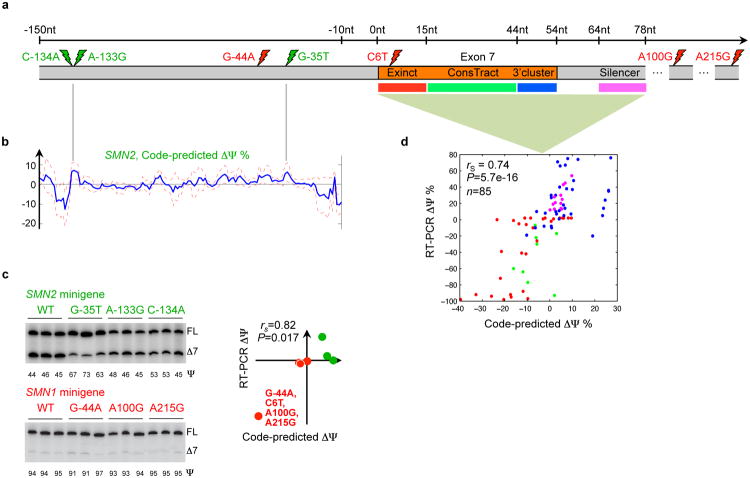
Figure 5. The mutational landscape of spinal muscular atrophy.
(a) Spinal muscular atrophy arises when there is homozygous loss of SMN1 function, but functional protein can be produced by modifying the regulation of SMN2, which differs from SMN1 in four nucleotides (red lightning bolts) and exhibits decreased inclusion of exon 7. (b) Three mutations that the splicing code predicts will increase exon 7 inclusion in SMN2 (green lighting bolts) were selected from predictions for all possible single-nucleotide substitutions 150nt into the intron. These were validated using RT-PCR (c), along with the predicted differences in SMN1 and SMN2 regulation due to three individual substitutions and all four substitutions. Predictions and RT-PCR data have a Spearman correlation of 0.82 (P=0.017, one-sided permutation test). (d) Predicted ΔΨ for 85 individual mutations located in four regions are plotted against RT-PCR-assessed values; the Spearman correlation is 0.74 (P=5.7e-16, one-sided permutation test).