Abstract
CATNAP (Compile, Analyze and Tally NAb Panels) is a new web server at Los Alamos HIV Database, created to respond to the newest advances in HIV neutralizing antibody research. It is a comprehensive platform focusing on neutralizing antibody potencies in conjunction with viral sequences. CATNAP integrates neutralization and sequence data from published studies, and allows users to analyze that data for each HIV Envelope protein sequence position and each antibody. The tool has multiple data retrieval and analysis options. As input, the user can pick specific antibodies and viruses, choose a panel from a published study, or supply their own data. The output superimposes neutralization panel data, virus epidemiological data, and viral protein sequence alignments on one page, and provides further information and analyses. The user can highlight alignment positions, or select antibody contact residues and view position-specific information from the HIV databases. The tool calculates tallies of amino acids and N-linked glycosylation motifs, counts of antibody-sensitive and -resistant viruses in conjunction with each amino acid or N-glycosylation motif, and performs Fisher's exact test to detect potential positive or negative amino acid associations for the selected antibody. Website name: CATNAP (Compile, Analyze and Tally NAb Panels). Website address: http://hiv.lanl.gov/catnap.
INTRODUCTION
Despite more than 30 years of focused scientific efforts worldwide, creating an effective HIV vaccine has proven difficult. HIV is extraordinary variable (1) and designing a vaccine able to stimulate immunological responses that will broadly cross-react with circulating variants is a challenge. Inducing neutralizing antibodies that can block viral infection of a target cell is considered essential for an effective vaccine. Several breakthrough experimental techniques developed in recent years allowed highly efficient interrogation of human memory B cells and plasma cells and consequently the isolation of many broadly neutralizing antibodies (bNAbs) (2). These bNAbs are able to neutralize multiple circulating HIV-1 strains, and large panels of pseudotyped viruses are used to assess the neutralization breadth and potency of these antibodies (3–7).
Neutralization panel data is usually available in the supplemental materials of the published studies and includes IC50, and for some studies IC80, neutralization values (the concentration at which infectivity is reduced by 50% or 80%, respectively (7)) for multiple antibodies and hundreds of viruses. Depending on the study, antibody structure and antibody sequences may also be available. The accumulation of these new monoclonal antibodies together with the vast related neutralization panel information requires storage, comparison and analysis tools. Several databases and servers to organize and access this data have become available in recent years (8–10). The standalone program AntibodyDatabase (8) provides an integrated platform for examining sequence, structure, and neutralization data in a holistic way; however this tool is not a web server. The bNAber database (9) collects neutralization scores and available antibody structures and sequences of the most important bNAbs, and has very useful visualization and analysis tools. The Neutralization-based Epitope Prediction (NEP) server predicts antibody-specific epitopes at the residue level based on neutralization panels of viral strains, using the user's data (10,11). These important web-based resources, however, do not contain the viral sequence data, and so do not readily enable the exploration of how HIV Envelope (Env) sequence variation is correlated with the neutralization sensitivity. The combination of published neutralization scores, antibody sequences, and viral sequences commonly used for the evaluation of neutralizing antibodies, together with initial analysis of bNAb associations with viral sequence mutations has first become available on the web through our new tool CATNAP (Compile, Analyze and Tally NAb Panels), available at the Los Alamos HIV Database. In addition, many large Env panels are published without accession numbers, and with wide discrepancies in sequence names used by different laboratories, making subsequent comparisons between studies and meta-analysis difficult. Given that Los Alamos HIV Database project has a mission of bringing together global HIV sequence and immunology data, we worked with the primary investigators to systematically determine the exact viruses used for neutralization studies in different laboratories. This has enabled an integrated view of HIV Envelope sequences, neutralizing antibody IC50 and IC80 data, and descriptions of HIV Env/antibody contact residues, collected from multiple studies and put into the framework of our database tools and web services. Links to Antibody/Env structures and complete information of antibody sequence data are also provided, and better visualization tools for this data are under development.
In this report, we focus on CATNAP, a web-based portal of neutralizing antibody IC50 and IC80 values in conjunction with viral data, inspired by studies by West et al. (8) and Gnanakaran et al. (12). CATNAP allows users to superimpose neutralization results and virus sequences from published sources and their own data, and perform initial analysis to find potential neutralization antibody signatures. This tool is a part of our larger neutralization antibody resources page at the Los Alamos HIV Database, which includes several other useful sources of information, such as a table of best neutralizing antibodies, a list of external tools for germline antibody reconstruction (13–18), an HIV genome browser, and the neutralizing antibody contexts and features database. The latter is compiled from multiple references and contains coordinates of important neutralizing antibody contact sites (3,4,19–28), mutations affecting neutralization sensitivity (22,29), CD4 contacts (4,26) cytoplasmic tail interactions (30), and antibody sensitivity signature predictions (8,10,11). CATNAP taps into the neutralizing antibody contexts and features database and links to our HIV Immunology and Sequence databases, allowing the rich data collected in these databases to be directly incorporated into the analysis.
MATERIALS AND METHODS
The neutralization panel data (IC50 and IC80 values for specific monoclonal antibodies and pseudotyped viruses) were collected from 49 published neutralization studies, mostly from tables in PDF format provided in the supplemental materials. The viral data was collected from Los Alamos HIV Database and in some cases personal communication with the authors, and required careful consideration and systematization to resolve sequence name ambiguities between different laboratories. Antibody sequences were downloaded from GenBank, and links to the structures in Protein Data Bank are provided. Standard statistics are applied to tally and analyze neutralization results. Specifically, antibody associations with viral mutations are evaluated by Fisher's exact test, counting the number of antibody-resistant or antibody-sensitive viruses (above or below threshold of detection) and the presence or absence of specific amino acids or N-glycosylation motif in the viral sequence alignment position (12).
RESULTS
Data retrieval
CATNAP includes neutralization data from published studies, as well as curated HIV-1 Env alignments corresponding to neutralization panels with carefully standardized virus sequence names. The current collection comprises 172 antibodies and 722 HIV-1 viruses (529 with sequences, and the remaining 193 viruses were published by companies, and so the sequences are proprietary) from 49 published studies, with more being added frequently.
The input page allows the user to retrieve details about selected HIV-1 Env sequences and antibodies used in neutralization panels, retrieve available details about the donor from whom the antibody was isolated, and select viruses and antibodies both individually and by study, for analysis (Figure 1). The antibody details option provides, for the specified antibody(s), links to the corresponding immunology database records, notes, references, links to crystal structures in the Protein Data Bank, antibody donor ID and clonal lineage, data from our Neutralizing Antibody Contexts and Features Database, and heavy and light chain antibody variable region mRNA sequences. The virus details option provides HIV subtype, sampling country, disease stage information, accession number, neutralization tier, and Los Alamos Database comments. (In some cases, the sequence of the virus used in experiments does not exactly correspond to the GenBank sequence. We resolved these issues to the extent possible via personal communication with the authors, and the comments associated with each sequence document whether the GenBank sequence or the unpublished one from the authors is used.) Importantly, a sequence name stored in CATNAP corresponds to the most frequently used name we identified in published neutralization studies. In many cases, however, different names are used in studies by different groups, and they frequently do not exactly correspond to the database sequence name. We provide a list of sequence name aliases and HIV database name for an easy one-to-one correspondence between the various short names, database names and accession numbers, thereby facilitating meta-studies. Finally, a set of accession numbers of the selected viruses can be automatically uploaded to the Sequence Search Interface of our HIV Sequence Database, to obtain information from many additional sequence database fields. The assay details option provides a downloadable table of available IC50 and IC80 data for selected viruses and antibodies from each study. When multiple studies assayed the same antibody–virus combination, both the original data and the geometric mean of the repeated data points is shown.
Figure 1.

Data retrieval. Examples of antibody and virus retrievable and downloadable details are shown. Green arrows and green text shown on Figures 1–3 represent comments added on the figure, but not present on the web site. (A) Antibody details include antibody structures in the Protein Data Bank; clickable donor ID from whom the antibody was isolated, which leads to donor information stored in the HIV database; Env positions related to antibody neutralization; and antibody variable chain sequences. PG9 is the antibody used in the example. The inset on the top of the figure shows an example of Env positions related to PG9 neutralization; Env positions shown in the inset have mutations that affect PG9 and related bNAb sensitivity. Inset on the right shows patient details for the donor from whom the antibody was isolated, and provides the link to HIV-1 sequences from that donor. (B) Virus details include virus name, subtype, country of isolation, patient health and risk status, GenBank accession number, neutralization tier, different names and aliases used in the literature, and our comments.
Analysis page
The analysis page superimposes virus data, neutralization data, and aligned viral sequences on one page. For each virus-antibody pair, the user can see virus information (tier, subtype, country, GenBank accession number, sequence aliases), neutralization values, both per study and the geometric means, and the virus sequences with highlighted N-linked glycosylation motifs (Figure 2). Analysis and additional information are provided under the superimposed data (Figures 2 and 3), and include several options.
Breadth and potency of each antibody/serum over the selected group of viruses are assessed, with potency calculated as a geometric mean of neutralization scores of both antibody-sensitive viruses and all viruses (Figure 2).
List of Env-antibody contact positions is displayed for the selected antibodies, with the link to another HIV database tool, QuickAlign, which aligns these positions to a large set of sequences from the HIV Sequence Database and shows sequence logos (amino acid compositions and frequencies) and summaries of how variable these positions are in HIV circulating sequences (Figure 2).
Antibody signature analysis options highlight a position of interest in the alignment (Figure 2), and for each sequence report the presence or absence of N-glycosylated motif for in this position (Figure 2), extract further information from the HIV databases and perform basic antibody signature analysis on this position (Figure 3). The extracted information includes entropy scores as a measure of amino acid variability in this position in M group, B clade and C clade database alignments, protein features such as functional domains associated with this position, antibody contacts and other NAb-associated features of this position, such as mutations affecting antibody sensitivity, signature predictions, and other features (Figure 3c). Statistical analysis displays the amino acid makeup of that position in the alignment (Figure 3a), the count of viruses with and without N-linked glycosylation motif (Figure 3b), the count of antibody sensitive and resistant viruses for each amino acid or N-glycosylation motif found in this position of the alignment, together with the corresponding Fisher's exact test results to detect a potential positive or negative amino acid signature for the selected antibody (12). In Figure 3b, for example, an N-glycosylation motif at Env position 160 is enriched in bNAb PG9-detected viruses (P < 2.2e−16), indicating, in agreement with published studies (8,31), an association of a glycosylation site at position 160 with increased susceptibility to PG9 neutralization. This analysis should be interpreted with caution, as the counts are not phylogenetically corrected, and significant correlations that are not directly related to antibody sensitivity may arise as a consequence of structure in the phylogenetic tree. Still such associations can be informative, particularly for hypothesis forming or validation of a pre-existing hypothesis. A signature analysis tool that incorporates a phylogenetic correction (12) is in progress at the database.
Figure 2.
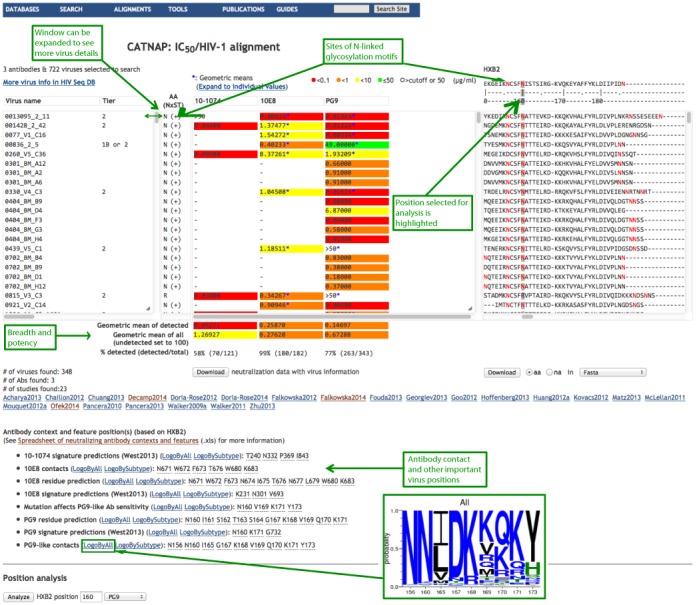
Analysis page. Three aligned windows are shown side-by-side. Each window has vertical and horizontal scroll bars, and each window can be expanded vertically and horizontally by clicking and dragging its right corner. All fields in each window are sortable, and all three windows are sorted simultaneously. For example, clicking on MAb 10E8 in the center window will sort all 3 windows by 10E8 neutralization scores. The left window lists virus names included in the analysis as well as neutralization tier, subtype, country, accession number, aliases (expandable data), and the presence of potential N-linked glycosylation motifs (denoted as NxST). The center window lists antibodies with their color-coded IC50 and/or IC80 neutralization scores. When multiple studies assayed the same antibody–virus combination, the geometric mean of the data points is shown and marked with an asterisk (*). The ‘Expand’ link shows the data points from multiple studies used to calculate the geometric means. A mouseover on each individual geometric mean will also show data-points together with the study references. The right window shows aligned Env sequences, with the HXB2 reference sequence on top. Potential N-glycosylation sites are highlighted in red. Additional information is provided under the three aligned windows above: breadth of neutralization and potency of each antibody over the selected group of viruses (potency is calculated two ways: for sensitive viruses that are below neutralization threshold of detection and for all tested viruses where resistant viruses are set to 100 μg/ml.); antibody contact HIV position(s) and a link to the sequence logo of these positions showing their variability in HIV-1 circulating sequences (see the inset). The option to analyze an individual HIV protein sequence position is on the bottom of the figure (this position is highlighted on the Env alignment in the right window with a gray vertical line). When a position of interest is entered, a new ‘AA NxST’ column appears between the virus and antibody windows. This column shows the amino acid found in each sequence at the position of interest, and the presence of an N-linked glycosylation motif in that position is indicated by ‘+’. N-linked glycosylation motifs are also highlighted in the alignment in red.
Figure 3.

Statistical analysis. (A) Counts of different amino acids at the chosen HXB2 position of ENV are shown, together with the counts of bNAb PG9 sensitive (detected, that is below threshold of detection) and resistant (undetected, that is above threshold of detection) viruses for each amino acid. (B) Counts of viruses with and without an N-linked glycosylation motif at the chosen HXB2 position of Env are shown. In (A) and (B), Fisher's exact test is based on a contingency table of ‘AA/not AA’ (or ‘NxST/not NxST’) and ‘virus sensitive/not sensitive’. (C) Information from the HIV database about the position of interest is shown, and includes entropy calculations using HIV Database tool Entropy and HIV-1 alignments for M group, and B and C subtypes, information about functional domains, and antibody features of this position, such as how mutations in this position affect various bNAbs neutralization sensitivity, signature predictions at this position, mutations affecting antibody binding, etc.
Custom CATNAP
Custom CATNAP allows users to superimpose and analyze any table of numerical data in conjunction with genetic sequences (Figure 2), and to look for associated genetic signatures (Figure 3). Examples include IC50 neutralization values for monoclonal antibodies, like those used in the basic CATNAP tool, or ID50 titers for plasma neutralization data, but the tool can be used for any numerical data (binding kinetic data, functional data) in conjunction with sequence data. As neutralization data frequently use cutoffs, with results below or above a threshold being considered undetected, for the statistical analysis the user should choose the type that describes the most potent scores in the data. For example, IC50 scores for the monoclonal antibodies reflect the concentration needed to achieve 50% neutralization, and so values with a ‘>’ symbol are considered undetected. ID50 scores for the plasma antibodies, on the other hand, reflect the number of dilutions required to reduce the infectivity by 50%, and the high scores are the most potent, so the values with a ‘<’ symbol are considered undetected. Like in basic CATNAP, potential genetic signatures are defined by Fisher's exact test (12). Users can use their own alignments, or can use a premade alignment that contains subsets of Env sequences in the CATNAP tool, including HIV-1 viral strains commonly used in neutralization panels.
DISCUSSION
Overall, we believe CATNAP is a useful tool for the HIV research community, particularly good for integrating and superimposing complete HIV sequence data and broadly neutralizing antibody data across many studies. It provides initial signature analysis on the fly, and it can be used to analyze custom data including genetic sequences in conjunction with numerical data. We consider CATNAP to be a foundation for a new neutralizing antibody relational database, and we plan to expand this resource to contain many more analysis tools and data tables, such as incorporating phylogenetic correction in evaluating antibody signatures in viral sequences (12), listing of germline antibody sequences, antibody variable sequence alignments, HIV-antibody 3D structure visualization tools, and within-patient studies with autologous viral and antibody sequence data.
FUNDING
National Institutes of Health (NIH) [contract AGRAAI1200700101000 HIV/SIV, Database and Analysis Unit (B.T.K., H.Y., J.M., B.F., K.Y.) and grant HIVRAD P01 AI100148 (P.J.B., A.P.W.)]; Bill & Melinda Gates Foundation [Collaboration for AIDS Vaccine Discovery, grant 1032144 (B.T.K., H.Y.) and grant 1040753 (P.J.B., A.P.W.)]. Funding for open access charge: NIH contract AGRAAI1200700101000 through Los Alamos National Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gaschen B., Taylor J., Yusim K., Foley B., Gao F., Lang D., Novitsky V., Haynes B., Hahn B.H., Bhattacharya T., et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 2.Burton D.R., Poignard P., Stanfield R.L., Wilson I.A. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.deCamp A., Hraber P., Bailer R.T., Seaman M.S., Ochsenbauer C., Kappes J., Gottardo R., Edlefsen P., Self S., Tang H., et al. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2014;88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seaman M.S., Janes H., Hawkins N., Grandpre L.E., Devoy C., Giri A., Coffey R.T., Harris L., Wood B., Daniels M.G., et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montefiori D.C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005 doi: 10.1002/0471142735.im1211s64. Chapter 12, Unit 12.11. [DOI] [PubMed] [Google Scholar]
- 8.West A.P. Jr, Scharf L., Horwitz J., Klein F., Nussenzweig M.C., Bjorkman P.J. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc. Natl. Acad. Sci. U.S.A. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eroshkin A.M., LeBlanc A., Weekes D., Post K., Li Z., Rajput A., Butera S.T., Burton D.R., Godzik A. bNAber: database of broadly neutralizing HIV antibodies. Nucleic Acids Res. 2014;42:D1133–D1139. doi: 10.1093/nar/gkt1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang G.Y., Acharya P., Schmidt S.D., Yang Y., Louder M.K., Zhou T., Kwon Y.D., Pancera M., Bailer R.T., Doria-Rose N.A., et al. Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. J. Virol. 2013;87:10047–10058. doi: 10.1128/JVI.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chuang G.Y., Liou D., Kwong P.D., Georgiev I.S. NEP: web server for epitope prediction based on antibody neutralization of viral strains with diverse sequences. Nucleic Acids Res. 2014;42:W64–W71. doi: 10.1093/nar/gku318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnanakaran S., Daniels M.G., Bhattacharya T., Lapedes A.S., Sethi A., Li M., Tang H., Greene K., Gao H., Haynes B.F., et al. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput. Biol. 2010;6:e1000955. doi: 10.1371/journal.pcbi.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kepler T.B. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giudicelli V., Brochet X., Lefranc M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 16.Brochet X., Lefranc M.P., Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaeta B.A., Malming H.R., Jackson K.J., Bain M.E., Wilson P., Collins A.M. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 18.Souto-Carneiro M.M., Longo N.S., Russ D.E., Sun H.W., Lipsky P.E. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 19.Liao H.X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diskin R., Scheid J.F., Marcovecchio P.M., West A.P. Jr, Klein F., Gao H., Gnanapragasam P.N., Abadir A., Seaman M.S., Nussenzweig M.C., et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pejchal R., Doores K.J., Walker L.M., Khayat R., Huang P.S., Wang S.K., Stanfield R.L., Julien J.P., Ramos A., Crispin M., et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falkowska E., Ramos A., Feng Y., Zhou T., Moquin S., Walker L.M., Wu X., Seaman M.S., Wrin T., Kwong P.D., et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 2012;86:4394–4403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blattner C., Lee J.H., Sliepen K., Derking R., Falkowska E., de la Pena A.T., Cupo A., Julien J.P., van Gils M., Lee P.S., et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scharf L., Scheid J.F., Lee J.H., West A.P. Jr, Chen C., Gao H., Gnanapragasam P.N., Mares R., Seaman M.S., Ward A.B., et al. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep. 2014;7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLellan J.S., Pancera M., Carrico C., Gorman J., Julien J.P., Khayat R., Louder R., Pejchal R., Sastry M., Dai K., et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou T., Georgiev I., Wu X., Yang Z.Y., Dai K., Finzi A., Kwon Y.D., Scheid J.F., Shi W., Xu L., et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou T., Xu L., Dey B., Hessell A.J., Van Ryk D., Xiang S.H., Yang X., Zhang M.Y., Zwick M.B., Arthos J., et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong P.D., Wyatt R., Robinson J., Sweet R.W., Sodroski J., Hendrickson W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Rourke S.M., Schweighardt B., Phung P., Mesa K.A., Vollrath A.L., Tatsuno G.P., To B., Sinangil F., Limoli K., Wrin T., et al. Sequences in glycoprotein gp41, the CD4 binding site, and the V2 domain regulate sensitivity and resistance of HIV-1 to broadly neutralizing antibodies. J. Virol. 2012;86:12105–12114. doi: 10.1128/JVI.01352-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postler T.S., Desrosiers R.C. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J. Virol. 2013;87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doria-Rose N.A., Georgiev I., O'Dell S., Chuang G.Y., Staupe R.P., McLellan J.S., Gorman J., Pancera M., Bonsignori M., Haynes B.F., et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]


