Summary
West Nile virus (WNV) causes an acute neurological infection attended by massive neuronal cell death. However, the mechanism(s) behind the virus-induced cell death is poorly understood. Using a library containing 77,406 sgRNAs targeting 20,121 genes, we performed a genome-wide screen followed by a second screen with a sub-library. Among the genes identified, seven genes, EMC2, EMC3, SEL1L, DERL2, UBE2G2, UBE2J1, and HRD1, stood out as having the strongest phenotype, whose knockout conferred strong protection against WNV-induced cell death with two different WNV strains and in three cell lines. Interestingly, knockout of these genes did not block WNV replication. Thus, these appear to be essential genes that link WNV replication to downstream cell death pathway(s). In addition, the fact that all of these genes belong to the endoplasmic reticulum-associated protein degradation (ERAD) pathway suggests that this might be the primary driver of WNV-induced cell death.
Introduction
West Nile virus (WNV) infection has emerged as a serious global health problem, and massive neuronal cell death induced by WNV is one of the major causes of mortality and morbidity. It has been shown that apoptosis occurs after WNV infection, and inhibition of apoptosis can reduce neuronal cell death (Kleinschmidt et al., 2007; Parquet et al., 2001; Samuel et al., 2007; Shrestha et al., 2003). It has also been shown that WNV-induced cell death can occur by either apoptosis or necrosis, depending on the dosage of infection (Chu and Ng, 2003). While the exact mechanism of how WNV triggers these downstream cell death pathways is not clear, it likely involves host factors, since WNV itself encodes only 10 proteins (Brinton, 2014). A common approach to identifying viral host factors is genome-wide functional screening in cultured cells.
Currently, such screens, including several to identify host factors for WNV and other flaviviruses (Gilfoy et al., 2009; Krishnan et al., 2008; Li et al., 2009; Sessions et al., 2009; Yasunaga et al., 2014), are mostly performed using RNAi. However, this approach has several pitfalls: 1) Gene silencing is generally not complete. 2) Silencing efficiency between different siRNAs or shRNAs targeting the same gene can vary dramatically. 3) Off-target effects can perturb unintended targets. Thus, it is not surprising that the results from different RNAi screens may not be concordant. For example, the overlap was poor between three independent genome-wide functional screens using siRNA performed to identify host genes linked to HIV replication (Brass et al., 2008; Konig et al., 2008; Zhou et al., 2008). Although hundreds of genes were identified in each screen, only three genes were identified in all three screens, and these did not include the obvious positive genes CD4 and CXCR4 (R4-tropic HIV challenge) (Bushman et al., 2009). This lack of concordance suggests that further improvement in genome-wide functional screening of cultured cells is needed.
The CRISPR-Cas9 system has recently been developed as a tool for gene editing in mammalian cells (Chang et al., 2013; Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013). This system allows easy knockout of both alleles in a substantial percentage of treated cells (Chang et al., 2013; Cho et al., 2013; Cong et al., 2013; Dicarlo et al., 2013; Gasiunas et al., 2012; Hwang et al., 2013; Jiang et al., 2013; Jinek et al., 2012; Jinek et al., 2013; Mali et al., 2013; Qi et al., 2013). Compared with RNAi-mediated gene silencing, the gene-silencing efficiency of each single-guide RNA (sgRNA) is uniform—in most cases, the target gene is completely eliminated. Thus, the CRISPR system has potential as an improved approach for performing genome-wide screening, as suggested by recent reports (Gilbert et al., 2014; Koike-Yusa et al., 2014; Konermann et al., 2014; Sanjana et al., 2014; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014). Here we have independently developed a method based on this system and identified cellular factors that appear to be essential factors that link WNV replication to downstream cell death pathway(s).
Results
The strategy for identifying genes essential for WNV-induced cell death
We designed a screening strategy to identify genes essential for WNV-induced killing, as illustrated in Figure 1. The exact procedure used is as follows: 1) An sgRNA library was synthesized as an oligo pool, with 77,406 unique sgRNAs covering 20,121 genes in the human genome (Dataset S1). To ensure complete gene disruption, the target sites were chosen in a region close to the translation initiation site, and most genes were covered four times. It is noteworthy that the sgRNAs start with either A or G, since our previous study showed that the preferred transcription initiation nucleotide for the U6 promoter can be either A or G (Ma et al., 2014). 2) The sgRNA oligo pool was PCR-amplified and cloned into the lentiviral vector, pLB. To test its quality, we PCR-amplified the library and analyzed the amplicons with deep sequencing. The results showed that at least 90% of the sgRNAs were retained in the plasmid library (Dataset S1). The library vector pool was then packaged with VSV-G pseudotyping to generate the lentiviral library. 3) The lentiviral sgRNA library was transduced into 293FT cells. To minimize the chance of inserting multiple sgRNAs into the same cell, we employed a low multiplicity of infection (MOI) to obtain a transduction rate of around 5%. To enrich for sgRNA-harboring cells, the transduced cells were sorted using an EGFP marker expressed by the lentiviral vector. 4) Cells harboring the sgRNA library were transfected with a plasmid that transiently expresses Cas9 to disrupt the particular gene targeted by the stably expressed sgRNA. The control groups were transfected with a plasmid that did not express Cas9. 5) The transfected cells were infected with WNV strain B956 and incubated for 12 days to select for cells resistant to virus-induced killing. Numerous colonies could be seen in Cas9-transfected cells, while there were very few colonies in the control group, suggesting that resistance was mostly due to Cas9-mediated gene disruption. 6) The sgRNAs in the surviving cells were PCR-amplified and deep sequenced using Illumina second-generation sequencing to identify the genes that had been targeted. The experiments were repeated three times independently.
Figure 1. Schematic of functional screening for genes that are essential for West Nile virus-induced cell killing.
For the first and second round of screen, the sgRNA library sequences can be found in Dataset S1 and S3,the sgRNAs identified in surviving cells can be found in Dataset S2 and S4 (GEO Accession: GSE69666) respectively; genes identified can be found in S2, S4 and S5; the genes with highest sgRNA reads was summarized in Table S1 and S3 respectively.
Different sizes of colonies, from big colonies with hundreds of cells to small colonies with only a few cells, were observed. Big colonies were likely derived from cells harboring sgRNAs whose target genes are essential for WNV-induced cell death but do not affect cell growth and therefore expand rapidly. As a consequence, the sgRNAs in these colonies should have a high number of reads. By contrast, small colonies were probably derived from cells harboring sgRNAs whose target genes are only partially required for WNV-induced cell death or are partially required for cell survival or growth, and the sgRNAs in these small colonies should have only a few reads. It is also possible that low knockout efficiency for an sgRNA, even one targeting a gene whose knockout confers strong resistance to WNV-induced killing, might limit the number of reads (the colonies will be big for these types of sgRNAs if the gene was knocked out in the founder cell, but because of the low knockout efficiency of the sgRNA, the number of colonies will be fewer). Other reasons, such as off-target effects, could also cause variability in colony size and sgRNA reads. However, in general, a high number of sgRNA reads indicates that these sgRNAs target genes whose knockout confers strong resistance to WNV-induced cell death but does not affect cell growth.
In total, 28,429 sgRNAs with reads numbering more than 10 in three independent experiments were identified (Dataset S2, GEO Accession: GSE69666). Multiple sgRNAs with a high number of reads were seen for three genes, EMC2, EMC3, and SEL1L in each of three independent experiments (Table S1). For example, one SEL1L sgRNA (sgRNA21067) had the highest number of total reads and another two (sgRNA45498 and sgRNA30740) also had high numbers of reads (1437 and 852, respectively), while two EMC3 sgRNAs (sgRNA59423 and sgRNA11892) ranked number 3 and 5, respectively, and two EMC2 sgRNAs (sgRNA70193 and sgRNA59282) ranked number 10 and 29, respectively (Dataset S2 and Table S1). Thus, these three genes might be truly essential to WNV-induced cell death. Similar multiple sgRNAs with high numbers of reads were not seen for most other genes, suggesting that some of these results represent “noise”. To test this hypothesis, we randomly chose five genes with different numbers of sgRNAs identified in three independent experiments, and two sgRNAs were designed for each gene (Table S2). EMC2 was included as a positive control. The sgRNA and Cas9-expressing plasmid were co-transfected with a plasmid expressing a puromycin-resistance gene, and the positively transfected cells were selected with puromycin. The genome-editing efficiency of each sgRNA was determined by amplification of target sites followed by deep sequencing. Most target sites were efficiently modified (Table S2). Indeed, only cells treated with EMC2 sgRNAs showed resistance to WNV-induced killing (Figure S1). In addition, only cells treated with EMC2 sgRNAs grew to confluence after 6 days of virus challenge, while cells treated with other sgRNAs finally died out (data not shown), suggesting that only EMC2 is a truly essential gene and that the other four genes represent noise. Thus, except for EMC2, EMC3, and SEL1L, which have multiple sgRNAs with high numbers of reads, most of the sgRNAs identified could be the result of a high noise level.
To improve data quality and distinguish essential WNV-induced cell death genes from noise, we performed a second round of screening using a sub-library constructed with sgRNAs targeting the genes identified in the first round that had more than five sgRNAs in three independent experiments (Dataset S2) and also some genes identified in a pilot screen (Dataset S3). A previous genome-wide screen using an siRNA library identified 283 host-susceptibility factors that are required for WNV replication (Krishnan et al., 2008). Since inhibition of WNV replication might also render the cells resistant to WNV-induced killing, we also included 50 genes with the highest scores from that screen (Krishnan et al., 2008). We designed a sub-library that contains 9,068 sgRNAs targeting 1,166 genes (eight sgRNAs for each gene, Dataset S3) and performed a second round of screening in the same way as in Figure 1.
Knockout of the seven genes confers almost complete protection against WNV-induced cell death
To identify key genes with strong phenotypes that are dispensable for cellular survival and yet essential for virus-induced host cell killing, we counted only sgRNAs with high numbers of reads (>2000). Six genes, SEL1L, UBE2J1, EMC3, EMC2, DERL2, and UBE2G2, stood out with the highest numbers of sgRNAs in three independent experiments (Table 1 and Dataset S4, GEO Accession: GSE69666) and appear to be truly essential genes, since multiple sgRNAs exhibiting high numbers of reads for these genes occurred in three independent experiments (Table S3 and Dataset S4). It appears that, for a particular gene, the knockout efficiency of different sgRNAs can vary significantly—the sgRNAs with high knockout efficiency tend to have many reads in all three experiments, while sgRNAs with low knockout efficiency tend to have few or no reads in all three experiments (Table S3). For example, only two sgRNAs (out of eight) for UBE2G2 have a high number of reads in three experiments, while the other six sgRNAs have a low number of reads (Table S3), which is consistent with previous studies (Koike-Yusa et al., 2014; Shalem et al., 2014; Wang et al., 2014). Thus, if we looked only at the total number of sgRNAs that appeared in three independent experiments, we might miss the truly essential genes. And, if a given sgRNA is enriched in three independent experiments, it is likely that the gene targeted by this sgRNA is a truly essential gene. According to this rule, we revisited the data from the first round and found that one of the sgRNAs targeting the Hrd1 (SYVN1) gene had a high number of reads in all three experiments, although the total number of sgRNAs identified for this gene was only three. In fact, this sgRNA had the second highest number of total reads (Dataset S2), suggesting that it might also be an essential WNV-induced cell death factor. Thus, we included this gene along with six other genes identified in the second round for validation.
Table 1.
The top genes in second-round screening.
| Gene Name | sgRNAs | Total sgRNAs | ||
|---|---|---|---|---|
| Exp #1 | Exp #2 | Exp #3 | ||
| SEL1L | 7 | 6 | 7 | 20 |
| UBE2J1 | 6 | 6 | 6 | 18 |
| EMC3 | 4 | 6 | 4 | 14 |
| EMC2 | 4 | 4 | 3 | 11 |
| DERL2 | 3 | 5 | 3 | 11 |
| UBE2G2 | 2 | 3 | 3 | 8 |
Only sgRNAs with more than 2000 reads were counted, and only genes with eight or more sgRNAs identified are shown. The raw sgRNA reads data can be found in Dataset S4, GEO Accession: GSE69666.
To validate the above-mentioned seven genes, we derived cells with each of these genes knocked out using two different sgRNAs for each gene. The genome-editing efficiency of the sgRNAs was first determined by deep sequencing of the target sites. A high allele-modification rate (more than 99% for most sgRNAs) was observed (Figure 2A), which is consistent with what was reported earlier (Shalem et al., 2014). We also determined the knockout efficiency using western blot. The results showed that most protein was eliminated (Figure 2B), which is consistent with deep sequencing data for the target sites, indicating that they were mutated in most cells. A small fraction of the indels actually caused in-frame mutations (Data no shown), and thus low levels of the targeted proteins were detected in most samples (Figure 2B).
Figure 2. The genes whose knockout confers resistance to WNV-induced cell death.
(A) The target site modification rate of the sgRNAs. The genes were mainly identified in the second round of screening (Dataset S4, GEO Accession: GSE69666). See also Table S2 showing similar rates of target site modification in another candidate genes validation experiment. Two sgRNAs were designed for each gene and inserted into a pX330 vector in which Cas9 is expressed (Cong et al., 2013). The sgRNA constructs and Cas9-expressing plasmid were co-transfected with a puromycin expression plasmid into 293FT cells, and the cells were selected with puromycin to eliminate the untransfected cells. The target site modification rate was determined by amplification of the target sites followed by deep sequencing. CBLL1 and HMGCR were two previously identified WNV host-susceptibility factors. (B) Western blotting to confirm the knockout efficiency in six of the seven genes in A. Actin or HSP90 is included as inner reference. (C) The cells derived from A were challenged with WNV (strain B956) at MOI=2 after puromycin selection. After incubating for 3 days, cell viabilities were evaluated with the MTT assay. The cell viability was normalized to the respective controls without WNV treatment. Error bar, 1 S.D. (n=3).
See also Figure S1 for similar results.
These cells were then challenged with WNV. A significant portion of the cells survived in EMC2 and EMC3 knockout cells, and most SEL1L, DERL2, UBE2G2, UBE2J1, and Hrd1 knockout cells survived, while very few control cells survived after 3 days of incubation (Figures 2C), suggesting that these genes are essential for WNV-induced-cell death. Two previously identified WNV host-susceptibility factors (HSFs), HMGCR (Mackenzie et al., 2007) and CBLL1 (Fernandez-Garcia et al., 2011; Krishnan et al., 2008), which are required for WNV replication, were included as positive controls but did not confer resistance when targeted (Figures 2C). In addition, cells treated with sgRNAs targeting the seven genes identified in the present study kept growing until confluent, while all of the cells treated with sgRNAs targeting HMGCR and CBLL1 died out over time (data not shown), suggesting that the host genes identified with our strategy are key WNV-induced cell death genes with strong phenotypes, whose knockout is sufficient to confer resistance to WNV-induced cell killing. It is not surprising that knockout of previously identified host factors, such as HMGCR and CBLL1, did not confer resistance, because the basis for their previous identification was different: their knockdown led to reduced WNV replication at certain time points (Krishnan et al., 2008; Mackenzie et al., 2007).
Genes with many sgRNAs with low numbers of reads
If sgRNAs with fewer reads are also counted (>50), many genes would have a substantial number of sgRNAs and, in some cases, a very high number of sgRNAs would be identified (Dataset S5). For example, DYNC2LI1 had 20 sgRNAs identified in three experiments, which is only slightly fewer than EMC2 (22 sgRNAs), SEL1L (22 sgRNAs), or UBE2J1 (21 sgRNAs) (Dataset S5). Having a high number of sgRNAs identified in three independent experiments suggests that these genes might also be truly essential genes required for WNV-induced cell death. However, the low number of reads suggests that the cells harboring these sgRNAs did not expand much, even when incubated for 12 days after WNV challenge. This is actually consistent with what we observed during screening: while big colonies with hundreds of cells could be observed, small colonies with only a few cells could also be seen (data not shown). The reason for this limited expansion might be either that knockout of these genes conferred only partial resistance or that the genes knocked out contribute to normal cell growth. These genes are not our main focus and will not be characterized further in this study. However, it is useful to know that these genes might be involved in WNV replication and/or in WNV-induced cell death.
Knockout of the seven genes did not block WNV replication but repressed WNV-induced cell death
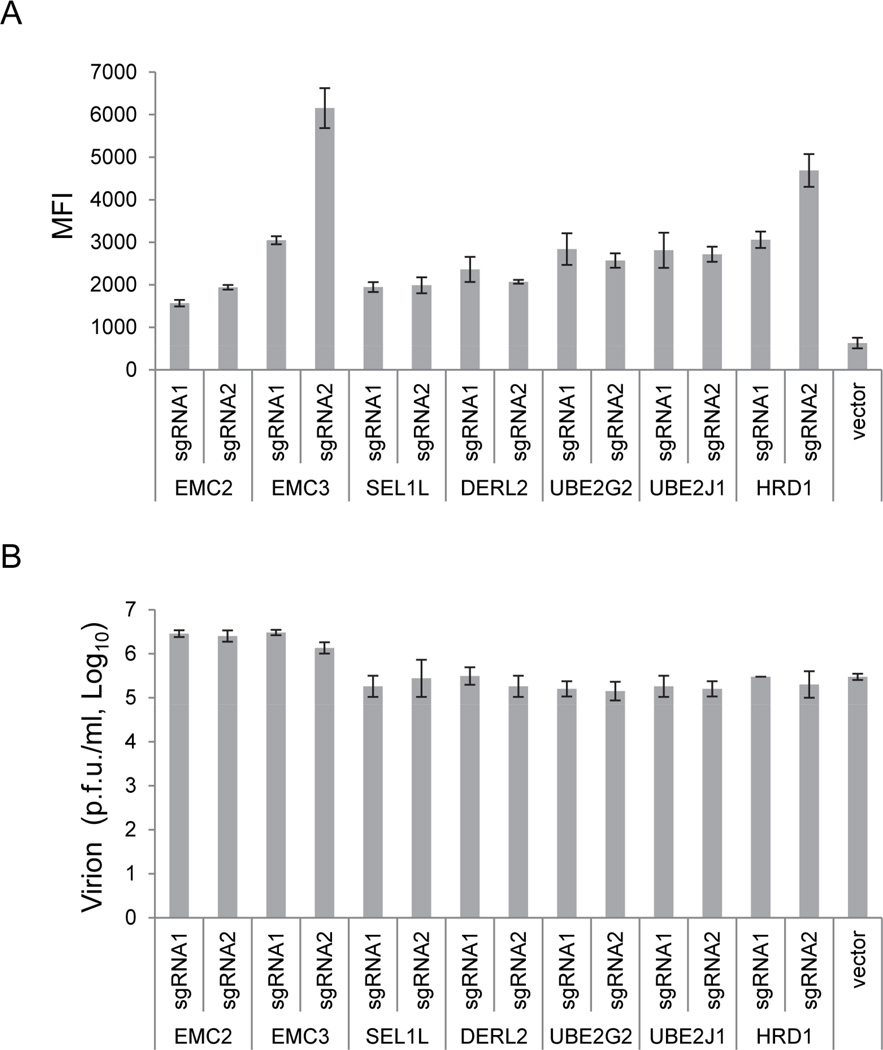
It is possible that the resistance to WNV-induced cell death in cells in which these seven genes were knocked out was due to repression of WNV replication. Therefore, we tested viral replication in the surviving cells after WNV challenge. After 72 hours of WNV challenge, while all of the cells in the control group had died out, the cells treated with sgRNAs targeting the seven genes had grown close to confluency (except EMC2 and EMC3). To test viral replication, the cells were stained with WNV envelope (ENV)-specific antibody and analyzed by flow cytometry. ENV accumulation was seen in all the knockout cells, albeit at varying levels. Interestingly, in all of these cells, the ENV level was higher than the peak level in WT cells (Figures 3A and S2). To confirm active viral replication, three days after infection the cells were reseeded into a new plate and the WNV titers in the supernatant determined at 72 hours by a plaque assay. The culture supernatant from all knockout cells had a viral titer close to the peak level of virus seen in WT cells at 36 hours (all infected WT cells died on day 3), suggesting that WNV production and release were not blocked in these cells (Figure 3B). Thus, knockout of these genes did not repress WNV replication but inhibited cell death induced by WNV replication.
Figure 3. Knockout of the genes (from Figure 2) did not block WNV replication.
(A) Cells derived as in Figure 2A were fixed and stained with WNV envelope (ENV) protein antibody 72 hours after WNV (strain B956) challenge and analyzed by flow cytometry. The mean fluorescence index (MFI) was used to represent the WNV ENV protein level, and see also Figure S2. Vector-transfected cells were included as a control, and the cells were fixed and analyzed at 24 hours post infection, which is at the peak of WNV replication in WT 293FT cells. Error bar, 1 S.D. (n=3). (B) Cells were reseeded into a new 24-well plate. Supernatants were collected 72 hours later, and viral titers were determined by plaque assay. The control reference level was the virus titer in the supernatant of WT cells infected with WNV at 36 hours, which is the peak level of virus in the supernatant. Error bar, 1 S.D. (n=3).
It is interesting that, although the cells were infected with WNV and constantly produced WNV, most appeared to have normal morphology and proliferated to the same extent as uninfected cells, even after a period of months (data not shown). However, EMC2- and EMC3-deficient cells were exceptional in this regard. Although these cells grew to confluency, a portion of dead cells was always observed (data not shown), probably because resistance was not complete or because knockout of EMC2 or EMC3 negatively affected cell viability. Taken together, these results suggest that knockout of these genes converts an acute infection into a chronic infection.
WNV replication in EMC2 or SEL1L knockout sublines
To understand how knockout of these genes affects WNV-induced cell death, we generated EMC2 and SEL1L knockout sublines in 293FT cells (Figure S3 A). It has been shown that, in mammalian cells, Sel1L controls the stability of the E3 ligase Hrd1, and is essential both in vivo and in vitro for cell survival (Sun et al. 2014). However, the SEL1L deficient 293FT cells grew as well as WT cells and appeared to have normal morphology (data not shown). Thus, it appears that dependence on SEL1L for survival is cell type-specific. Consistent with the sgRNA transfection data (Figures 2C), EMC2-deficient cells were partially resistant to WNV-induced killing, while SEL1L-deficient cells were fully resistant to WNV-induced killing (Figures 4A). To exclude the possibility that off-target effects caused the resistance, we performed a rescue experiment by transfecting the EMC2 knockout 293FT subline with an EMC2 expression plasmid. The killing of cells by WNV could indeed be restored by expression of EMC2 (Figure 4B), suggesting that EMC2 knockout and not off-target knockout of unintended genes is responsible for resistance to WNV-induced cell killing. We were not able to perform the same experiment for SEL1L, because transfection of SEL1L-expressing plasmid caused cell death.
Figure 4. WNV replication in EMC2 or SEL1L knockout sublines.
(A) WNV resistance in EMC2- or SEL1L-deficient sublines, which were generated as described in Figure S4. The cells were challenged with WNV (strain B956), and the cell viability was measured with the MTT assay at different time points and normalized to the respective controls without WNV treatment. Error bar, 1 S.D. (n=3). (B) Phenotypic rescue of EMC2 knockout cells. The cells were transfected with EMC expression plasmid and subjected to WNV challenge, and images were taken 72 hours later. Scale bar, 400 µm. WT, wild type cells; KO, knockout cells. (C) Viral replication in EMC2- or SEL1L-deficient cells. The cells were challenged with WNV and fixed and stained with anti-WNV ENV antibody at the indicated time points. The percentage of WNV ENV-positive cells was determined by FACS, and the raw data is found in Figure S4. Error bar, 1 S.D. (n=3). (D) The WNV released in the supernatant. The supernatants were collected at the indicated time points from WNV challenged cells. The virion was titrated by plaque assay and expressed as plaque-forming units per ml (p.f.u./ml). Error bar, 1 S.D. (n=3). (E) The accumulation of WNV ENV protein in WNV-infected cells at the indicated time points. The protein level was represented by the mean fluorescence intensity (MFI), and only ENV-positive cells were considered for calculation. The raw data can be found in Figures S4. Error bar, 1 S.D. (n=3). (F) WNV ENV, nonstructural protein 3 (NS3) and capsid protein levels in EMC- or SEL1L-deficient cells at the indicated time points determined by western blotting. Numbers below each lane indicate the densitometry value in percentage relative to Actin.
Next, we determined the extent of viral replication in knockout cells. The FACS analysis results appear to be consistent with our earlier data in that knockout of either of these two genes did not block viral ENV protein synthesis and virus release into the supernatant, although it appears that viral replication was somewhat delayed (Figures 4C, D). Moreover, viral ENV protein accumulated to similar or even higher levels in EMC2- or SEL1L-deficient cells than in WT cells (Figures 4E). It is interesting that the virus ENV protein level declined after reaching a peak at 24 hours in WT cells, while the ENV protein appeared to accumulate steadily in both knockout cells (Figure 4E). We also checked the protein level of virus envelope, capsid, and nonstructural protein 3 (NS3) with western blotting. Consistent with the flow cytometry data, the ENV level reached a peak at 24 hours and declined at 36 hours in WT cells, while the protein continued to accumulate over time to reach a level higher in knockout cells than WT cells at 36 hours (Figure 4F). Interestingly, both the NS3 and capsid protein levels were significantly lower in knockout cells. Thus, knocking out EMC2 or SEL1L appears to have different effects on ENV compared with capsid and NS3 proteins.
Resistance to the WNV-NY99 strain and Saint Louis encephalitis virus (SLEV)
The WNV-NY99 strain is pathogenic and most prevalent in the USA. Thus, we tested resistance to this strain in cells in which the seven genes had been knocked out individually. The results were consistent with those for the B956 strain, in which knockout of these seven genes conferred resistance to cell death (Figure 5A).
Figure 5. Validating the identified genes with different WNV strains, Saint Louis encephalitis virus (SLEV), as well as Neuro-2a and HeLa cells.
The 293FT knockout cells prepared as in Figure 2 were infected with WNV-NY99 (A) or SLEV (B) at MOI=2. The resistance was determined as in Figure 2C. Neuro-2a and HeLa cells were treated as in Figure 2A to generate knockout cells. The knockout efficiency was determined by western blotting, as shown in Figure S4. The knockout cells were infected with the WNV B956 strain (C, Neuro-2a; E, Hela) or the NY99 strain (D, Neuro-2a; F, Hela), and the resistance was determined as in Figure 2C.
We also tested resistance of the knockout cells to SLEV, which is a member of the flavivirus family and is closely related to WNV. As shown in Figure 5B, the knockout cells showed resistance to SLEV-induced cell death, although to a lesser extent than WNV (Figure 5B).
Validating the identified genes in other cell lines
To exclude the possibility that the identified genes work only in 293FT cells, we tested the effect of knocking out the genes in HeLa cells and also in a more relevant neuronal cell line, Neuro-2a. The cells were transfected with sgRNAs and Cas9-expressing plasmids and selected with puromycin, and the knockout efficiency was confirmed by western blotting (Figure S3 B, C). The cells were challenged with WNV B956 or NY99 strains. For Neuro-2a cells, knockout of these genes conferred resistance to both strains compared to vector control (Figure 5C and D). For Hela cells, knockout of the seven genes also conferred resistance to B956 strain-induced cell death; however, knockout of EMC2 and EMC3 conferred stronger resistance than the other five genes (Figure 5E). For the NY99 strain, only knockout of EMC2 and EMC3 conferred close to 100% resistance to WNV-induced cell death, while knockout of the other five genes conferred minimal or no resistance to WNV-induced cell death compared to vector control (Figure 5F). It is interesting that knockout of CBLL1 conferred minimal but significant resistance to WNV-induced cell death in HeLa cells, in which CBLL1 was previously identified in a genome-wide siRNA library screening (Krishnan et al., 2008). Thus, CBLL1 can serve as a positive control in HeLa cells.
Discussion
Using an independently designed CRISPR-Cas9-based screening method, we identified seven genes that are essential for WNV-induced cell death.
Unlike previously reported CRISPR-Cas9-based screening in which Cas9 was stably expressed (Koike-Yusa et al., 2014; Sanjana et al., 2014; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014), we expressed Cas9 transiently. The reason for using transient expression of Cas9 was that it might reduce any off-target effects that might be amplified by stable expression of Cas9. However, it has been shown that prolonged expression of Cas9 and sgRNA can significantly increase the knockout efficiency (Shalem et al., 2014). Thus, transient expression of Cas9 might have decreased the sensitivity of our system.
Although in the first round of screening we were able to identify several genes, such as SEL1L, EMC2, and EMC3, that are essential for WNV-induced cell death (Table S1, Dataset S2), it is clear that some other genes probably remained unidentified due to the high noise level. While such an unusually high noise level was observed in both rounds of screening for WNV host factors (Dataset S2 and S4), this was not observed in our previous screening for HIV host factors (data not shown). Therefore, the high level of noise might not be a problem with our screening strategy per se. A possible explanation for the high noise level observed in WNV host factor screening is that traces of genomic DNA were somehow left behind by the cells killed by WNV replication. Because millions of cells were used before virus challenge, contamination with even 0.1% of the genomic DNA (integrated with different sgRNAs) would be enough to cause a high noise level. However, by using a sub-library targeting the genes enriched in the first round of screening, we were able to effectively distinguish true hits from noise, and all of the top candidates were validated as essential WNV-induced cell death genes. In a sense, the second-round screening itself served as a large-scale validation of the genes enriched in the first round. Thus, our approach provides a general strategy to increase the specificity and sensitivity of CRISPR-mediated screening, even in the setting of a high noise level.
Defining a good criterion for choosing the sgRNAs and genes from the screening data is critical to improving the data quality. Based on our data, it appears that an sgRNA with a high number of reads across independent experiments is a reliable indication that the targeted gene is essential for WNV-induced cell death. For example, only one sgRNA was identified for the HRD1 gene in the first round of screening; however, this sgRNA had a high number of reads in all three independent experiments. And indeed, HRD1 was validated with two individual sgRNAs to be a truly essential gene for WNV-induced cell death. Thus, reproducibility across biological replicates appears to be a reliable criterion for choosing sgRNAs.
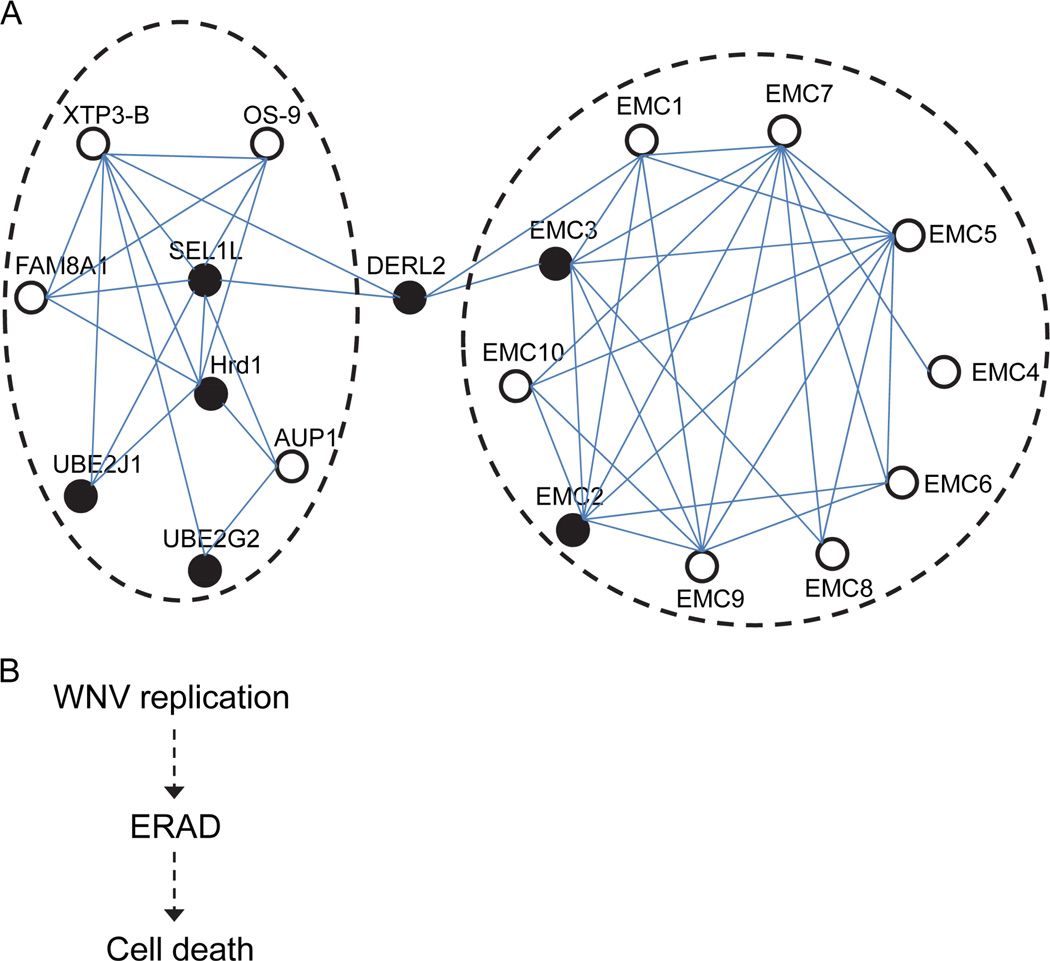
Among the genes identified, seven genes stood out as having the strongest phenotype, whose knockout conferred strong protection against WNV-induced cell death (Figure 2). Interestingly, all of these genes belong to the ERAD pathway and are members of two complexes that interact with each other. While SEL1L, Hrd1, UBE2J1, DERL2, and UBE2G2 belong to the SEL1L–Hrd1 complex, EMC2 and EMC3 belong to the mammalian ER membrane complex (mEMC), a newly discovered complex that is poorly understood (Christianson et al., 2012). These two complexes appear to interact with each other through DERL2 (Christianson et al., 2012) (Figure 6A). It is unlikely to be a coincidence that all of the top genes identified belong to the same complex. Rather, it suggests that our screening strategy with two rounds of screening can identify most, if not all, of the genes involved in a biological process.
Figure 6. The proteins encoded by the seven genes belong to the ERAD pathway and might be the missing connection between WNV replication and the ensuing cell death.
(A) The essential WNV-induced-cell-death factors identified belong to two protein complexes that interact through DERL2. Solid black circles, the identified host factors. Open circles, other components of the two complexes. (B) ERAD might be the missing connection between WNV replication and ensuing cell death.
ERAD is a mechanism for degrading misfolded or misassembled proteins in the ER (Christianson and Ye, 2014). Flaviviruses, including West Nile virus, are known to replicate in close association with ER-derived membrane structures and to induce ER stress (Brinton, 2014), and some components of the ERAD pathway have been shown to play a role in flavivirus replication (Krishnan et al., 2008; Li et al., 2014; Saeed et al., 2011; Sharma et al., 2014) as well as the replication of other viruses (Bennett et al., 2013; Bernasconi et al., 2012; Reggiori et al., 2010; van den Boomen et al., 2014). It is known that WNV replication induces cell death; however, how exactly WNV replication triggers this is unknown. Until now, involvement of the ERAD pathway has been unsuspected. Our study showed that seven genes of the ERAD pathway are essential for activation of the downstream cell death pathway. Knockout of these genes did not prevent WNV replication or release but did block WNV replication-induced cell death (Figures 2–4). Thus, our results suggest that ERAD might be the primary driver of WNV replication-induced cell death and might be the missing connection between WNV replication and ensuing cell death (Figure 6B).
The ER membrane protein complex (EMC) was first identified in yeast, and it appears that loss of EMC leads to accumulation of misfolded proteins, suggesting that they might also be components of ERAD (Jonikas et al., 2009). More recently, Christianson et al. showed that the EMC was also present in mammalian cells and interacted with other ERAD components through DERL2 and UBAC2 (Christianson et al., 2012). However, the exact function of the EMC is poorly understood. Our study revealed that two components of the EMC, EMC2 and EMC3, play important roles during WNV-induced cell death. Knockout of these genes confers strong resistance to different WNV strains and SLEV in different cell lines (Figures 2C and 5). Moreover, it appears that the effect of knocking out EMC2 and EMC3 is different with the other five genes. The effect of EMC2 and EMC3 knockout was weaker in 293FT cells (Figures 2C and 5A) but stronger in HeLa cells (Figure 5E, F, suggesting that the mechanism by which these two groups of genes regulate WNV-induced cell death might be different. It will be interesting to elucidate the exact role of the EMC in WNV-induced cell death.
A previous genome-wide screen using an siRNA library identified 283 host susceptibility factors (HSFs) whose silencing reduced WNV replication. Among these 283 genes, 10 are components of the ERAD pathway. Three of the10, HRD1, Derlin2, and UBE2J1, were also identified in our screening as essential genes whose knockout blocks WNV-induced cell death. It is noteworthy that the previous RNAi study screened for genes whose knockdown caused diminished WNV replication at 24 hours, while we screened for genes whose knockout blocked WNV-induced cell death.
The effect of knocking out the genes we identified was validated with different WNV strains in different cell lines (Figures 2C and 5A, C, D, E, F). In addition to WNV, knockout of these genes also conferred resistance to SLVE (Figure 5B). Thus, the seven genes that we have identified might have a broader role in virus-induced cell death.
In conclusion, seven ERAD family members appear to be essential for WNV infection to cause cell death and in the absence of them, although viral replication continues cells survive. Since massive neuronal cell death is one of the major causes for mortality in WNV infection, ERAD proteins we identified might provide novel therapeutic targets to prevent WNV-induced mortality.
Experimental Procedures
Cells and viruses
293FT, and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) with high glucose. The medium was supplemented with 10% fetal bovine serum (FBS, Life Technologies) and penicillin-streptomycin (Life Technologies). All cells were maintained at 37°C and 5% CO2 in a humidified incubator. The WNV-B956 strain was grown and titrated as previously described (Kumar et al., 2006). The WNV-NY99 strain and SLEV were obtained from ATCC and grown in C6/36 cells. The titers were determined by plaque assay in BHK-21cells.
sgRNA library construction
The sequences of human protein-coding transcripts were downloaded from Ensembl (Biomart 70). The sgRNA target sites were selected based on the following rules: they must start with a G or A (both nucleotides can initiate transcription from the U6 promoter, data not shown) and have a PAM motif 20 nt downstream. Two target sites per exon were chosen from the first two exons immediately after the start codon. If not enough target sites were found in these two exons, sites were chosen from the following exons until there were four target sites for each transcript. To all target site oligos, the sequence TCTACAGTCCGACGATCATGCAT was added to the 5’ end, and the sequence GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGG CACCGAGTCGGTGCTTT was added to the 3’ end. The library was synthesized as an oligo pool (CustomArray, Inc), which was purified on a 15% urea gel and PCR-amplified with the primers: TCTACAGTCCGACGATCATGCAT (forward) and ATCGAACGTTGTCAGCATCTCGAGAAAAAAGCACCGACTCGGTG (reverse). The amplified oligos were digested with NsiI followed by Pfx50 (Life Technologies) treatment for 10 min at 68°C to generate blunt ends and then digested with XhoI. The oligos were then ligated with the pLB vector digested with HpaI and XhoI. The ligated plasmids were transformed into E. coli and cultured overnight. By sampling 0.1% of the transformations in the plates, we estimated that there were 1.52×106 colonies in total, which should cover the library ~20 times.
Lentiviral library packaging
The library was packaged with the ViraPower lentivirus expression system. 293FT cells were seeded in 150-cm2 culture dishes at 1×107 cells per dish. After overnight culture, the medium was replaced with DMEM medium containing 2.5% FBS. A mixture containing 10 µg of library plasmid and 30 µg of ViraPower packaging plasmid mix was transfected using the classic Hepes-calcium phosphate transfection method. Six hours later, the supernatants were changed back to culture medium. Forty-eight hours after transfection, culture supernatants were collected, clarified by centrifugation, filtered with 0.45µm filters, and aliquoted for −80°C storage until use.
Lentiviral transduction
293FT cells (4×107) were transduced at MOI=0.05. The transduction rate was ~5%, as analyzed by FACS at 72 hours after transduction. The cells were then sorted on a FACSAria II cell sorter, and it was confirmed that >90% of the sorted cells were GFP-positive. The sorted cells were kept in culture under the same conditions as the original 293FT cells. Approximately 5×106 cells were recovered, which was expanded to 2×107 cells in the next step.
Screening for genes essential for WNV-mediated killing
Cells (2×107) harboring the sgRNA library were transfected with the pX261–dU6 plasmid using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s instructions. The transfected cells were infected with the WNV-B956 strain at MOI=2 and incubated for 12 days.
sgRNA recovery
Genomic DNA was extracted from the surviving cells, and sgRNA sequences were amplified by nested PCR with the first pair of primers, GATAGGCTTGGATTTCTATAAG (forward) and CTGCTGGAATCTCGTGAAG (reverse), followed by amplification with the second pair of primers, AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNN NCTTGGAGAAAAGCCTTGTT (forward) and CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGA TCGCTATTTCTAGCTCTAAAAC (reverse, where XXXXXX indicates index sequences) to add an index sequence and sequences required for Illumina second-generation sequencing technology. After sequencing with MiSeq, the sgRNA sequences were recovered by decoding with the index sequence and removing the 5’ and 3’ sequences to identify the genes that are essential for virus-induced cell killing.
Validation of the genes identified
sgRNAs were designed as oligos and inserted into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (Addgene plasmid 42230) at the BbsI site (Cong et al., 2013). The oligo sequences are listed in Table S4. The constructs were co-transfected using Lipofectamine 2000 into 293FT or HeLa cells with a puromycin-expressing plasmid, pX261–dU6, which was modified from the pX261-U6-DR-hEmx1-DR-Cbh-NLS-hSpCas9-NLS-H1-shorttracr-PGK-puro plasmid (Addgene plasmid 42337) by deleting the fragment harboring the U6 promoter and crRNA. Puromycin was added 1 day after transfection at 3 µg/ml for 293FT cells or HeLa cells or Neuro-2a cells and incubated for 2 days. After removing puromycin, the cells were allowed to recover for 3–6 days before WNV challenge. The knockout efficiency was determined by amplifying the target sites with the primers in Table S5, followed by deep sequencing of 293FT cells and western blotting for all three cells. The recovered cells were challenged with WNV at MOI=2. After incubating for 3 days for 293FT or Nuero-2a cells and 2 days for HeLa cells, the cell viability was evaluated with the MTT assay as per the manufacturer’s instructions (Promega).
Supplementary Material
Highlights.
A knockout screening method based on CRISPR-Cas9
Two rounds of screening improve sensitivity and specificity
Seven genes of the ERAD pathway are essential for WNV-induced cell death
These genes connect WNV replication to ensuing cell death
Acknowledgements
This work was supported partially by NIH/NIHLB grant 1R21HL116268 for P.S. and H.W., NIH R01GM113188 for L.Q., and NIH/NIAID grant 1R56AI114357 for H.W.. We thank Drs. Feng Zhang, George Church, and Linda S Wicker for providing us with plasmids.
Footnotes
Author Contributions
H.W., H.M., and Y.D designed the study. H.M., Y.D., Y.W., G.J., E.A., J.Z., S.A., J.C., and G.S. performed experiments. H.W., H.M., Y.D., N.M. and L.Q. analyzed data. H.W., H.M., and N.M. wrote the manuscript.
References
- Bennett SM, Jiang M, Imperiale MJ. Role of cell-type-specific endoplasmic reticulum-associated degradation in polyomavirus trafficking. Journal of virology. 2013;87:8843–8852. doi: 10.1128/JVI.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Molecular cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Brinton MA. Replication cycle and molecular biology of the West Nile virus. Viruses. 2014;6:13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS pathogens. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in Zebrafish embryos. Cell research. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nature cell biology. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JJ, Ng ML. The mechanism of cell death during West Nile virus infection is dependent on initial infectious dose. The Journal of general virology. 2003;84:3305–3314. doi: 10.1099/vir.0.19447-0. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic acids research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia MD, Meertens L, Bonazzi M, Cossart P, Arenzana-Seisdedos F, Amara A. Appraising the roles of CBLL1 and the ubiquitin/proteasome system for flavivirus entry and replication. Journal of virology. 2011;85:2980–2989. doi: 10.1128/JVI.02483-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfoy F, Fayzulin R, Mason PW. West Nile virus genome amplification requires the functional activities of the proteasome. Virology. 2009;385:74–84. doi: 10.1016/j.virol.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt MC, Michaelis M, Ogbomo H, Doerr HW, Cinatl J., Jr Inhibition of apoptosis prevents West Nile virus induced cell death. BMC microbiology. 2007;7:49. doi: 10.1186/1471-2180-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature biotechnology. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014 doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Lee SK, Shankar P, Manjunath N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS medicine. 2006;3:e96. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang YY, Chiu S, Hu Z, Lan KH, Cha H, Sodroski C, Zhang F, Hsu CS, Thomas E, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS pathogens. 2014;10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wu Y, Dang Y, Choi JG, Zhang J, Wu H. Pol III Promoters to Express Small RNAs: Delineation of Transcription Initiation. Molecular therapy Nucleic acids. 2014;3:e161. doi: 10.1038/mtna.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie JM, Khromykh AA, Parton RG. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell host & microbe. 2007;2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parquet MC, Kumatori A, Hasebe F, Morita K, Igarashi A. West Nile virus-induced bax-dependent apoptosis. FEBS letters. 2001;500:17–24. doi: 10.1016/s0014-5793(01)02573-x. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Verheije MH, Cali T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell host & microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M, Suzuki R, Watanabe N, Masaki T, Tomonaga M, Muhammad A, Kato T, Matsuura Y, Watanabe H, Wakita T, et al. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. The Journal of biological chemistry. 2011;286:37264–37273. doi: 10.1074/jbc.M111.259085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Morrey JD, Diamond MS. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. Journal of virology. 2007;81:2614–2623. doi: 10.1128/JVI.02311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M, Bhattacharyya S, Nain M, Kaur M, Sood V, Gupta V, Khasa R, Abdin MZ, Vrati S, Kalia M. Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes. Autophagy. 2014;10 doi: 10.4161/auto.29455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B, Gottlieb D, Diamond MS. Infection and injury of neurons by West Nile encephalitis virus. Journal of virology. 2003;77:13203–13213. doi: 10.1128/JVI.77.24.13203-13213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boomen DJ, Timms RT, Grice GL, Stagg HR, Skodt K, Dougan G, Nathan JA, Lehner PJ. TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proceedings of the National Academy of Sciences of the United States of America. 2014 doi: 10.1073/pnas.1409099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga A, Hanna SL, Li J, Cho H, Rose PP, Spiridigliozzi A, Gold B, Diamond MS, Cherry S. Genome-wide RNAi screen identifies broadly-acting host factors that inhibit arbovirus infection. PLoS pathogens. 2014;10:e1003914. doi: 10.1371/journal.ppat.1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell host & microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.