | 0.107 | 8e-06* |
| PC2 (Memory) | 3380 | 0.124 | 0.102 | 0.1 |
| PC3 (Social Cognition) | 3380 | 0.149 | 0.104 | 0.08 |
Note: V(G)/Vp=Estimate of the percentage of phenotypic variation attributable to the additive influence of common single nucleotide polymorphisms; SE=standard error of V(G)/Vp; p= [p(V(G)/Vp=0)]; WRAT= Wide Range Achievement Test; blue shading= executive function; beige= memory; green= complex cognition; purple=social cognition; PC=principal component; gray= reading;
= p<0.05 following conservative correction for 17 independent tests.
Bivariate GCTA Analysis
First, we estimated bivariate correlations between each of the variables with nominally significant (p<0.05) univariate heritability (Table 2). The genetic correlations ranged from -0.25 (emotion identification and nonverbal reasoning; s.e. 0.20; p(rg=0)=0.09) to 1.00 (WRAT and language reasoning; s.e. 0.15; p(rg=0)=2e-07). More than forty percent of variation in the genetic correlations (40.8%) was explained by the phenotypic correlations (p=0.01), which led the variables to cluster genetically in a manner similar to that predicted by the phenotypic factor and principal component analyses. As shown in Figure S3, the genetic correlations were, on average, greater than the phenotypic correlations, reflecting substantial pleiotropy across cognitive traits.
Table 2.
Genetic correlations between variables with nominally significant univariate heritability.
| r(g) (SE) | WRAT | Spatial Reasoning | Nonverbal Reasoning | Language Reasoning | Verbal Memory |
|---|---|---|---|---|---|
| Spatial Reasoning | 0.43 (0.16) p=0.01 | ||||
| Nonverbal Reasoning | 0.23 (0.16) p=0.1 | 0.67 (0.15) 0.0003 | |||
| Language Reasoning | 1.00 (0.15) 2e-07 | 0.72 (0.17) p=0.0007 | 0.43 (0.16) p=0.02 | ||
| Verbal Memory | 0.48 (0.20) p=0.01 | 0.57 (0.23) p=0.009 | 0.53 (0.21) p=0.008 | 0.41 (0.23) p=0.05 | |
| Emotion Identification | 0.40(0.16) p=0.01 | 0.26 (0.18) p=0.08 | -0.25 (0.20) p=0.09 | 0.45 (0.20) p=0.01 | 0.17 (0.22) p=0.2 |
Note: WRAT=Wide Range Achievement Test; SE= standard error of genetic correlation; p= probability of no genetic correlation between the variables.
Second, we correlated the individual tasks (e.g. language reasoning) with the common factor score. These correlations indicate the strength of the genetic relationship between the variable and the factor, and the extent of genetic influences on the variable that are distinct from those on the factor (Table S4). With the exception of the association between emotion identification and the factor score (rg=0.18; s.e. 0.18; p(rg=0)=0.2), each of the genetic correlations exceeded 0.5. However, of the six item-to-factor genetic correlations, four were significantly different from one (p(rg=1)<0.05). This further suggests that while many of the genetic influences on cognitive ability are shared, cognitive domains often also have unique genetic influences.
Genomic partitioning
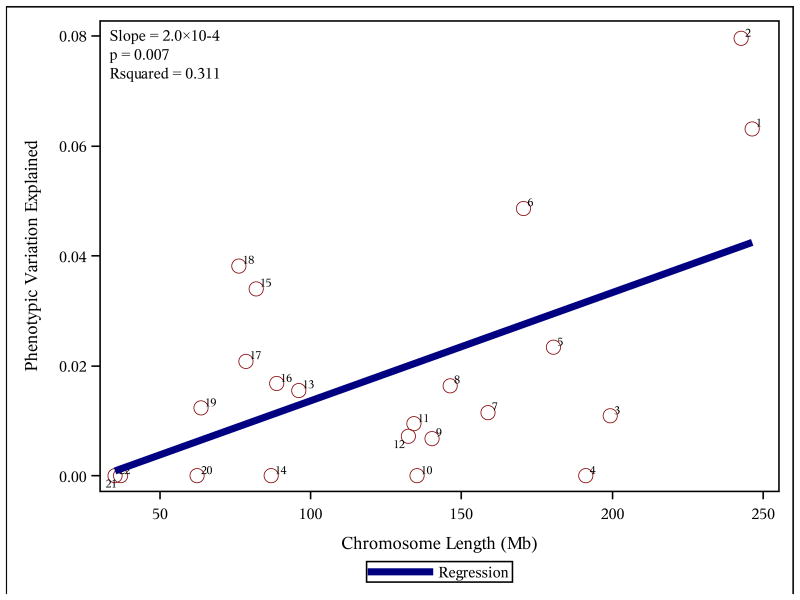
We estimated the association between chromosome length and phenotypic variation explained per chromosome for the common factor score, which suggested that the factor is highly polygenic. Chromosome length explained 31.1% (p=0.007) of variation in GRM prediction (Figure 1). The polygenic model is further supported by the regional analyses. SNPs in both genic and intergenic space were significantly associated with variation in cognitive ability (p(joint association with genic and intergenic regions)=0.009-0.08; Figure S4).
Figure 1. Genetic influences on complex cognition are distributed across the genome.
Note: Mb=megabases; Rsquared= phenotypic variation attributable to chromosome length.
Discussion
These analyses suggest that many cognitive domains are influenced by common genetic variants, and that the domains' degree of genetic overlap is strongly predicted by their phenotypic associations. The more genetically proximal traits in the analysis (e.g. language reasoning and reading) are better candidates for joint interrogation in future genetic and neurobiological studies; the traits with limited association (e.g. emotion identification and nonverbal reasoning) are more likely to reflect largely independent physiological influences.
Several aspects of these analyses may introduce variation between the findings reported here and heritability assessed in a different population. First, SNP-based heritabilities, like any association metric, are influenced by the approach taken to assess the phenotype. These results accordingly reflect both the common genetic influences on cognitive domains, as well as the domains' methods of measurement in the PNC. The measures in this analysis that have lower reliability may accordingly have higher estimated heritability in a sample in which the construct has been assessed differently. While the measures used in this analysis have been previously validated (28), it will be important to evaluate the genetic influences on these constructs using additional assessment approaches.
In a pediatric population, age of the participants is also likely to introduce variation in SNP-based heritabilities. An association between age and the heritability of IQ has been shown in pediatric samples (29); specifically, genetic variation appears to have a greater influence on IQ as children mature. The present analyses should accordingly be interpreted in the context of the participants' age distribution, and mean age of 13.7 years. Despite the broad age distribution of the PNC sample, the univariate heritability results we present for the common factor score, and the domains that are most correlated with intelligence (e.g. reasoning), are similar to those reported for IQ in pediatric populations of similar mean age (5).
The extent to which the genetic correlations may be associated with age is less clear. The phenotypic correlations were strongly associated with the genetic correlations in this analysis; age-based stability in the genetic correlations is accordingly suggested by the consistent phenotypic associations we report across age groups. Further, both twin and molecular genetic studies have suggested that, despite increasing heritability estimates, the genetic influences on intelligence are largely stable over time (6, 30). If the genetic influences on cognition are themselves stable, it would follow for the genetic correlations to be relatively stable as well.
While this study introduces the largest dataset available with this array of cognitive assessments, it was limited by the size of the standard errors in both the univariate heritability and genetic correlation analyses. In particular, the strong relationship between the phenotypic and genetic correlations suggests a nonrandom ranking of genetic correlations. However, the standard errors associated with the genetic correlation estimates make them difficult to compare statistically, either independently or as a set. After multiple testing correction, it is unlikely that many of the genetic correlations would be significantly different from others within Table 2. The results should accordingly be primarily used to highlight pairs of variables with high degrees of genetic sharing (e.g. reading and language reasoning) or variables with strong common genetic influence (e.g. the reasoning and executive function principal component).
These analyses were also limited by the need to reduce the sample to individuals of European ancestry. Future studies will be needed to assess the extent to which the present findings extend to non-European populations. Further analysis will also be necessary to replicate the present findings and examine these patterns for age and sex effects. This sample was not large enough to stratify by age or sex, as we would not have had the statistical power necessary to meaningfully compare any group-specific estimates.
These analyses are among the first to use a molecular genetic approach to evaluate a system of complex cognitive traits in a pediatric sample. The development of genetic relationship matrices among cognitive and behavioral phenotypes introduces an opportunity to more effectively study those traits, and gain insight into the neuropsychiatric disorders existing at the tail of the impairment distributions (31).
Supplementary Material
Acknowledgments
We greatly appreciate the efforts of the participants and families involved in the Philadelphia Neurodevelopmental Cohort. The PNC was supported by NIH RC2 grants MH089983, MH089924. EBR was funded by 1K01MH099286-01A1 from the National Institutes of Mental Health.
Footnotes
Supplementary information is available at Molecular Psychiatry's website.
Conflict of Interest: The authors declare no competing financial interests.
References
- 1.Wadsworth SJ, Corley RP, DeFries JC. Cognitive abilities in childhood and adolescence. In: Finkel D, Reynolds CA, editors. Behavior Genetics of Cognition Across the Lifespan. NY: Springer New York; 2014. pp. 3–40. [Google Scholar]
- 2.Plomin R, DeFries JC, Knopik VS, Neiderhiser JM. Behavioral Genetics. 6th. NY, NY: Worth; 2013. General Cognitive Abilities. [Google Scholar]
- 3.Plomin R, DeFries JC, Knopik VS, Neiderhiser JM. Behavioral Genetics. 6th. NY, NY: Worth; 2013. Specific Cognitive Abilities. [Google Scholar]
- 4.Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16(10):996–1005. doi: 10.1038/mp.2011.85. Epub 2011/08/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ, et al. Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.184. Epub 2013/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482(7384):212–5. doi: 10.1038/nature10781. Epub 2012/01/20. [DOI] [PubMed] [Google Scholar]
- 7.Plomin R, Haworth CM, Meaburn EL, Price TS, Davis OS. Common DNA markers can account for more than half of the genetic influence on cognitive abilities. Psychol Sci. 2013;24(4):562–8. doi: 10.1177/0956797612457952. Epub 2013/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. Epub 2010/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenwood TA, Braff DL, Light GA, Cadenhead KS, Calkins ME, Dobie DJ, et al. Initial heritability analyses of endophenotypic measures for schizophrenia: the consortium on the genetics of schizophrenia. Arch Gen Psychiatry. 2007;64(11):1242–50. doi: 10.1001/archpsyc.64.11.1242. Epub 2007/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stins JF, de Sonneville LM, Groot AS, Polderman TC, van Baal CG, Boomsma DI. Heritability of selective attention and working memory in preschoolers. Behav Genet. 2005;35(4):407–16. doi: 10.1007/s10519-004-3875-3. Epub 2005/06/23. [DOI] [PubMed] [Google Scholar]
- 11.Rietveld MJ, Hudziak JJ, Bartels M, van Beijsterveldt CE, Boomsma DI. Heritability of attention problems in children: longitudinal results from a study of twins, age 3 to 12. J Child Psychol Psychiatry. 2004;45(3):577–88. doi: 10.1111/j.1469-7610.2004.00247.x. Epub 2004/04/02. [DOI] [PubMed] [Google Scholar]
- 12.Lundstrom S, Chang Z, Rastam M, Gillberg C, Larsson H, Anckarsater H, et al. Autism spectrum disorders and autisticlike traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry. 2012;69(1):46–52. doi: 10.1001/archgenpsychiatry.2011.144. Epub 2012/01/04. [DOI] [PubMed] [Google Scholar]
- 13.Gur RE, Nimgaonkar VL, Almasy L, Calkins ME, Ragland JD, Pogue-Geile MF, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164(5):813–9. doi: 10.1176/ajp.2007.164.5.813. Epub 2007/05/04. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–51. doi: 10.1038/nrg3240. Epub 2012/07/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trzaskowski M, Davis OS, DeFries JC, Yang J, Visscher PM, Plomin R. DNA evidence for strong genome-wide pleiotropy of cognitive and learning abilities. Behav Genet. 2013;43(4):267–73. doi: 10.1007/s10519-013-9594-x. Epub 2013/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovas Y, Plomin R. Generalist genes: implications for the cognitive sciences. Trends Cogn Sci. 2006;10(5):198–203. doi: 10.1016/j.tics.2006.03.001. Epub 2006/04/04. [DOI] [PubMed] [Google Scholar]
- 17.Trzaskowski M, Shakeshaft NG, Plomin R. Intelligence indexes generalist genes for cognitive abilities. Intelligence. 2013;41(5):560–5. doi: 10.1016/j.intell.2013.07.011. Epub 2014/02/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45(9):984–94. doi: 10.1038/ng.2711. Epub 2013/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, Nurnberger JI, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371–9. doi: 10.1016/S0140-6736(12)62129-1. Epub 2013/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gur RC, Richard J, Calkins ME, Chiavacci R, Hansen JA, Bilker WB, et al. Age group and sex differences in performance on a computerized neurocognitive battery in children age 8-21. Neuropsychology. 2012;26(2):251–65. doi: 10.1037/a0026712. Epub 2012/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TD, Smith BL. Relationship between the Wide Range Achievement Test 3 and the Wechsler Individual Achievement Test. Psychol Rep. 1998;83(3 Pt 1):963–7. doi: 10.2466/pr0.1998.83.3.963. Epub 1999/01/29. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43(6):519–25. doi: 10.1038/ng.823. Epub 2011/05/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43(10):969–76. doi: 10.1038/ng.940. Epub 2011/09/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. Epub 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inc SI. SAS/STAT® 9.3 User's Guide. Care, NC: SAS Institute Inc; 2011. [Google Scholar]
- 26.Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, et al. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44(3):247–50. doi: 10.1038/ng.1108. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–5. doi: 10.1038/nature11011. Epub 2012/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gur RC, Richard J, Hughett P, Calkins ME, Macy L, Bilker WB, et al. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. Journal of neuroscience methods. 2010;187(2):254–62. doi: 10.1016/j.jneumeth.2009.11.017. Epub 2009/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haworth CM, Wright MJ, Luciano M, Martin NG, de Geus EJ, van Beijsterveldt CE, et al. The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry. 2010;15(11):1112–20. doi: 10.1038/mp.2009.55. Epub 2009/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoekstra RA, Barterls M, Boomsma DI. Longitudinal genetic study of verbal and nonverbal IQ from early childhood to young adulthood. Learning and Individual Differences. 2007;17:97–114. [Google Scholar]
- 31.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–8. doi: 10.1038/nrg2670. Epub 2009/10/28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.