Summary
Two decades after the discovery that 20% of familial amyotrophic lateral sclerosis (ALS) cases were linked to mutations in the superoxide dismutase-1 (SOD1) gene, a substantial proportion of the remainder of cases of familial ALS have now been traced to an expansion of the intronic hexanucleotide repeat sequence in C9orf72. This breakthrough provides an opportunity to re-evaluate longstanding concepts regarding the cause and natural history of ALS, coming soon after the pathological unification of ALS with frontotemporal dementia through a shared pathological signature of cytoplasmic inclusions of the ubiquitinated protein TDP-43. However, with profound clinical, prognostic, neuropathological, and now genetic heterogeneity, the concept of ALS as one disease appears increasingly untenable. This background calls for the development of a more sophisticated taxonomy, and an appreciation of ALS as the breakdown of a wider network rather than a discrete vulnerable population of specialised motor neurons. Identification of C9orf72 repeat expansions in patients without a family history of ALS challenges the traditional division between familial and sporadic disease. By contrast, the 90% of apparently sporadic cases and incomplete penetrance of several genes linked to familial cases suggest that at least some forms of ALS arise from the interplay of multiple genes, poorly understood developmental, environmental, and age-related factors, as well as stochastic events.
Introduction
In the autumn of 2011, a monumental discovery changed the way we understand amyotrophic lateral sclerosis (ALS). Some 150 years after Charcot provided the first description of ALS, a genetic variant was discovered that meant neuroscientists must revisit what they knew, or think they knew, about the disorder. Understanding of the cause of this therapeutically resistant neurodegenerative disorder has entered a new phase with the discovery of linkage to an expansion of an intronic hexanucleotide repeat in the previously unknown gene C9orf72. 1 and 2 At the most basic level, this breakthrough provides incontrovertible evidence that ALS and frontotemporal dementia are manifestations of a clinicopathological spectrum. Most tantalising however is the emergence of a unifying pathogenic theme of nuclear protein mishandling in ALS. The apparent RNA binding function of the C9orf72 expansion adds to known mutations in other key RNA-processing and proteosomal genes linked to ALS, notably the transactive-region DNA-binding protein gene TARDBP, 3 the fused in sarcoma gene FUS (also known as translocated in liposarcoma, TLS), 4 and the ubiquilin-2 gene UBQLN2, 5 whose proteins are widely expressed in human cells, with fundamental physiological roles.
The recent C9orf72 genetic breakthrough further reinforces the concept that ALS pathogenesis involves multiple pathways. 6 To date, models of disease have largely centred around mutant superoxide dismutase-1 (SOD1), discovered 20 years ago 7 ( figure 1). This genetic discovery heralded development of the first animal model of ALS, 8 and the availability of a coherent strategy for development of successful therapy. Recent seminal findings including a recognition of the importance of TDP-43 deposition in most forms of ALS and some forms of frontotemporal dementia, 9 although not in SOD1-related disease, 9 and the likely role of RNA dysregulation in disease pathogenesis, suggest that translation of findings from the SOD1 model to benefit patients living with ALS might be more restricted than previously thought. Fundamental aspects of ALS, not least its striking clinical heterogeneity, still puzzle clinicians as much now as at the turn of the 20th century and remain the subject of debate. The juxtaposition of the new and exciting discovery of a major ALS gene, with the emergent problems in the translation of findings from the SOD1 mouse model, provided the backdrop to our debate, held as part of the 22nd International Symposium on ALS/MND with the specific aim of highlighting areas that with future research and prioritisation might lead to the development of treatments. To better guide this therapeutic potential, we considered the clinical scope of ALS, dealing with concepts of heterogeneity, while also dissecting arguments concerning focality of disease onset, methods to identify patterns of disease spread, and predictors of prognosis, with implications for the design of future clinical trials. Recent genetic discoveries and controversies concerning heritability and the current models of ALS were incorporated into the discussion, in the context of emergent technology, with consideration of priorities for future ALS research.
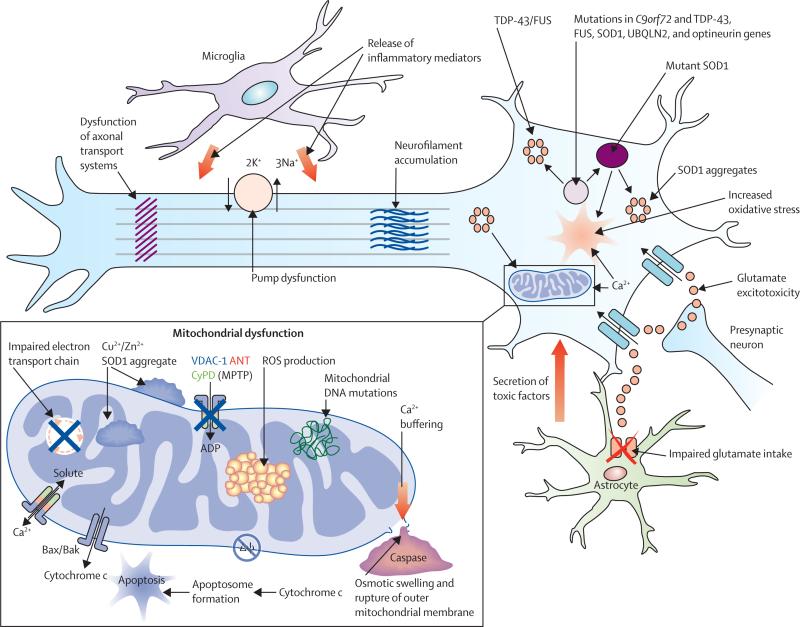
Figure 1. Proposed mechanisms underlying neurodegeneration in ALS.
Many of these pathways are mechanisms of cell death common to a range of neurological disorders, although in the case of amyotrophic lateral sclerosis (ALS), have been derived from studies undertaken predominantly using the SOD1 mouse model. Pathophysiological mechanisms involving more recent genetic discoveries, particularly the C9orf72 hexanucleotide repeat expansion, have yet to be elucidated. Neurodegeneration in ALS might reflect combinations of glutamate excitoxicity, generation of free radicals, mutant SOD1 enzymes, along with mitochondrial dysfunction and disruption of axonal transport processes through accumulation of neurofilament intracellular aggregates. Mutations in several ALS-related genes are associated with the formation of intracellular aggregates, which appear harmful to neurons. Activation of microglia results in secretion of proinflammatory cytokines resulting in further toxicity. Failure of cytoplasmic mitochondria induces increased susceptibility to glutamate-mediated excitotoxicity, perturbations in motor neuronal energy production, and apoptosis. Mitochondrial dysfunction is associated with increased production of reactive oxygen species (ROS). Cytoplasmic aggregates of SOD1 might directly inhibit conductance of VDAC1, thereby reducing the supply of ADP to mitochondria for ATP generation. Δψ=mitochondrial membrane potential.
Has the concept of ALS as one disease become untenable?
From a clinical viewpoint, ALS has been identified by most neurologists as a single entity because it characteristically involves a recognised pattern of progressive motor neuronopathy, with respiratory failure as the mode of death for most patients. This core phenotype is confidently diagnosed by an experienced clinician when progression is rapid, although the mean diagnostic delay has stubbornly remained around 1 year from symptom onset,10 reflecting the reality of more slowly progressing cases. Although ALS is a clinically recognisable condition, there are now strong reasons to consider the classic phenotype as a manifestation of a wider aetiopathogenic spectrum.
Although median survival is 3 years from symptom onset, the survival curve for the remaining cases tapers out to 20 years or more. Many of the long-term survivors appear to have either pure upper motor neuron (termed primary lateral sclerosis) or lower motor neuron (termed progressive muscular atrophy) involvement.11 and 12 There is an urgent need for reliable subclassification that goes beyond the number of body territories with combined clinical upper motor neuron and lower motor neuron signs that underpin the current diagnostic criteria.13 There is increasing frustration with this diagnostic framework (panel 1), which was developed principally as a research aid, since a substantial proportion of patients die with a diagnostic label of “possible ALS”14 even though the clinical progression is the same as that of “definite ALS”, but with simply fewer demonstrable upper motor neuron signs.
Panel 1.
Diagnostic criteria for amyotrophic lateral sclerosis (ALS) derived from El Escorial13 and modified Airlie House criteria14
| Diagnosis of ALS requires: |
| (1) The presence of: (a) evidence of lower motor neuron degeneration by clinical, electrophysiological, or neuropathological examination; (b) evidence of upper motor neuron degeneration by clinical examination; and (c) progression of the motor syndrome within a region or to other regions, as determined by history or examination; and |
| (2) The absence of: (a) electrophysiological and pathological evidence of other disease processes that might explain the signs of lower or upper motor neuron degeneration; and (b) neuroimaging evidence of other disease processes that might explain the observed clinical and electrophysiological signs. |
| El Escorial criteria |
| Definite ALS: upper and lower motor neuron signs in three regions. |
| Probable ALS: upper and lower motor neuron signs in at least two regions with upper motor neuron signs rostral to lower motor neuron signs. |
| Possible ALS: upper and lower motor neuron signs in one region, upper motor neuron signs alone in two or more regions, or lower motor neuron signs above upper motor neuron signs. |
| Suspected ALS: lower motor neuron signs only in two or more regions. |
| Airlie House (modified) criteria |
| Clinically definite ALS: clinical evidence alone of upper and lower motor neuron signs in three regions. |
| Clinically probable ALS: clinical evidence alone of upper and lower motor neuron signs in at least two regions with some upper motor neuron signs rostral to (above) the lower motor neuron signs. |
| Clinically probable–laboratory-supported ALS: clinical signs of upper and lower motor neuron dysfunction are in only one region, or upper motor neuron signs alone in one region with lower motor neuron signs defined by electromyography criteria in at least two limbs, together with proper application of neuroimaging and clinical laboratory protocols to exclude other causes. |
| Possible ALS: clinical signs of upper and lower motor neuron dysfunction in only one region, or upper motor neuron signs alone in two or more regions; or lower motor neuron signs rostral to upper motor neuron signs and the diagnosis of clinically probable-laboratory-supported ALS cannot be proven. |
| Suspected ALS: this category has been deleted from the revised El Escorial criteria. |
At least 16 genes of major (typically dominant) effect are associated with ALS.15 Recognisable phenotypes have been reported for some known mutations (table 1), such as the SOD1 A4V variant, which produces a rapidly progressive, primarily lower motor neuron syndrome; 16 and FUS mutations linked to a juvenile onset, primarily lower motor neuron syndrome. 17 However, perhaps the most striking development has been the recent recognition of a characteristic phenotype associated with the relatively common C9orf72 hexanucleotide repeat expansion. Patients with the expansion have a younger age of onset, shortened survival, increased rates of cognitive and behavioural impairment, and a strong family history of neurodegenerative disease. 18 and 19 Postmortem studies also suggest that C9orf72-associated ALS has a different pathological signature to other forms of ALS.
Table 1.
Genes (in order of discovery) associated with dominant mutations linked to adult-onset motor-predominant syndromes, showing the range of clinical phenotypes and principal neuropathological protein aggregates
| Clinical involvement | Neuropathological protein aggregates | |
|---|---|---|
| SOD1 | LMN, UMN | Cytoplasmic inclusions of SOD1, or neurofilaments (eg, I113T); generally no TDP-43 aggregates |
| VAPB | LMN, UMN | Probable TDP-43 aggregates |
| ANG | LMN, UMN, FTD | TDP-43 aggregates |
| TARDBP | LMN, UMN, FTD | TDP-43 aggregates |
| FIG4 | UMN | Not known |
| FUS | LMN, UMN, FTD | FUS aggregates, no TDP-43 aggregates |
| OPTN | LMN, UMN, FTD | TDP-43 aggregation (Glu478Gly) |
| C9orf72 | LMN, UMN, FTD | TDP-43 aggregates, but also pathognomonic TDP-43-negative aggregates in cerebellar and hippocampal neurons (molecule currently unknown) |
| UBQLN2 | LMN, UMN, FTD | Ubiquilin-2, also TDP-43-positive and FUS-positive |
Heterogeneity in neuropathology can also be discerned. Cytoplasmic inclusions of TDP-43 are found in nearly all autopsied cases of sporadic ALS, and suggest a consistent final common pathway. However, such inclusions are notably absent in ALS associated with pathogenic mutations of SOD1, 20 and the pathophysiological processes underlying this and other abnormal protein aggregation have not been fully elucidated. Expansions of the C9orf72 hexanucleotide repeat have a characteristic neuropathological signature, with TDP-43 inclusions but in a different distribution including hippocampal and cerebellar regions, and with prominent staining for p62, 21 supporting the view that different primary pathophysiological processes have a final common, clinically indistinguishable, endpoint. As such, the clinical, pathological, and genetic frameworks traditionally used to explain disease heterogeneity remain incomplete, with the corollary that therapeutic interventions might need to be tailored across different subgroups of patients, on the basis of the prevailing upstream pathological events. Understanding of the physiological roles of TDP-43 and C9orf72 will be an essential first step.
Although cognitive and behavioural impairment are now recognised features of ALS, not all patients become impaired. Population based studies suggest that 14% of incident cases have frontotemporal dementia, and a further 30% have evidence of cognitive impairment, mostly but not exclusively in executive dysfunction.22 One of the strongest predictors of cognitive or behavioural impairment in ALS is the presence of the C9orf72 repeat expansion. Taken together, these findings point perhaps to a distinct phenotype in ALS that includes roughly 10% of all cases. Patients with early features of executive impairment progress more rapidly, whereas those who are cognitively normal at the time of diagnosis have a better prognosis and largely do not exhibit features of cognitive decline on serial testing. 23 Furthermore, cognitive impairments common to classic ALS have been notably less prevalent in progressive muscular atrophy 24 and primary lateral sclerosis. 25 Such observations might reflect an increased resistance in some individuals to the penetration of pathology beyond its initial clinical focus, with obvious therapeutic value if the biological substrate for this resistance could be identified.
Going forward, clinical and genetic studies will require stratification with respect to the presence or absence of the C9orf72 expansion and deeper analysis of clinical phenotyping and genotyping will clearly be needed, including development of a greater understanding of the extreme phenotypes. This process has recently been commenced in several population cohorts with C9orf72-related repeats, 18 and has shown distinct clinical and family signatures. Separately, studies of the typically more slowly progressive clinical phenotypes such as primary lateral sclerosis 26 and progressive muscular atrophy 27 have also been undertaken, although major gaps persist in our understanding of molecular changes associated with these disorders, in addition to delineation of the extent and timing of upper motor neuron versus lower motor neuron involvement. 28 Moreover, there is a particular lack of detailed clinicopathological studies in primary lateral sclerosis, 29 regarded by some as a separate disease altogether. The development and application of novel in-vivo neurophysiological and neuroimaging methods to specific clinical subtypes 30 and 31 have the potential to circumvent the end-stage pathology perspective of traditional histopathology.
Should ALS even be regarded as a neuromuscular disease?
Many ALS physicians are also specialists in neuromuscular disease, and the characteristic presence of progressive muscle wasting, weakness, and fasciculations clearly reflects degeneration of anterior horn cells and lower motor neurons. The electromyogram is an adjunct to clinical evaluation, and the Awaji criteria, based on features of denervation and reinnervation, have recently been established as a useful diagnostic aid.32 However, as originally recognised by Charcot, corticospinal tract involvement is nearly always seen pathologically, even in cases without any clinical evidence of upper motor neuron involvement. The emergence of a close biological relation between ALS and frontotemporal dementia argues strongly in favour of a realignment of specialist expertise towards neurodegenerative and behavioural clinics, rather than concentration exclusively within the neuromuscular domain. And although the transgenic SOD1 model might have some value for understanding of the process of anterior horn cell degeneration, the extramotor manifestations of the disease, including significant cognitive and behavioural impairments, command attention that is not provided by existing animal models.
Further to such discussion, a causal primacy of lower motor neuron over upper motor neuron degeneration33 remains an issue of debate. Many of the initial pathological changes in models of ALS occur in the peripheral motor system, supporting a dying-back view of pathogenesis.34 However, ALS can also be viewed primarily as a disease of the upper motor neurons, or corticomotoneurons, which connect monosynaptically with anterior horn cells,35 supported by the clinical observation that the oculomotor, abducens, and Onuf's motor nuclei, which all lack direct corticomotorneuron connections, are strikingly resistant to degeneration.
At the cortical level, premotor and motor regions might seem dominant, although frontal and anterior temporal regions, in addition to subcortical pathways, are key structures. Because motor planning, motor abstraction, emotional expression, and language, which are important aspects of human neocortical evolution, are all affected in ALS, these might hold clues to selective vulnerability and disease progression.36 As such, future studies need to consider ALS as a systems degeneration, rather than focusing on individual neuronal populations. Advanced MRI is uniquely able to study the brain at such a network level. Application of graph theory to structural data concerning white matter tract integrity,37 and independent component-type analysis to cerebral functional data,38 in parallel with detailed clinical and neuropsychological characterisation, offers a comprehensive in-vivo approach to probing disease activity in ALS.39 and 40
Can apparently sporadic ALS be inherited?
In populations of European origin, only 5–10% of patients with ALS report a family history of the disease (figure 2). It seems likely that there is a complex set of genetic determinants, each with potentially small but significant effect.15 Standard practice has been to reassure patients without an apparent family history of ALS. However, early studies of the C9orf72 hexanucleotide repeat expansion suggest that the repeat expansion is also present in a sizeable minority of apparently sporadic cases, 19 and 41 and careful interrogation of kindreds of these patients, supported by death certification, shows increased rates of neurodegenerative conditions, most notably frontotemporal dementia. Such findings suggest that a family history of frontotemporal dementia, which might have been misclassified as Alzheimer's disease, must now be considered in a revised definition of familial ALS.
Figure 2. The current macrogenetic landscape in ALS, highlighting the increasingly blurred boundary between familial and apparently sporadic disease.
Only 5–10% of cases of amyotrophic lateral sclerosis (ALS) presenting to specialists report a positive family history of ALS or frontotemporal dementia (FTD). At least two-thirds of such cases can now be linked to one of four major genes, the largest group being those with expansions of the hexanucleotide repeat sequence in C9orf72. However, this expansion and mutations of the other genes identified are also detectable in a small but significant proportion of the 90–95% of cases of apparently sporadic ALS reporting no family history.
Furthermore, the definition of familial ALS is increasingly being recognised as inconsistent.42 The definition is based on the presence of known disease in other members of a kindred. In this context the size of the kindred also becomes important in determining heritability. Specifically, the presence of a small number of first-degree and second-degree relatives might erroneously lead to a diagnosis of sporadic ALS, whereas large extended kindreds might serve to increase the risk of a second case of ALS occurring by chance alone. Perhaps more reassuringly for patients with sporadic disease is the limited evidence obtained from clinic-based populations to date, suggesting that although there is approximately an eight-fold increase to first-degree relatives of patients with ALS, the absolute increase in risk seemed to be very small in a follow-up study over 16 years.43
As with previous familial linkage to ALS, there appears to be incomplete penetrance of the C9orf72-related repeat expansion. Incomplete family histories and small family sizes might blur the distinction between sporadic and familial ALS further, suggesting that the reassuring figures for recurrence within offspring cannot be applied across all patients. 44 A clearer definition of familial ALS, followed by more extensive analyses of disease penetrance, might ultimately lead to a change in the routine counselling of patients. Whether the routine screening of all apparently sporadic cases of ALS for the known familial genetic abnormalities is the optimum approach at present, with the potential to register psychological harm, needs urgent debate.
The explosion in ALS genetics has challenged other long held doctrines relating to known genes of major effect, and evidence of an oligogenic cause in familial and sporadic ALS further compounds the problem of ALS heritability. Until recently, there had been an assumption that single genes of major effect with incomplete penetrance accounted for most cases of familial ALS. As each new ALS gene has been identified, ALS DNA banks have been screened to identify pathogenic variants in the new gene. DNA samples containing known variants are then excluded from further analyses. However, emerging evidence suggests a more complex genetic cause, in which the phenotypic variability might be due to a small number of genetic variants that co-occur with greater frequency than would be expected by chance. Recently described examples include FUS and TARDBP mutations in combination with ANG mutations and C9orf72 repeat expansions combined with TARDBP, SOD1, and FUS mutations. 45 If replicated, these findings will have important implications for the design and interpretation of ALS genetic studies. Whether this explanation accounts for the substantial phenotypic variations within C9orf72 kindreds remains to be established.
That the most important gene in ALS was identified with conventional genetic approaches, rather than the newer technologies of genome-wide association studies (GWAS) and next-generation sequencing perhaps seems ironic. Whereas the chromosome 9 locus was identified a decade before its characterisation in sporadic ALS,46 the intronic repeat expansion could never have been identified with more modern sequencing methods because of the insensitivity of these technologies to detect very large intronic repeat expansions.
The history of GWAS in ALS started with fairly small studies (table 2) that initially had negative results or results that were not replicated in independent studies.47 and 50 Through international collaborations and data sharing, ongoing combined analysis of available data has now provided consistent results. Nevertheless, by contrast with other neurodegenerative diseases such as Parkinson's or Alzheimer's disease, the results of GWAS looking for common variability have been disappointing in ALS, again suggesting that ALS is not one clinical entity, but is heterogeneous by nature. The best example of consistent GWAS results to date was the identification of the chromosome 9 locus, which appears aberrant in 5–8% of sporadic ALS. The larger GWAS effectively fine mapped the 9p locus from several megabases to a 110 kb region containing three genes, allowing the C9orf72 discovery. Nevertheless, continuing international collaboration will be needed to generate sample sizes that are comparable with other complex diseases, such as diabetes and inflammatory bowel disease. Also, the combined analysis of GWAS data of ALS with other neurodegenerative diseases (eg, Alzheimer's disease, Parkinson's disease, and frontotemporal dementia) will probably be of great value.
Table 2.
Genome-wide association studies in amyotrophic lateral sclerosis
| n (cases/controls) | Genome-wide significant* result in or near gene | Independent samples or overlap | Remarks | |
|---|---|---|---|---|
| Schymick et al 200747 | 276/271 | None | Overlap | .. |
| Van Es et al 200748 | 461/450 | None | Overlap | SNP in or near ITPR2 replicated in third phase of study |
| Cronin et al 200849 | 958/932 | None | Overlap | .. |
| Van Es 200850 | 461/450 | DPP6 | Overlap | Genome-wide significant in GWAS data plus replication cohort |
| Simpson et al 200951 | 781/702 | ELP3 | Unknown | Different kind of genetic markers used (microsatellites) |
| Chio et al 200952 | 553/2338 | None | Overlap | Original SNPs in or near DPP6, ITPR2, FGGY not replicated |
| Landers et al 200953 | 1821/2258 | None | Overlap | .. |
| Van Es et al 200954 | 4855/14953 | C9orf72 (9p21.2), UNC13A (19p13.3) | Overlap | .. |
| Li et al 200955 | 58/52 | None | Independent | SNP in or near DPP6 not replicated |
| Shatunov et al 201056 | 4312/8425 | C9orf72 (9p21.2), UNC13A (19p13.3) | Overlap | .. |
| Laaksovirta et al 201057 | 442/521 | C9orf72 (9p21.2), SOD1 | Independent | .. |
| Iida et al 201158 | 92/233 | None | Independent | Stepwise case-control association study, combined analysis genome-wide significant at SNP in or near ZNF512B; SNPs in or near DPP6, ITPR2, FGGY, UNC13A not replicated |
| Kwee et al 201259 | 442/348† | None | Independent | Original SNP in DPP6 not replicated, other SNP in DPP6 (p=0·0004) with same direction; ITPR2 not replicated; other SNP in ELP3 (p=0·0004); SNP in UNC13A, odds ratio 1·2 (same direction; p=0·04) |
| ALSGEN Consortium 201260 | 4243/5112 | UNC13A (recessive model) | Overlap | No replication in samples independent from previous studies of DPP6 (69% power), ITPR2 (30% power), UNC13A (17% power) |
Most replication attempts in these genome-wide association studies (GWAS) were underpowered because of a considerable overlap between samples included, so for these genes larger independent replication studies are needed to definitively assess their role. Additionally, when susceptibility genes also have a detrimental effect on survival, population-based cohorts will be needed that automatically will include patients with short survival in order to avoid underestimation of allele frequencies. Finally, this table clearly shows the need for combined analysis of available data to arrive at the so-called latest and greatest GWAS in amyotrophic lateral sclerosis, such as in the ALSGEN Consortium publication. Even more GWAS datasets are available now and will be in the near future. SNP=single nucleotide polymorphism.
p<5·0×10–8.
Also second cohort (n=5909).
In view of the recent findings of intronic variation and repeated sequences in ALS, and the notion that rare genetic variants as opposed to the common genetic variants underlie pathogenesis, high-throughput exome sequencing and analysis of non-coding DNA by whole-genome sequencing in large samples are now needed to fully elucidate the genetic contribution to ALS pathogenesis.61 As has been the case for other diseases such as breast cancer, the discovery of one gene that affects a large minority of patients will probably help to identify other genes, since all datasets will now need to be reanalysed after removal of people with the C9orf72 expansion.
Is there a premorbid phenotype that reflects vulnerability to ALS?
Despite conflicting results regarding premorbid athleticism from case-control studies,62 and 63 a clinical impression remains that patients who develop ALS frequently have a higher than average level of physical activity or fitness, perhaps best exemplified by the American baseball player Lou Gehrig. Whether the findings of reduced coronary artery disease premorbidly in patients with ALS,64 and a cardioprotective vascular profile of patients65 and their first degree relatives,66 are significant in terms of premorbid fitness or athleticism remains uncertain. Several occupations likely to be linked with high physical activity and fitness have been associated with increased incidence of ALS, including professional footballers in Italy, the military, and manual workers.67 and 68 Separately, a lower than average body-mass index appears more commonly associated with ALS, linked to the observation of hypermetabolism that often develops in ALS. The biological processes accounting for these changes remain unclear. Mitochondrial impairment has been long considered as a possible pathogenic mechanism in neurodegeneration, and increasing the efficiency of oxidative phosphorylation remains one of the proposed mechanisms of the drug dexpramipexole, recently the subject of a phase 3 trial for ALS.69
It has been argued that physical activity might be harmful to a minority of the population who carry a complex genetic profile that adversely affects the metabolic response to vigorous muscle activity, including oxidative stress.70 Many patients with ALS specifically ask whether they can safely continue to exercise regularly without fear of accelerating their disease.70 Although low-grade exercise programmes might even have some benefit,71 there is no firm evidence that exercise exerts a harmful effect, although avoidance of very strenuous activity seems reasonable.
An alternative formulation, which is not mutually exclusive, is that athleticism or fitness in patients with ALS is a reflection of a shared set of genetic determinants that predispose to neurodegeneration, without there being a direct causal link between ALS and exercise.62 In a cortically focused model of pathogenesis there might even be preferred loci for neurodegeneration,72 and 73 perhaps reflected in regional involvement,74 and an athletic profile might then also reflect a distinct motor system architecture. There would be an evolutionary advantage for a configuration associated with fitness, that is perhaps only rendered disadvantageous for a minority as a result of the relatively recent increases in life expectancy. Potential evolutionary trade-offs have been noted in other neurodegenerative disorders.75
When does ALS begin?
Longitudinal studies in presymptomatic individuals carrying pathogenic SOD1 mutations have shown apparently abrupt changes in both lower motor neuron loss 76 and the development of cortical hyperexcitability 77 within a year of symptom onset. Whether the nervous system of people who later develop ALS was normal immediately before disease onset or whether, like in Alzheimer's and Parkinson's diseases, a more prolonged presymptomatic phase of disease might have been detectable, seems uncertain. In support of a prolonged presymptomatic phase, changes in parenchymal metabolites (N-acetylaspartate in relation to choline and myoinositol) were detected in the spinal cord of presymptomatic individuals carrying pathological SOD1 mutations long before clinical disease onset. 78 The intriguing suggestion that the risk of ALS develops from factors operating at the time of conception or perhaps during early development warrants further consideration. 79
Detailed study of presymptomatic individuals carrying pathogenic mutations could provide unique insight into the nature of the earliest events before the tipping point of symptom onset. This pursuit is likely to be relevant to the long-term strategy of primary prevention, although these studies will be necessarily long term and costly. Although there might be ethical concerns associated with testing and studying of asymptomatic family members, a robust framework for this process has recently been established.80
How does ALS spread?
Typically, ALS has a striking focal clinical presentation, although this view must be interpreted with the understanding that most biological systems have a degree of redundancy and also in view of emerging data covering presymptomatic changes. For example, at least a third of muscle fibres have already been lost before clinical wasting occurs in ALS.81 Nonetheless, post-mortem studies suggest that the pathological changes in ALS radiate from an area in which maximum upper motor neuron and lower motor neuron dysfunction aggregate together in the spinal region of first symptom onset.82 Spread of symptoms then appears to occur via an anatomically contiguous space within both upper motor neuron and lower motor neuron systems, although the rate might appear to differ within these two anatomical axes. Separately, a complete model of the ALS system failure requires incorporation of a third extramotor axis; namely, the substrate for cognitive and behavioural impairments.
Irrespective of the basis for the regional pattern of involvement in ALS, important and unexplained differences remain in sex and age, particularly in relation to the site of disease onset. For example, respiratory-onset disease tends to be more prevalent in men,83 and a predilection for isolated corticobulbar disease has been noted in elderly women.84 Bulbar-onset ALS is strikingly under-represented in young-onset patients,85 and the biological reasons for these phenomena require dedicated investigation.86 By contrast, regional onset could be purely stochastic82 or perhaps related to patterns of use or development.74 The convergence of upper motor neuron and lower motor neuron features might then reflect the topography of muscle innervation. Separately, there are also physiological differences between short and long axons that must be considered, including axonal membrane conductances and the Na+/K+ pump, the dysfunction of which might be relevant to pathogenesis.87
The development of advanced neuroimaging techniques has allowed novel assessment of structural and functional connections within the brain of patients with ALS that has further guided concepts about disease spread, individual susceptibility, and prognosis. Cortical correlates for regional symptoms can be shown with volumetric MRI.88 Neuronal connections are organised in networks and appear to have their own characteristics. Properties include the overall connectivity of the network, clustering of network nodes, the efficiency of the network, and the degree to which there are identifiable hubs.89 Recent investigations concerning functional connectivity in ALS have identified that patients with a strong connectedness of the motor network have a fast progression of disease, suggesting a potential role for assessment of the functional organisation of the motor network as a marker of disease progression.39 and 90 Positive correlation might also suggest patterns of disease spread along functional connections of the motor network in ALS, by contrast with the theory that ALS solely affects a fixed set of primary motor connections.40 These more widespread effects on the brain network suggest that focal damage in primary motor regions in ALS might ultimately manifest in connectivity disturbances elsewhere in the brain, with patterns of change potentially serving as biomarkers for disease progression. At the synaptic level, parallels have also been drawn for a prion-like spread of abnormal protein conformation and propagation of cellular toxicity in ALS,91 and 92 perhaps involving exosomes or autophagy.93
How can the course of ALS be predicted and the design of therapeutic clinical trials improved?
Prediction of the rate of progression in a heterogeneous disorder such as ALS has clear value in care-planning, but also in the assessment of potential therapeutic agents. Typical inclusion criteria for therapeutic trials have tended to be based on time from symptom onset (typically with a cutoff less than 3 years) and a minimum level of respiratory function as measured by vital capacity. Within only these criteria there will inevitably be a wide range of prognoses, thereby greatly reducing statistical power.
However, broad phenotypes with consistent prognostic patterns can be identified in a population-based context. These patterns include rapid decline in patients with respiratory-onset disease,83 reduced survival in those with executive impairment,23 and slower progression in those with upper motor neuron-predominant disease (consistent with the extreme example of primary lateral sclerosis).12 and 94 Consistent regional (generally contiguous) patterns of symptom spread are also recognisable,95, 96 and 97 and simple clinical parameters at diagnosis can allow robust stratification of prognosis for clinic-based patients.98 and 99 In view of these developments, a formal staging system for ALS has been proposed that attempts to better incorporate clinical phenotype.100 Stratification by rate of disease progression (time to spread to a new region) and degree of cognitive impairment, rather than the traditional divisions by site of onset or upper motor neuron- dominance versus lower motor neuron dominance, might yet prove more useful for future clinical trial enrolment.
At the individual patient level, prognostication soon after symptom onset remains difficult; for instance, it is difficult to distinguish typical bulbar-onset ALS (with progression to limb weakness) from a more atypical, isolated bulbar onset, which might remain isolated sometimes for several years before the involvement of other body regions (sometimes termed progressive bulbar palsy).101 Intrinsic heterogeneity among trial participants can obscure treatment effects that might be more obvious in specific clinical (or genetic) subgroups, and various strategies have been proposed to improve efficiency of clinical trials in ALS.102 Subgroup analysis of patients involved in previous clinical trials might have value; for example, some have suggested that riluzole could have an effect in delaying cognitive involvement.
Pending validation of the numerous and varied candidate biomarkers (summarised in panel 2),103, 104 and 105 the revised ALS functional rating score (ALSFRS-R; panel 3) is a frequently used outcome measure in therapeutic trials.106 This score has good intrarater and inter-rater reliability, and prognostic value as a single measurement at the point of diagnosis as well as an indicator of the rate of change.107 However the slope of its decline might not be consistently linear, with deviations noted in early and late stages of the disease,108 so that statistical assumptions based on a linear model in trials involving patients early in their disease might be incorrect. Patient dropouts due to disease progression further complicate the design of ALS clinical trials, in addition to inherent and progressive problems for patients to attend the protocol centres, leading to potentially critical loss of statistical power. A method to combine survival and ALSFRS-R into one primary outcome measure was developed as the primary endpoint for the failed phase 3 trial of dexpramipexole.109 There is also increasing momentum to consider a further adjustment of the ALSFRS-R to include assessment of executive function, although further data are needed to validate available cognitive tools for ALS.
Panel 2.
| CSF |
| • Neurofilaments |
| • Cystatin C |
| • TDP-43 |
| • Neuroinflammatory molecules |
| Blood |
| • Neurofilaments |
| • TDP-43 |
| • Neuroinflammatory molecules |
| MRI |
| • Reduced motor cortical density or thickness |
| • Fractional anisotropy of the corticospinal tract or corpus callosum |
| • Functional connectivity of the cortex |
| • Altered motor cortical metabolites |
| Neurophysiology |
| • Neurophysiological index |
| • Motor unit nerve estimation |
| • Electrical impedance myography |
| • Threshold-tracking transcranial magnetic stimulation |
Panel 3.
Amyotrophic lateral sclerosis functional rating scale, revised106
| 1 Speech |
| 4 Normal speech |
| 3 Detectable disturbance |
| 2 Intelligible without repeating |
| 1 Speech with non-verbal communication |
| 0 Loss of useful speech |
| 2 Salivation |
| 4 Normal |
| 3 Slight, but definite excess of saliva |
| 2 Moderate excessive saliva, minimum drooling |
| 1 Marked excessive of saliva, some drooling |
| 0 Marked drooling, needs constant tissue |
| 3 Swallowing |
| 4 Normal eating habits |
| 3 Early eating problems, occasional choking |
| 2 Dietary consistency changes |
| 1 Needs supplemental tube feeding |
| 0 Nil orally |
| 4 Handwriting |
| 4 Normal |
| 3 Slow or sloppy, all words legible |
| 2 Not all words legible |
| 1 Able to grip pen, but cannot write |
| 0 Unable to grip pen |
| 5 Cutting food and handling utensils |
| 4 Normal |
| 3 Slow and clumsy, but no help needed |
| 2 Can cut most foods, although clumsy and needs some help |
| 1 Food must be cut by someone else |
| 0 Needs to be fed |
| 6 Dressing and hygiene |
| 4 Normal |
| 3 Independent, but decreased efficiency |
| 2 Some help with closures and fasteners |
| 1 Provides minimum assistance to caregiver |
| 0 Unable to perform any task |
| 7 Turning in bed |
| 4 Normal |
| 3 Slow and clumsy |
| 2 Can turn alone with difficulty |
| 1 Can initiate but cannot turn or adjust sheets |
| 0 Total dependence |
| 8 Walking |
| 4 Normal |
| 3 Early ambulation difficulties |
| 2 Walks with assistance |
| 1 Non-ambulatory, functional movement |
| 0 No purposeful leg movement |
| 9 Climbing stairs |
| 4 Normal |
| 3 Slow |
| 2 Mild unsteadiness or fatigue |
| 1 Needs assistance |
| 0 Cannot do |
| 10 Dyspnoea |
| 4 None |
| 3 Occurs when walking |
| 2 Occurs when eating, bathing, or dressing |
| 1 Occurs at rest |
| 0 Considerable difficulty |
| 11 Orthopnoea |
| 4 None |
| 3 Some difficulty, does not routinely use more than two pillows |
| 2 Needs extra pillows to sleep |
| 1 Only sleeps sitting up |
| 0 Unable to sleep |
| 12 Respiratory insufficiency |
| 4 None |
| 3 Intermittent use of non-invasive ventilation |
| 2 Continuous use of non-invasive ventilation at night |
| 1 Continuous use of non-invasive ventilation, day and night |
| 0 Mechanical ventilation via tracheostomy |
Motor unit number estimation, axonal excitability, electrical impedence myography, the neurophysiological index (NI), and cortical excitability all have potential as alternative outcome measures.105 The neurophysiological index has been shown to have good intrarater reliability and sensitivity to change in ALS, including the differentiation of fast and slow progressing patients.110 Longitudinal analysis supports incorporation of the NI into future therapeutic trials.111 More generally, that future trials must routinely incorporate candidate biomarkers if the historical reliance on survival as the primary outcome measure is to be overcome is widely agreed.
Has the SOD1 mouse model outlived its usefulness?
The transgenic mouse model overexpressing SOD1 contains multiple copies of mutant human DNA. The model has been used extensively to dissect the likely pathogenic mechanisms of mutant SOD1 and to evaluate the possible benefits of new therapeutic agents. There has been a disappointing lack of drugs that meaningfully affect disease progression in the model and also in the translation of any modest disease-delaying benefits from mouse to human ALS. The repeated failures of phase 2 and 3 trials in human beings based on SOD1 mouse studies have led to calls for a reappraisal of the usefulness of the model.
Although the SOD1 mouse recapitulates several core clinical features of ALS—namely, a predictable and progressive loss of motor neurons, associated with muscle weakness commencing at approximately 90 days, and premature death—the pathological cascade plays out over a short period of time. 8 Therein might lie an inherent problem in the attempt to translate the results of therapeutic challenges to the human disease, in which symptoms do not emerge for several decades and where the disease develops through a combination of factors that are beyond genetic mutations and not captured by any model to date. Separately, therapeutic agents that have been tested and judged beneficial in the mouse model have been administered well before symptom onset.
A further failure of translation might partly relate to errors in early mouse studies, which were often inadequately powered and confounded by factors including differential effects of sex, failure to control for variations in copy number, differences in animal husbandry, and variability in mouse strain genetic backgrounds, leading to potentially erroneous conclusions. However, to address these issues, two recent workshops have generated guidelines that could largely resolve the confounders of early mouse studies, thereby rendering comparative analysis more robust.112, 113, 114 and 115
Proponents of the mouse model point out that translation from mouse to human being will always be imperfect because of cross-species differences in neuroanatomy, pharmacokinetics, and dynamics, as well as longevity. They also point to the reproducible murine phenotype and the similarities in neuropathology, including the early and selective degeneration of anterior horn cells. Conversely, some issues need to be further considered including the requirement for large copy numbers of mutant DNA to generate a phenotype, the absence of phenotype in the mouse with highly penetrant human mutations that lead to a severe phenotype in human beings (eg, the A4V mutation), and the observation that SOD1-related ALS is not associated with TDP-43 inclusions—the pathological signature common to nearly all other forms of human ALS, including those associated with hexanucleotide repeat expansions in C9orf72. Although recent genetic advances have led to the development of new animal models based on transgenic TDP-43 and FUS mutants, no other model identified to date has recapitulated the clinical disease phenotype to the extent of the SOD1 mouse, despite the aforementioned caveats.
Novel approaches to understanding of ALS pathogenesis
The cost of whole-genome sequencing is falling exponentially to a level that will soon make this approach feasible for a large number of patient samples. An internationally collaborative approach to sequence whole genomes from large cohorts of well characterised patients with ALS seems the next logical step. As a result, the characterisation of the genetic architecture of ALS might soon be complete, but is likely to be only the first step towards identification of key pathways amenable to therapeutic intervention. A limitation of the downstream transcriptomic approach is the difficulty in distinguishing primary (initiating) and secondary (responsive) changes in gene expression from the mass of data generated, although this approach holds promise.116 High-throughput proteome-wide studies are still beyond the horizon.
The bioinformatics task presented by the enormous volume and complexity of both genomic and RNA expression data has become the immediate challenge. It requires the neuroscience research community to engage those with the mathematical expertise needed to meaningfully interrogate and interpret such datasets, in conjunction with more sophisticated clinical phenotyping of patients.
Although animal modelling for ALS remains challenging, and rodent models have limitations, new models that engage technological developments hold promise for rapid assessment of therapeutic potential. Such models include the Caenorhabditis elegans worms, established for high-throughput, large-scale functional validation of candidate protective genes, including lines expressing forms of TDP-43 or FUS. 117, 118 and 119 Selection of nematode promoters can now also be used to express these proteins in neurons or more specifically only in motor neurons. In addition to the development of phenotypic assays, such studies could also unlock a holy grail in ALS research—the mechanism of disease spread at a molecular level.
The development of induced pluripotent stem cell technology offers an additional and potentially cost-effective approach as a platform for early modelling pathology and testing therapeutic candidates (so-called disease-in-a-dish). It marks a significant divergence from the traditional view of stem cells as a potential therapy for ALS.120 One of the major challenges will be to overcome the great heterogeneity across cell colonies, and to model the in-vivo network environment of such cells, which might be as important as any individual cellular dysfunction observed in isolation.121 Emerging themes might be common to other neurodegenerative disorders, in particular protein degradation pathways,6 and 122 making overlap in treatment strategy between ALS and other disorders increasingly likely.123
Conclusion
The enormous advances in scientific research during the past two decades have revealed the complexity of ALS pathogenesis (figure 3), which might have been underestimated at the time of the discovery of SOD1. Developments in molecular biology, particularly the discovery of C9orf72, when combined with new approaches in clinical neurophysiology and neuroimaging have accentuated rather than lessened the importance of detailed clinical assessment. The multiple pathogenic processes support the view of multiple routes to a common endpoint of progressive upper motor neuron and lower motor neuron loss, which might mean that successful treatment will ultimately involve multiple targets.
Figure 3. Hypothesised sequence of events in the pathogenesis of the ALS syndrome.
The early substrate for amyotrophic lateral sclerosis (ALS; A) is likely to involve genetic, developmental, and environmental factors. For most apparently sporadic cases, multiple genetic factors with small individual effects might in part affect development and maturation of the nervous system. This process might result in a motor system architecture that is more permissive to pathological changes later in life, for example. Environmental triggers might then operate on an already primed system. Several molecular mechanisms for neurodegeneration have been postulated (B), common themes including excitotoxicity (or loss of inhibitory neuronal buffering) and oxidative stress. A common endpoint seems to be protein misaggregation, in which several recent genetic discoveries have implicated abnormal RNA biology. ALS is a syndrome with cases showing variable involvement of upper and lower motor neurons, and extramotor cognitive impairment (C), in which there can be significant variation in rates of progression. Although there are extremes of relatively isolated involvement of each of these compartments, termed primary lateral sclerosis (PLS), progressive muscular atrophy (PMA), and frontotemporal dementia (FTD), all of these overlap with classic ALS on post-mortem neuropathological examination. A greater understanding of the nature of the apparent barriers between upper, lower, and extramotor neuronal populations might ultimate offer a disease-modifying strategy.
The search for biomarkers in ALS is one of several research priorities (panel 4). Despite a large number of negative trials, ALS remains attractive to biotechnology companies with an array of approaches that are increasingly based on combating hypothesis-led mechanisms. Examples include small molecules to potentially interfere with protein aggregation, gene therapy to replace specific mutations, stem cell therapy to support glial as well as motor neuronal cells, and oligonucleotides to interfere with abnormal RNA processing. With improved understanding of the chemical structure of the drugs most likely to be effective, there is also potential for the repositioning of compounds held in pharmaceutical company drug libraries, abandoned for their original purposes, but in which extensive pharmacological and toxicity studies have often been completed.
Panel 4.
Priority areas for amyotrophic lateral sclerosis (ALS) research
| • Understanding of the development of cortical and spinal motor neuron populations, their interaction with each other, and how it changes with normal ageing and in ALS |
| • Factors determining rate of disease progression |
| • Development of in-vitro and in-vivo models that accurately mimic ALS-related protein misfolding, aggregation, and accumulation |
| • Identification of the complete ALS-related genome |
| • Identification of the earliest, presymptomatic characteristics of ALS through the collaborative identification and study of relatives of patients with familial ALS |
| • Identification and standardisation of a biomarker for presymptomatic or early ALS and incorporation (with a view to validation) of at least one biomarker candidate in future therapeutic trials |
| • High-throughput compound screens in cellular and animal models of ALS (including zebrafish and worm models) |
| • Identification of possible environmental factors through large-scale international collaboration |
Advances in the understanding of pathogenesis in ALS have occurred despite limitations in our understanding of fundamental aspects of the disorder and its taxonomy. These advances, coupled with the positive effects of the globalisation of scientific discussion and debate, will drive the emergence of the therapeutic era of ALS within the first half of this century.
Search strategy and selection criteria
We searched PubMed (1966, to Dec 31, 2012), Embase (1980, to Dec 31, 2012), and the Cochrane Library using the search terms “amyotrophic lateral sclerosis” or “motor neuron disease” in combination with “diagnosis”, “epidemiology”, “fronto-temporal dementia”, “imaging”, “neurophysiology”, “management”, and “neuroprotection”. Further articles were included from reference lists, review articles, and major textbook chapters. Abstracts and reports from relevant meetings were also included. The final reference list was generated on the basis of originality and relevance to the topics covered in this report. Emphasis was placed on publications from the past 5 years, but did not exclude commonly referenced and highly regarded older publications.
Acknowledgements
The Fort Denison consensus meeting was supported by Neuroscience Research Australia and the University of New South Wales. MCK receives funding support from the National Health and Medical Research Council. MRT is supported by the Medical Research Council & Motor Neuron Disease UK Lady Edith Wolfson Clinician Scientist Fellowship. OH received funding from the Health Seventh Framework Programme (FP7/2007-2013) under grant agreement No 259867, ALSA (the ALS Association), HRB (the Health Research Board) Clinical Scientist Award, and Research Motor Neuron (previously named Motor Neuron Disease Research Foundation). BJT is supported in part by the Intramural Research Programs of the NIH, National Institute on Aging (Z01-AG000949-02). We are grateful to Olaf Ansorge for his expert advice on table 1. Helpful discussion with Linda Greensmith and Denise Figlewicz regarding the SOD1 mouse model of ALS is gratefully acknowledged.
Footnotes
Conflicts of interest
MB receives research funding from MDA, ALSA, FDA, and NIH and provides consulting services for Cytokinetics and Asublo. OH has received honoraria for consulting work for Biogen Idec and Cytokinetics. AC serves on scientific advisory boards for Cytokinetics and Biogen Idec. BRB receives research funding from Carolina ALS Research Fund, MDA, ALSA, NINDS, Biogen Idec, Cytokinetics, Neuraltus, and GlaxoSmithKline. MRT received payment from Biogen Idec for a consultancy on ALS staging. RGM provided consulting services for Cytokinetics, Asubio, Biogen, and Knopp and received research support as principal investigator for Neuraltus, and was on speakers' bureau for Avanir. MCK, MS, KT, BJT, LHVdB, JHV, SV, MdC, PGI, CL, HM, GN, JR, and PJS declare that they have no conflicts of interest.
References
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothstein JD. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann Neurol. 2009;65(suppl 1):S3–S9. doi: 10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 7.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 8.Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85:94–134. doi: 10.1016/j.pneurobio.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell JD, Callagher P, Gardham J, et al. Timelines in the diagnostic evaluation of people with suspected amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND)—a 20-year review: can we do better? Amyotroph Lateral Scler. 2010;11:537–541. doi: 10.3109/17482968.2010.495158. [DOI] [PubMed] [Google Scholar]
- 11.Turner MR, Parton MJ, Shaw CE, Leigh PN, Al-Chalabi A. Prolonged survival in motor neuron disease: a descriptive study of the King's database 1990–2002. J Neurol Neurosurg Psychiatry. 2003;74:995–997. doi: 10.1136/jnnp.74.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chio A, Calvo A, Moglia C, Mazzini L, Mora G. Phenotypic heterogeneity of amyotrophic lateral sclerosis: a population based study. J Neurol Neurosurg Psychiatry. 2011;82:740–746. doi: 10.1136/jnnp.2010.235952. [DOI] [PubMed] [Google Scholar]
- 13.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 14.Traynor BJ, Codd MB, Corr B, Forde C, Frost E, Hardiman OM. Clinical features of amyotrophic lateral sclerosis according to the El Escorial and Airlie House diagnostic criteria: a population-based study. Arch Neurol. 2000;57:1171–1176. doi: 10.1001/archneur.57.8.1171. [DOI] [PubMed] [Google Scholar]
- 15.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 16.Juneja T, Pericak-Vance MA, Laing NG, Dave S, Siddique T. Prognosis in familial amyotrophic lateral sclerosis: progression and survival in patients with glu100gly and ala4val mutations in Cu,Zn superoxide dismutase. Neurology. 1997;48:55–57. doi: 10.1212/wnl.48.1.55. [DOI] [PubMed] [Google Scholar]
- 17.Baumer D, Hilton D, Paine SM, et al. Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology. 2010;75:611–618. doi: 10.1212/WNL.0b013e3181ed9cde. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chio A, Calvo A, Mazzini L, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–1989. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie IR, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 21.Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9ORF72 -linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 22.Phukan J, Elamin M, Bede P, et al. The syndrome of cognitive impairment in amyotrophic lateral sclerosis: a population-based study. J Neurol Neurosurg Psychiatry. 2012;83:102–108. doi: 10.1136/jnnp-2011-300188. [DOI] [PubMed] [Google Scholar]
- 23.Elamin M, Phukan J, Bede P, et al. Executive dysfunction is a negative prognostic indicator in patients with ALS without dementia. Neurology. 2011;76:1263–1269. doi: 10.1212/WNL.0b013e318214359f. [DOI] [PubMed] [Google Scholar]
- 24.Raaphorst J, de Visser M, van Tol MJ, et al. Cognitive dysfunction in lower motor neuron disease: executive and memory deficits in progressive muscular atrophy. J Neurol Neurosurg Psychiatry. 2011;82:170–175. doi: 10.1136/jnnp.2009.204446. [DOI] [PubMed] [Google Scholar]
- 25.Grace GM, Orange JB, Rowe A, Findlater K, Freedman M, Strong MJ. Neuropsychological functioning in PLS: a comparison with ALS Can. J Neurol Sci. 2011;38:88–97. [PubMed] [Google Scholar]
- 26.Gordon PH, Cheng B, Katz IB, et al. The natural history of primary lateral sclerosis. Neurology. 2006;66:647–653. doi: 10.1212/01.wnl.0000200962.94777.71. [DOI] [PubMed] [Google Scholar]
- 27.Visser J, de Jong JM, de Visser M. The history of progressive muscular atrophy: syndrome or disease? Neurology. 2008;70:723–727. doi: 10.1212/01.wnl.0000302187.20239.93. [DOI] [PubMed] [Google Scholar]
- 28.Van den Berg-Vos RM, Visser J, Kalmijn S, et al. A long-term prospective study of the natural course of sporadic adult-onset lower motor neuron syndromes. Arch Neurol. 2009;66:751–757. doi: 10.1001/archneurol.2009.91. [DOI] [PubMed] [Google Scholar]
- 29.Pringle CE, Hudson AJ, Munoz DG, Kiernan JA, Brown WF, Ebers GC. Primary lateral sclerosis. Clinical features, neuropathology and diagnostic criteria. Brain. 1992;115:495–520. doi: 10.1093/brain/115.2.495. [DOI] [PubMed] [Google Scholar]
- 30.Vucic S, Kiernan MC. Abnormalities in cortical and peripheral excitability in flail arm variant amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2007;78:849–852. doi: 10.1136/jnnp.2006.105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwata NK, Kwan JY, Danielian LE, et al. White matter alterations differ in primary lateral sclerosis and amyotrophic lateral sclerosis. Brain. 2011;134:2643–2655. doi: 10.1093/brain/awr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa J, Swash M, de Carvalho M. Awaji criteria for the diagnosis of amyotrophic lateral sclerosis: a systematic review. Arch Neurol. 2012 doi: 10.1001/archneurol.2012.254. http://dx.doi.org/10.1001/archneurol.2012.254 published online Aug 13. [DOI] [PubMed]
- 33.Chou SM, Norris FH. Amyotrophic lateral sclerosis: lower motor neuron disease spreading to upper motor neurons. Muscle Nerve. 1993;16:864–869. doi: 10.1002/mus.880160810. [DOI] [PubMed] [Google Scholar]
- 34.Dadon-Nachum M, Melamed E, Offen D. The “dying-back” phenomenon of motor neurons in ALS. J Mol Neurosci. 2010;43:470–477. doi: 10.1007/s12031-010-9467-1. [DOI] [PubMed] [Google Scholar]
- 35.Eisen A, Kim S, Pant B. Amyotrophic lateral sclerosis (ALS): a phylogenetic disease of the corticomotoneuron? Muscle Nerve. 1992;15:219–224. doi: 10.1002/mus.880150215. [DOI] [PubMed] [Google Scholar]
- 36.Eisen A. Amyotrophic lateral sclerosis—evolutionary and other perspectives. Muscle Nerve. 2009;40:297–304. doi: 10.1002/mus.21404. [DOI] [PubMed] [Google Scholar]
- 37.Sporns O. The human connectome: a complex network. Ann NY Acad Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- 38.Smith SM, Fox PT, Miller KL, et al. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douaud G, Filippini N, Knight S, Talbot K, Turner MR. Integration of structural and functional magnetic resonance imaging in amyotrophic lateral sclerosis. Brain. 2011;134:3470–3479. doi: 10.1093/brain/awr279. [DOI] [PubMed] [Google Scholar]
- 40.Verstraete E, Veldink JH, Mandl RC, van den Berg LH, van den Heuvel MP. Impaired structural motor connectome in amyotrophic lateral sclerosis. PLoS One. 2011;6:e24239. doi: 10.1371/journal.pone.0024239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrne S, Elamin M, Bede P, Hardiman O. Absence of consensus in diagnostic criteria for familial neurodegenerative diseases. J Neurol Neurosurg Psychiatry. 2012;83:365–367. doi: 10.1136/jnnp-2011-301530. [DOI] [PubMed] [Google Scholar]
- 43.Hanby MF, Scott KM, Scotton W, et al. The risk to relatives of patients with sporadic amyotrophic lateral sclerosis. Brain. 2011;134:3454–3457. doi: 10.1093/brain/awr248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbot K. Familial versus sporadic amyotrophic lateral sclerosis—a false dichotomy? Brain. 2011;134:3429–3431. doi: 10.1093/brain/awr296. [DOI] [PubMed] [Google Scholar]
- 45.van Blitterswijk M, Vlam L, van Es MA, et al. Genetic overlap between apparently sporadic motor neuron diseases. PLoS One. 2012;7:e48983. doi: 10.1371/journal.pone.0048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosler BA, Siddique T, Sapp PC, et al. Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA. 2000;284:1664–1669. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- 47.Schymick JC, Scholz SW, Fung HC, et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007;6:322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- 48.van Es MA, Van Vught PW, Blauw HM, et al. ITPR2 as a susceptibility gene in sporadic amyotrophic lateral sclerosis: a genome-wide association study. Lancet Neurol. 2007;6:869–877. doi: 10.1016/S1474-4422(07)70222-3. [DOI] [PubMed] [Google Scholar]
- 49.Cronin S, Berger S, Ding J, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17:768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- 50.van Es MA, van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2008;40:29–31. doi: 10.1038/ng.2007.52. [DOI] [PubMed] [Google Scholar]
- 51.Simpson CL, Lemmens R, Miskiewicz K, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiò A, Schymick JC, Restagno G, et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:1524–1532. doi: 10.1093/hmg/ddp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landers JE, Melki J, Meininger V, et al. Reduced expression of the kinesin-associated protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2009;106:9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 55.Li XG, Zhang JH, Xie MQ, et al. Association between DPP6 polymorphism and the risk of sporadic amyotrophic lateral sclerosis in Chinese patients. Chin Med J (Engl) 2009;122:2989–2992. [PubMed] [Google Scholar]
- 56.Shatunov A, Mok K, Newhouse S, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laaksovirta H, Peuralinna T, Schymick JC, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iida A, Takahashi A, Kubo M, et al. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet. 2011;20:3684–3692. doi: 10.1093/hmg/ddr268. [DOI] [PubMed] [Google Scholar]
- 59.Kwee LC, Liu Y, Haynes C, et al. A high-density genome-wide association screen of sporadic ALS in US veterans. PLoS One. 2012;7:e32768. doi: 10.1371/journal.pone.0032768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.The ALSGEN Consortium Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging. 2013;34:357.e7–357.e19. doi: 10.1016/j.neurobiolaging.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schymick JC, Talbot K, Traynor BJ. Genetics of sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2007;16:R233–R242. doi: 10.1093/hmg/ddm215. [DOI] [PubMed] [Google Scholar]
- 62.Scarmeas N, Shih T, Stern Y, Ottman R, Rowland LP. Premorbid weight, body mass, and varsity athletics in ALS. Neurology. 2002;59:773–775. doi: 10.1212/wnl.59.5.773. [DOI] [PubMed] [Google Scholar]
- 63.Veldink JH, Kalmijn S, Groeneveld GJ, Titulaer MJ, Wokke JH, van den Berg LH. Physical activity and the association with sporadic ALS. Neurology. 2005;64:241–245. doi: 10.1212/01.WNL.0000149513.82332.5C. [DOI] [PubMed] [Google Scholar]
- 64.Turner MR, Wotton C, Talbot K, Goldacre MJ. Cardiovascular fitness as a risk factor for amyotrophic lateral sclerosis: indirect evidence from record linkage study. J Neurol Neurosurg Psychiatry. 2012;83:395–398. doi: 10.1136/jnnp-2011-301161. [DOI] [PubMed] [Google Scholar]
- 65.Sutedja NA, van der Schouw YT, Fischer K, et al. Beneficial vascular risk profile is associated with amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82:638–642. doi: 10.1136/jnnp.2010.236752. [DOI] [PubMed] [Google Scholar]
- 66.Huisman MH, de Jong SW, Verwijs MC, et al. Family history of neurodegenerative and vascular diseases in ALS: a population-based study. Neurology. 2011;77:1363–1369. doi: 10.1212/WNL.0b013e318231530b. [DOI] [PubMed] [Google Scholar]
- 67.Chio A, Benzi G, Dossena M, Mutani R, Mora G. Severely increased risk of amyotrophic lateral sclerosis among Italian professional football players. Brain. 2005;128:472–476. doi: 10.1093/brain/awh373. [DOI] [PubMed] [Google Scholar]
- 68.Weisskopf MG, O'Reilly EJ, McCullough ML, et al. Prospective study of military service and mortality from ALS. Neurology. 2005;64:32–37. doi: 10.1212/01.WNL.0000148649.17706.D9. [DOI] [PubMed] [Google Scholar]
- 69.Cheah BC, Kiernan MC. Dexpramipexole, the R(+) enantiomer of pramipexole, for the potential treatment of amyotrophic lateral sclerosis. IDrugs. 2010;13:911–920. [PubMed] [Google Scholar]
- 70.Kiernan MC. Amyotrophic lateral sclerosis and the neuroprotective potential of exercise. J Physiol. 2009;597:3759–3760. doi: 10.1113/jphysiol.2009.177022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de Almeida JP, Silvestre R, Pinto AC, de Carvalho M. Exercise and amyotrophic lateral sclerosis. Neurol Sci. 2012;33:9–15. doi: 10.1007/s10072-011-0921-9. [DOI] [PubMed] [Google Scholar]
- 72.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62:42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bak TH, Chandran S. What wires together dies together: verbs, actions and neurodegeneration in motor neuron disease. Cortex. 2012;48:936–944. doi: 10.1016/j.cortex.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 74.Turner MR, Wicks P, Brownstein CA, et al. Concordance between site of onset and limb dominance in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2010;82:853–854. doi: 10.1136/jnnp.2010.208413. [DOI] [PubMed] [Google Scholar]
- 75.Turner M, Goldacre R, Goldacre M. Reduced cancer incidence in Huntington's disease: record linkage study clue to an evolutionary trade-off? Clinical Genet. 2012 doi: 10.1111/cge.12010. http://dx.doi.org/10.1111/cge.12010 published online Sept 28. [DOI] [PubMed]
- 76.Aggarwal A, Nicholson G. Detection of preclinical motor neurone loss in SOD1 mutation carriers using motor unit number estimation. J Neurol Neurosurg Psychiatry. 2002;73:199–201. doi: 10.1136/jnnp.73.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vucic S, Nicholson GA, Kiernan MC. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain. 2008;131:1540–1550. doi: 10.1093/brain/awn071. [DOI] [PubMed] [Google Scholar]
- 78.Carew JD, Nair G, Andersen PM, et al. Presymptomatic spinal cord neurometabolic findings in SOD1-positive people at risk for familial ALS. Neurology. 2011;77:1370–1375. doi: 10.1212/WNL.0b013e318231526a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vivekananda U, Manjalay ZR, Ganesalingam J, et al. Low index-to-ring finger length ratio in sporadic ALS supports prenatally defined motor neuronal vulnerability. J Neurol Neurosurg Psychiatry. 2011;82:635–637. doi: 10.1136/jnnp.2010.237412. [DOI] [PubMed] [Google Scholar]
- 80.Benatar M, Wuu J. Presymptomatic studies in ALS: rationale, challenges, and approach. Neurology. 2012;79:1732–1739. doi: 10.1212/WNL.0b013e31826e9b1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wohlfart G. Collateral regeneration from residual motor nerve fibers in amyotrophic lateral sclerosis. Neurology. 1957;7:124–134. doi: 10.1212/wnl.7.2.124. [DOI] [PubMed] [Google Scholar]
- 82.Ravits JM, La Spada AR. ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology. 2009;73:805–811. doi: 10.1212/WNL.0b013e3181b6bbbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shoesmith CL, Findlater K, Rowe A, Strong MJ. Prognosis of amyotrophic lateral sclerosis with respiratory onset. J Neurol Neurosurg Psychiatry. 2007;78:629–631. doi: 10.1136/jnnp.2006.103564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Logroscino G, Traynor BJ, Hardiman O, et al. Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry. 2010;81:385–390. doi: 10.1136/jnnp.2009.183525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Turner MR, Barnwell J, Al-Chalabi A, Eisen A. Young-onset amyotrophic lateral sclerosis: historical and other observations. Brain. 2012;135:2883–2891. doi: 10.1093/brain/aws144. [DOI] [PubMed] [Google Scholar]
- 86.Grinberg LT, Rueb U, Heinsen H. Brainstem: neglected locus in neurodegenerative diseases. Front Neurol. 2011;2:42. doi: 10.3389/fneur.2011.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vucic S, Krishnan AV, Kiernan MC. Fatigue and activity dependent changes in axonal excitability in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2007;78:1202–1208. doi: 10.1136/jnnp.2006.112078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bede P, Bokde A, Elamin M, et al. Grey matter correlates of clinical variables in amyotrophic lateral sclerosis (ALS): a neuroimaging study of ALS motor phenotype heterogeneity and cortical focality. J Neurol Neurosurg Psychiatry. 2012 doi: 10.1136/jnnp-2012-302674. http://dx.doi.org/10.1136/jnnp-2012-302674 published online Oct 20. [DOI] [PubMed]
- 89.van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verstraete E, van den Heuvel MP, Veldink JH, et al. Motor network degeneration in amyotrophic lateral sclerosis: a structural and functional connectivity study. PLoS One. 2010;5:e13664. doi: 10.1371/journal.pone.0013664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147:498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kanouchi T, Ohkubo T, Yokota T. Can regional spreading of amyotrophic lateral sclerosis motor symptoms be explained by prion-like propagation? J Neurol Neurosurg Psychiatry. 2012;83:739–745. doi: 10.1136/jnnp-2011-301826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Swash M. How does ALS spread between neurones in the CNS? J Neurol Neurosurg Psychiatry. 2013;84:116–117. doi: 10.1136/jnnp-2012-303992. [DOI] [PubMed] [Google Scholar]
- 94.Sabatelli M, Zollino M, Luigetti M, et al. Uncovering amyotrophic lateral sclerosis phenotypes: clinical features and long-term follow-up of upper motor neuron-dominant ALS. Amyotroph Lateral Scler. 2011;12:278–282. doi: 10.3109/17482968.2011.580849. [DOI] [PubMed] [Google Scholar]
- 95.Turner MR, Brockington A, Scaber J, et al. Pattern of spread and prognosis in lower limb-onset ALS. Amyotroph Lateral Scler. 2010;11:369–373. doi: 10.3109/17482960903420140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fujimura-Kiyono C, Kimura F, Ishida S, et al. Onset and spreading patterns of lower motor neuron involvements predict survival in sporadic amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2011;82:1244–1249. doi: 10.1136/jnnp-2011-300141. [DOI] [PubMed] [Google Scholar]
- 97.Gargiulo-Monachelli GM, Janota F, Bettini M, Shoesmith CL, Strong MJ, Sica RE. Regional spread pattern predicts survival in patients with sporadic amyotrophic lateral sclerosis. Eur J Neurol. 2012;19:834–841. doi: 10.1111/j.1468-1331.2011.03616.x. [DOI] [PubMed] [Google Scholar]
- 98.Haverkamp LJ, Appel V, Appel SH. Natural history of amyotrophic lateral sclerosis in a database population. Validation of a scoring system and a model for survival prediction. Brain. 1995;118:707–719. doi: 10.1093/brain/118.3.707. [DOI] [PubMed] [Google Scholar]
- 99.Turner MR, Bakker M, Sham P, Shaw CE, Leigh PN, Al-Chalabi A. Prognostic modelling of therapeutic interventions in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2002;3:15–21. doi: 10.1080/146608202317576499. [DOI] [PubMed] [Google Scholar]
- 100.Roche JC, Garcia-Rojas R, Scott KM, et al. A proposed staging system for amyotrophic lateral sclerosis. Brain. 2012;135:847–852. doi: 10.1093/brain/awr351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burrell JR, Vucic S, Kiernan MC. Isolated bulbar phenotype of amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12:283–289. doi: 10.3109/17482968.2011.551940. [DOI] [PubMed] [Google Scholar]
- 102.Cudkowicz ME, Katz J, Moore DH, et al. Toward more efficient clinical trials for amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2010;11:259–265. doi: 10.3109/17482960903358865. [DOI] [PubMed] [Google Scholar]
- 103.Turner MR, Agosta F, Bede P, Govind V, Lule D, Verstraete E. Neuroimaging in amyotrophic lateral sclerosis. Biomark Med. 2012;6:319–337. doi: 10.2217/bmm.12.26. [DOI] [PubMed] [Google Scholar]
- 104.Turner MR, Kiernan MC, Leigh PN, Talbot K. Biomarkers in amyotrophic lateral sclerosis. Lancet Neurol. 2009;8:94–109. doi: 10.1016/S1474-4422(08)70293-X. [DOI] [PubMed] [Google Scholar]
- 105.Bowser R, Turner MR, Shefner J. Biomarkers in amyotrophic lateral sclerosis: opportunities and limitations. Nat Rev Neurol. 2011;7:631–638. doi: 10.1038/nrneurol.2011.151. [DOI] [PubMed] [Google Scholar]
- 106.Cedarbaum JM, Stambler N, Malta E, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III) J Neurol Sci. 1999;169:13–21. doi: 10.1016/s0022-510x(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 107.Kimura F, Fujimura C, Ishida S, et al. Progression rate of ALSFRS-R at time of diagnosis predicts survival time in ALS. Neurology. 2006;66:265–267. doi: 10.1212/01.wnl.0000194316.91908.8a. [DOI] [PubMed] [Google Scholar]
- 108.Gordon PH, Cheng B, Salachas F, et al. Progression in ALS is not linear but is curvilinear. J Neurol. 2010;257:1713–1717. doi: 10.1007/s00415-010-5609-1. [DOI] [PubMed] [Google Scholar]
- 109.Cudkowicz M, Bozik ME, Ingersoll EW, et al. The effects of dexpramipexole (KNS-760704) in individuals with amyotrophic lateral sclerosis. Nat Med. 2011;17:1652–1656. doi: 10.1038/nm.2579. [DOI] [PubMed] [Google Scholar]
- 110.Swash M, de Carvalho M. The neurophysiological index in ALS. Amyotroph Lateral Scler Other Motor Neuron Disord. 2004;5(suppl 1):108–110. doi: 10.1080/17434470410020067. [DOI] [PubMed] [Google Scholar]
- 111.Cheah BC, Vucic S, Krishnan AV, Boland RA, Kiernan MC. Neurophysiological index as a biomarker for ALS progression: validity of mixed effects models. Amyotroph Lateral Scler. 2011;12:33–38. doi: 10.3109/17482968.2010.531742. [DOI] [PubMed] [Google Scholar]
- 112.Ludolph AC, Bendotti C, Blaugrund E, et al. Guidelines for the preclinical in vivo evaluation of pharmacological active drugs for ALS/MND: report on the 142nd ENMC international workshop. Amyotroph Lateral Scler. 2007;8:217–223. doi: 10.1080/17482960701292837. [DOI] [PubMed] [Google Scholar]
- 113.Ludolph AC, Bendotti C, Blaugrund E, et al. Guidelines for preclinical animal research in ALS/MND: a consensus meeting. Amyotroph Lateral Scler. 2010;11:38–45. doi: 10.3109/17482960903545334. [DOI] [PubMed] [Google Scholar]
- 114.Benatar M. Lost in translation: treatment trials in the SOD1 mouse and in human ALS. Neurobiol Dis. 2007;26:1–13. doi: 10.1016/j.nbd.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 115.Scott S, Kranz JE, Cole J, et al. Design, power, and interpretation of studies in the standard murine model of ALS. Amyotroph Lateral Scler. 2008;9:4–15. doi: 10.1080/17482960701856300. [DOI] [PubMed] [Google Scholar]
- 116.Brockington A, Ning K, Heath PR, et al. Unravelling the enigma of selective vulnerability in neurodegeneration: motor neurons resistant to degeneration in ALS show distinct gene expression characteristics and decreased susceptibility to excitotoxicity. Acta Neuropathol. 2013;125:95–109. doi: 10.1007/s00401-012-1058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:219–231. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 118.Kraemer BC, Zhang B, Leverenz JB, Thomas JH, Trojanowski JQ, Schellenberg GD. Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc Natl Acad Sci USA. 2003;100:9980–9985. doi: 10.1073/pnas.1533448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Miyasaka T, Ding Z, Gengyo-Ando K, et al. Progressive neurodegeneration in C. elegans model of tauopathy. Neurobiol Dis. 2005;20:372–383. doi: 10.1016/j.nbd.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 120.Thomson SR, Wishart TM, Patani R, Chandran S, Gillingwater TH. Using induced pluripotent stem cells (iPSC) to model human neuromuscular connectivity: promise or reality? J Anat. 2012;220:122–130. doi: 10.1111/j.1469-7580.2011.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bilican B, Serio A, Barmada SJ, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci USA. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tashiro Y, Urushitani M, Inoue H, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem. 2012;287:42984–42994. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]