Abstract
Vaccines have drastically reduced the mortality and morbidity of many diseases. However, vaccines have historically been developed empirically, and recent development of vaccines against current pandemics such as HIV and malaria has been met with difficulty. The advent of high-throughput technologies, coupled with systems biological methods of data analysis, has enabled researchers to interrogate the entire complement of a variety of molecular components within cells, and characterize the myriad interactions among them in order to model and understand the behavior of the system as a whole. In the context of vaccinology, these tools permit exploration of the molecular mechanisms by which vaccines induce protective immune responses. Here we review the recent advances, challenges, and potential of systems biological approaches in vaccinology. If the challenges facing this developing field can be overcome, systems vaccinology promises to empower the identification of early predictive signatures of vaccine response, as well as novel and robust correlates of protection from infection. Such discoveries, along with the improved understanding of immune responses to vaccination they impart, will play an instrumental role in development of the next generation of rationally designed vaccines.
Keywords: Vaccines, systems biology, systems vaccinology
Introduction
Since Edward Jenner’s discovery in the late 1700’s that inoculation with the cowpox virus provided protection from smallpox infection, vaccines have emerged as one of the greatest public health tools in history. The last 60 years have established a golden age in the field of vaccinology, marked by events such as the eradication of smallpox by 1980 [1] and the development of polio vaccines in the 1950s, which have lead to near-eradication of the disease [2]. Despite the great success of these and other vaccines, there remain significant challenges for development of new vaccines against current global pandemics such as HIV and malaria. Among the many problems facing this field are: (i) most currently used vaccines were designed largely empirically. As a result there is little or no understanding of what the correlates and mechanisms of protection are for many vaccines. For example, although the two commercially available types of influenza vaccine, trivalent inactivated (TIV) and live attenuated (LAIV), provide similar levels of protection from infection [3], they generate significantly different immune responses. TIV induces higher levels of IgG antibody secreting cells (ASCs) in the blood as well as higher levels of serum antibodies than LAIV in adults. This is likely due to the different routes of administration, as LAIV, which is administered as an intranasal spray, is thought to produce a more local response in the nasal mucosa and upper respiratory tract, including IgA (mucosal) antibodies and cellular immune responses. As a result, the correlate of protection for TIV is generally considered to be serum antibodies, while the correlate of protection for LAIV is less clear [3]. (ii) The path to licensure of candidate vaccines involves very lengthy and expensive phase IIB and III clinical trials to assess their efficacy and safety. These trials involve thousands of subjects and can cost hundreds of millions of dollars to complete. As a result, very few vaccine concepts are tested in phase III trials. For example, during the past 30 years, only 4 HIV-1 vaccine concepts have been tested for clinical efficacy [4], and despite repeated failures, the correlates and mechanisms of protective immunity against HIV remain poorly understood.
The conventional immunological methods, such as ELISA, ELISPOT, flow cytometry, etc., used to study vaccines have played a valuable role in the field of vaccinology, and will remain essential in evaluating responses to vaccination in the future. However these approaches are generally only able to analyze a single or small number of components of the immune system at a given time, and are insufficient to analyze the full complexity of the structure and dynamics of the human immune system as a whole. This represents a critical obstacle towards understanding the molecular mechanisms by which vaccines generate protective immune responses and identifying meaningful correlates of protection.
To address this issue, vaccinologists have turned to systems biology. By examining how coordinated interactions at a molecular level give rise to immune responses, systems biology approaches enable a holistic view of the immune system and its many components. This developing field provides many promising tools to overcome the challenges facing current vaccine development. Enabling researchers to evaluate the immune responses of fewer subjects in a more in-depth and detailed fashion has the potential to dramatically improve our understanding of the mechanisms of protection of novel vaccines and decrease the length and costs of current clinical trials.
Systems Vaccinology
Within the past 20 years, advances in high-throughput technologies have granted researchers the ability to interrogate the properties and abundances of entire classes of molecular components within the cell. For example, development of lower cost next-generation sequencing technology has facilitated the growth of transcriptomics, which seeks to measure the expression of all RNA transcripts within a cell or population of cells. By sequencing and mapping mRNA transcripts, RNA-sequencing enables the accurate quantification of gene expression as well as simultaneous identification of RNA structure such as transcription start site and exon usage/splice junctions, the regulation of which has been shown to play an important role in many biological processes, including within the immune system [5, 6]. Simultaneously, in the growing domain of metabolomics, analytical chemistry techniques such as liquid chromatography-mass spectrometry (LC-MS) have been harnessed to identify and quantify the set of metabolites within cells or tissues [7]. Changes in metabolic activity are an important component of both innate and adaptive immune responses [8], such as the recognized role that lipid metabolism plays during inflammation [8–10].
Systems vaccinology is an emerging field that applies such ‘omics’ technologies, in combination with bioinformatics tools such as transcriptional network analysis and predictive modeling, to study immune responses to vaccination [11–13]. As a systems-based approach, it aims to use data generated through high-throughput measurements in the context of vaccination to characterize the interactions between individual components of the immune system in the interest of understanding and predicting behavior of the system as whole. This includes analysis of transcriptional, signaling, and metabolic pathways whose activity is perturbed in the various cells of the immune system in response to vaccination, as well as identification of molecular signatures that are predictive of various measurements of protection from infection. The knowledge obtained through these analyses can aid in the rational design of new vaccines that generate long-lasting protection and induce improved responses in populations with diminished immune function such as the elderly.
5 year historic perspective
The first examples of the use of such approaches to study responses to vaccination were performed on the yellow fever vaccine [14, 15]. This vaccine contains a live-attenuated strain (YF-17D) of the yellow fever virus, which induces potent and long-lived CD8+ T cell and neutralizing antibody responses [16, 17]. By combining high-throughput measurements such as microarray gene expression profiling and multiparameter flow cytometry with computational modeling, we were able to detect a regulated network of interferon and innate antiviral genes that were induced post-vaccination in peripheral blood mononuclear cells (PBMCs) [14]. An independent YF-17D study by Gaucher et al. revealed induction of similar transcriptional responses to vaccination, including type I interferon and inflammatory pathways [15]. In addition to examining innate immune pathways activated by vaccination, we successfully identified unique gene signatures that were capable of accurately predicting the CD8+ T cell and neutralizing antibody responses, respectively [14]. The predictive CD8+ T cell signature contained complement protein C1qB and eukaryotic translation initiation factor 2 alpha kinase 4, which is an orchestrator of the integrated stress response. Meanwhile the B cell growth factor receptor TNFRSF17 was among the genes included in the antibody response signature. This work demonstrated for the first time that the immunogenicity of a vaccine could be successfully predicted using early transcriptional measurements within 1 week of vaccination.
Following these initial studies, systems biology approaches have been used to examine immune responses to vaccines against a wide range of pathogens, including influenza [18, 19], malaria [20], smallpox [21], and HIV [22]. In particular, as YF-17D is a live-attenuated vaccine that induces an acute viral infection, the study of influenza vaccination (TIV) enabled investigation into to whether or not similar methods could be used to identify molecular signatures predictive of response to an inactivated vaccine. We identified transcriptional signatures related to the expansion of plasmablasts and the unfolded protein response within B cells on day 7 post-vaccination that correlated with and were predictive of day 28 influenza-specific antibody responses [18]. Indeed, these findings were consistent with studies by Bucasas et al. [19] and Obermoser et al. [23]. Interestingly, TNFRSF17, which was predictive of antibody responses to YF-17D, also appeared in the signatures predictive of TIV response [18]. Recently, Tsang et al. [24], and Furman et al. [25] have extended this approach to search for baseline signatures capable of discriminating between high and low responders to vaccination. However, possibly due to limited sample sizes and weaker signal at baseline, neither study was able to successfully predict antibody response using baseline transcriptional measurements alone. Instead, Tsang et al. utilized cell subset frequencies, while Furman et al. generated a model based on transcriptional modules, serum cytokines, cell subset frequencies, and pre-existing antibody titers. Additionally, as these studies were conducted using cohorts from an individual flu season, the effect of changes in influenza strains included in the TIV vaccine on the performance of these models remains to be examined. To address this question, we are performing a comprehensive analysis of over 400 adults vaccinated with seasonal TIV during 5 consecutive influenza seasons (Nakaya et al., manuscript in preparation). This analysis is an important step towards generating robust and clinically relevant signatures that can be used to predict the efficacy of vaccines in clinical trials.
Among the vaccines under investigation, malaria is one for which a human challenge model exists, allowing for identification of subjects who are protected from or susceptible to infection [26]. Vahey et al. used this model, in which subjects vaccinated with the RTS,S malaria vaccine were challenged using mosquitos infected with an antimalarial-sensitive strain of Plasmodium, to identify transcriptional signatures in PBMCs capable of discriminating between protected and nonprotected vaccinees [20]. Protected subjects had increased expression 2 weeks post-vaccination (but before challenge) of genes involved in the immunoproteasome degradation pathway, involved in MHC peptide processing, compared to subjects who developed parasitemia [20]. These systems-based approaches can be used to identify early signatures of efficacy of other vaccines for which human challenge models have been developed, such as typhoid [27, 28] and Shigella [29], enabling rapid evaluation of novel candidate vaccines in future clinical trials.
A critical question is the degree to which transcriptional responses to vaccination were conserved across vaccines, and whether or not there are universal signatures capable of predicting antibody responses to vaccination. To this end, we used a systems-based approach to compare signatures induced by different types of vaccines [YF-17D, LAIV, TIV, the carbohydrate meningococcal vaccine (Menimmune), and the conjugate meningococcal vaccine (Menectra)] [30]. We observed that while recall antibody responses to inactivated vaccines (e.g. seasonal influenza vaccine, diphtheria toxoid component of the conjugate meningococcal vaccine) were associated with transcriptional modules related to plasmablast differentiation, the antibody responses of live-attenuated vaccines (e.g. yellow fever vaccine) were highly correlated with modules involving innate immunity and type I interferon responses. Meanwhile, antibody responses to polysaccharide components of the meningococcal vaccines were associated with increased proinflammatory cytokines as well as activation of antigen-presenting cells [30]. Thus, these preliminary results suggest that vaccine-induced signatures of immunity are significantly dependent on the class of vaccine in question.
Finally, systems approaches could also potentially be used to identify signatures of vaccine safety. Adverse reactions to vaccination pose major regulatory challenges to vaccine development. Relatively mild adverse reactions such as transient fever, local swelling at the injection site are fairly common and occur within a few hours or days after vaccination. The more serious adverse reactions such as the often fatal viscerotropic disease caused by yellow fever vaccination are very rare (1 in 250,000 cases [16]. The ability to identify signatures at baseline that would be predictive of such adverse reactions would thus represent an important advance, however a major obstacle to this is the rarity of such serious adverse events. In this context, detailed immunological characterization of a patient who developed viscerotropic disease after yellow fever vaccination, revealed a 200-fold expansion of CD14+CD16+ inflammatory monocytes during the period of disease, and a 20-fold elevation in the numbers of such cells even during the convalescent phase, long after the virus had been cleared [63]. This raises the possibility that the persistently elevated levels of the CD14+CD16+ inflammatory monocytes may have predisposed this subject towards viscerotropic disease after vaccination with the yellow fever vaccine. The further evaluation of this possibility is stymied by the rarity of these adverse reactions, and the difficulty in obtaining samples to perform detailed analysis. Future efforts should thus be aimed at creating a clinical infrastructure that would facilitate the acquisition of samples from vaccine adverse reactions at multiple time points, including the convalescent phase.
Challenges
Systems vaccinology analyses face challenges from both the biological and technological perspectives (Fig. 1). These problems must be tackled simultaneously in order to achieve a deeper understanding of how the immune system as a whole responds to vaccination. This is why it is important that systems vaccinology be carried out through collaboration between vaccinologists, immunologists and bioinformaticians. As biological knowledge and insight as well as rigorous and sound data analysis are both required for successful discovery, these two groups should work in close contact and coordinate throughout each phase of the scientific process, including hypothesis generation, experimental design, execution, and data analysis.
Figure 1.
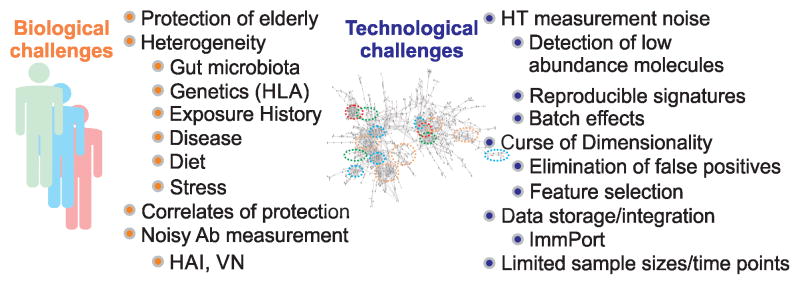
Biological and technological challenges in systems vaccinology.
Biological Challenges
A major factor complicating the analysis of responses to vaccination is the sheer diversity within the human population. The state of the immune system is a function of a host of variables, including genetics [38], previous infections/vaccinations, age [34], chronic diseases, and environmental factors such as diet [39], gut microbial composition [40], stress [41], and physical activity [42]. While the role of genetics in creating susceptibility to cancer and many heritable diseases has been well established, its effect on vaccine response is less clear. A recent study of immune function in twins by Brodin et al. found that while a majority of immune variation is due to non-heritable factors, certain responses, such as IL-2 and IL-7 induced STAT5 phosphorylation in T cells, were highly heritable [43]. In addition, candidate gene studies as well as genome-wide association studies are beginning to be used to identify polymorphisms in genes associated with improved or diminished vaccine responses [44]. Another source of genetic immune variation in humans is that of HLA alleles. As a key step in generating adaptive immune responses involves antigen processing and presentation to T and B cells, the capability of the HLA molecule to bind peptides derived from processed infectious agents or vaccines affects the ability of the immune system to successfully respond to infection or vaccination. Therefore, recent studies have examined the impact that variation in HLA alleles has on antibody responses to vaccines against influenza [45, 46], measles [47], smallpox [48, 49], and hepatitis B [50].
Although human diversity is a challenging aspect of immunology and vaccinology, systems biology approaches are particularly well suited to characterize and account for the complex inter-individual differences within a population [13]. Regression models and machine learning algorithms are able to analyze the effect of many phenotypic variables on responsiveness to vaccination simultaneously. As our understanding of how factors such as genetics, age, and gut microbiome affect immune responses improves, it is reasonable to believe that we will see development of specific vaccine formulations tailored to specific populations such as the elderly or immunocompromised that are designed to stimulate the deficient components of the response unique to each group.
In particular, effect of age on vaccine response is a critical issue. While vaccines are largely successful in preventing infection among the young and in healthy adults, many vaccines, such as the influenza vaccine [31], zoster vaccine, and pneumococcal vaccine [32] show reduced effectiveness in the elderly. This represents a pressing problem for vaccine development, as the world population is rapidly aging, with an expected doubling in the proportion of the population over 60 from 10% in 2000 to 21.8% in 2050 [33]. The mechanisms responsible for the decrease in vaccine efficacy in the elderly are just beginning to be examined. For example, the lower efficacy of TIV in elderly compared to young adults has been associated with the some of the changes resulting from natural age-associated deterioration of immune functions (immunosenescence) [34]. At a cellular level, this includes a reduction in number of antibody-secreting cells (ASC) [35], loss of the less differentiated influenza-specific memory CD8+ T cells [36] and lower frequency of effector memory CD4+ T cells [37]. However, the causes of reduced vaccine efficacy in the elderly at a molecular level remain largely unknown. We are currently performing a comparative analysis of young and elderly (>65 years) subjects vaccinated with TIV, using miRNA and gene expression profiling, to explore the age-dependent regulation of transcriptional responses to vaccination (Nakaya et al., manuscript in preparation).
Meanwhile, for vaccines stimulating recall responses such as the influenza and zoster vaccines, history of exposure to the pathogen is very important, as pre-existing antibodies are associated with a decreased response to vaccination [51, 52]. As these antibodies may be neutralizing viral particles from the vaccine and preventing an otherwise capable immune system from generating a response, this factor should be considered when examining the mechanisms of response. While several groups have attempted various methods to normalize the measure of antibody response to account for variations in baseline antibody levels [19, 24], there is not yet a consensus approach.
Finally, a considerable challenge for developing successful vaccines is to identify reliable correlates of protection from infection. While antigen-specific antibody titers are established as the primary correlate of protection for most vaccines [53], emerging research suggests that in many cases other measures may be equally if not more indicative of protected status. For example, there has been recent work suggesting that serum antibody titers may not be predictive of risk of influenza infection in the elderly [54]. Instead, McElhaney et al. found that strong influenza-specific T cell responses (identified by high IFN-γ:IL-10 ratios following ex vivo stimulation of PBMCs with influenza virus) were indicative of a reduction in risk of illness in elderly subjects [54]. In addition, there is evidence that T cell responses may play an important role in preventing infection for many of the vaccines under current development such as HIV [55], tuberculosis [56], and malaria [57].
Even when antigen-specific antibody titers are a robust correlate of protection, the techniques used to measure them may not be reliable. Two of the most widely used assays to measure influenza-specific antibodies, hemagglutination inhibition (HAI) and virus neutralization (VN) have been shown to have significant reproducibility issues. In a comparison of antibody titer measurements on identical samples across several laboratories, HAI and VN assays had geometric coefficients of variation ranging from 138–261% and 256–369%, which corresponded to fold differences of 16–128 and 91–724 respectively [58]. For the VN assay, 21% of replicate measurements within the same laboratory differed by more than 2-fold [58]. Many studies of influenza vaccination use the FDA criteria of seroconversion [18, 59, 60] (>4 fold increase in HAI titer post-vaccination) to categorize subjects into ‘responders’ and ‘nonresponders’ to vaccination; these results demonstrate that this classification may vary greatly between laboratories. As the accuracy of the endpoint measurement is crucial for obtaining meaningful analysis in any study, improvements in standardization of these assays will be of great benefit to the field.
Alternatively, systems biology approaches can be harnessed to establish novel correlates of protection. Currently, nested case-control studies are used in clinical trials to retrospectively compare subjects who developed infection with subjects that remained uninfected and identify distinguishing biomarkers [61, 62]. The development of experimental challenge models (such as those available for malaria, typhoid, Shigella, and dengue fever as described above) provides a complementary approach to enable detection of subjects protected from or vulnerable to infection. Systems level profiling of these subjects can then be used to reveal reliable molecular markers of vaccine-induced protection.
Technological Challenges
A distinct feature of a true systems analysis of vaccine response is the integration of diverse measurement types. Due to the high coverage and relative ease of measurement provided by microarrays and RNA-sequencing, most of the initial work in systems vaccinology has focused on transcriptional responses to vaccination. However, modulation of gene expression represents only one of many methods by which biological systems respond to perturbation. Among other factors, changes in protein, metabolite, and signaling molecule abundances, post-translational modifications, and shifts in cell subset populations all contribute to the immune response post-vaccination. As the technologies capable of detecting these additional layers of biological regulation improve, future systems vaccinology analyses should incorporate these measurements in order to develop a more complete picture of the biological mechanisms of vaccine-induced immunity.
As with any type of assay, measurements made using high-throughput technologies suffer from varying degrees of noise. This creates difficulty in reliable detection of weak signals, such as those coming from low-expressed transcripts or metabolites present in limited amounts. Unfortunately, the abundance of a particular molecule is not necessarily an indicator of its importance, and these difficult to detect components often have significant biological impact. Primary examples of this are transcription factors, which are often present in very low numbers but are important regulators of many biological processes [64]. Measurement noise, along with batch effects (particularly in the case of microarrays) [65], can also reduce the reproducibility of results. This is an important obstacle that needs to be overcome in order for laboratory findings such as predictive signatures of vaccine efficacy to be translated into tools with clinical diagnostic value.
Another challenge common to many areas of medical research is the limited number of samples and time points available in a given dataset due to ethical and logistical constraints. As sample collection and processing is both taxing on trial participants and costly, studies are often restricted to a small group of subjects who are evaluated at a handful of time points. While small sample sizes create obvious problems in statistical hypothesis testing, the limited availability of time course data also poses a significant obstacle to successful kinetic modeling of immune responses to vaccination. As immune responses involve coordinated interactions between diverse cell types across multiple tissues, understanding of cellular and transcriptional dynamics post-vaccination is an important goal of systems vaccinology. However, while transcriptional responses can take place within minutes to hours [66], in clinical studies sample collection is often only feasible on the time scale of days to weeks. This is complicated by the fact that while immune cells migrate between the blood, the site of infection/vaccination, and the lymph nodes, analysis is often performed only on immune cells within peripheral blood. These limitations, combined with the inter-individual variability in responses, make it extremely difficult to capture the entirety of transcriptional and cellular events that occur in vivo post-vaccination.
In addition to limited number of samples, in some cases (e.g. pediatric studies) researchers are also constrained by the small sample volume that can be collected from subjects. Typical guidelines for blood sample volume limits range from 1–5% of total blood volume, in children this equates to 1–3 mL/kg of body weight [67]. The available blood drawn from neonates may therefore be <5 mL. As the concentration of PBMCs is typically around 1 million per mL of blood, a neonatal blood sample may only contain a few million PBMCs. This limits the potential analyses that can be performed, as assays such as flow cytometry and ELISPOT normally utilize larger numbers of cells. Development of more sensitive technologies that require fewer cells or less starting material, such as RNA-sequencing in place of microarrays, will enable improved analysis of vaccine response in these populations.
Unlike conventional techniques, which generate a limited number of measurements, high-throughput technologies produce tens or hundreds of thousands of measurements per sample. These developments have resulted in an explosion of datasets where the number of features (or dimension, p) is much larger than the sample size (n), known as ‘large p, small n’ or ‘high dimension, low sample size’ (HDLSS) data. Under these conditions, traditional statistical and computational algorithms perform poorly due to the resulting sparsity of the dataset; this is known as the ‘curse of dimensionality’. As a result, an entire branch of statistics has emerged to handle this type of data and reduce spurious results [68]. In order to avoid being swamped by false positives, suitable corrections for multiple hypothesis testing must be used to control false discovery rates [69, 70]. Machine learning techniques [71] such as logistic regression, Bayesian networks, support vector machines (SVMs), and artificial neural networks (ANNs) are being used to predict antibody responses to vaccination based on expression values of sets of genes (or other measurements such as cell population frequencies) made shortly after vaccination (or even potentially at baseline). However, a large majority of the measurements made using high-throughput technologies are not relevant to the particular biological processes being examined, and these algorithms are not equipped to identify the subset of features (genes, metabolites, etc.) that are most predictive of responsiveness to vaccination. Therefore, an additional step known as feature selection must be performed, in order to reduce the computational complexity of the problem and to prevent overfitting (in which the algorithm performs well on training data but poorly on independent testing data). Various approaches to this task have been developed, such as random forests [72], filter and wrapper approaches [73], and Bayesian methods [74], however this is an area of ongoing research. This is a critical step, because the identification of a small number of predictive features is necessary for clinical diagnostic applications, and the identity of these features provides insight into the underlying biology of the immune response to vaccination.
Finally, the generation of such large amounts of information requires appropriate computational resources as well as data management and storage. Creation of relevant and centralized databases such as the Immunology Database and Analysis Portal (ImmPort) [75] (http://immport.niaid.nih.gov/) will promote data sharing between groups and facilitate integration of diverse data types. Standardization of measurement techniques as well as data formatting should be encouraged, as this will empower the meta-analysis of multiple related studies, enabling discovery of novel associations which were previously undetectable.
Drowning in a Sea of Big Data
The revolution in high-throughput technologies has created a dramatic change in the landscape of biological research: with such large amounts of information generated with each experiment, we are now ‘drowning in a sea of big data.’ A cornerstone of successful systems vaccinology is the ability to extract meaningful knowledge from this wealth of data, and then to harness this knowledge to generate improved biological understanding of the immune system’s response to vaccination. In the first transition, from data to knowledge, statistical approaches [76] are generally used to identify sets of genes, metabolites, etc. that undergo significant changes during system perturbation (e.g. before and after vaccination). However, a simple list of differentially expressed genes or metabolites is not highly informative, and may contain false positives. One approach to improve this type of analysis is to integrate expression measurements with a priori knowledge about the interactions and co-expression of genes within a given biological context. One such bioinformatics tool is gene-set enrichment analysis [77], which identifies enrichment of biologically relevant groups of genes (modules) within gene lists ranked by response to perturbation. This can be performed using manually curated sets of biological pathways, such as the Kyoto Encyclopedia of Genes and Genomes [78] and Reactome [79] databases, or through extraction of gene modules from existing data by searching for groups of co-expressed genes across experimental conditions or biological contexts [30, 80, 81]. MSigDB, a popular gene set database, contains collections of both types [82]. Examining changes in activity of pathways or modules rather than individual genes reduces noise and appearance of false positives while simultaneously providing improved biological meaning and functional context to the data analysis.
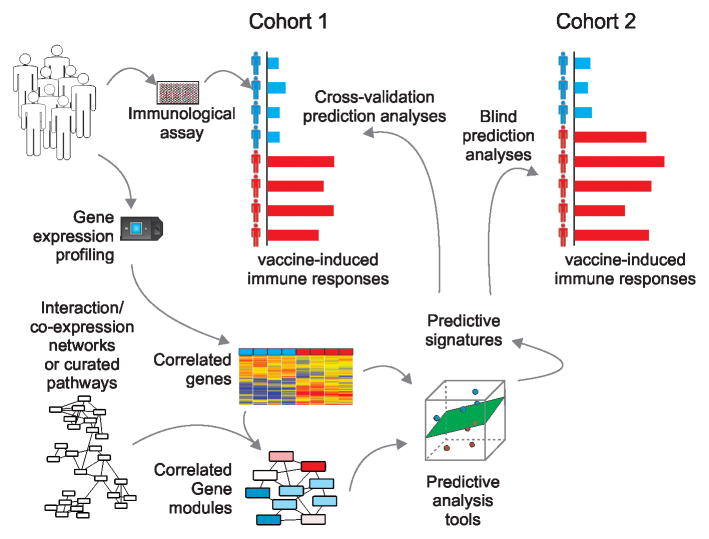
Differential transcriptional pathways and networks can also be coupled with feature selection and machine learning algorithms (such as those mentioned in the previous section) to generate biologically informative predictive signatures of vaccine response (Fig. 2). These signatures can be used to assess vaccine efficacy and identify subjects at risk for infection. In addition, analysis of predictive pathways can yield insight into the mechanisms responsible for deficient immune responses. Once predictive signatures are generated using the data at hand, they should be validated in independent cohorts to ensure their robustness across diverse populations.
Figure 2. Predictive analysis of vaccine response.
The goal of predictive analyses is to identify early transcriptional signatures capable of predicting vaccine-induced immune responses. Integration of transcriptomic measurements with gene co-expression networks or biological pathways provides improved functional understanding of the immune processes activated in response to vaccination. These gene or module-level features can then be used as inputs to feature selection and machine learning algorithms. Such tools enable identification of molecular signatures that are predictive of responses to vaccination, such as antibody or T cell responses. Once predictive signatures are verified through blind prediction of independent datasets, they can be employed in a clinical setting to rapidly identify vaccinees with deficient responses and assess vaccine efficacy.
While the knowledge of biological pathways activated or suppressed in response to vaccination is useful in its own right, achieving deeper understanding and control over the mechanisms by which vaccines induce protective immune responses requires human knowledge and insight coupled with traditional methods of experimental validation. Ideally, observations made through high-throughput data analysis can help generate biological hypothesis that are then tested in animal models such as knockout mice. Recently our group has used this approach to identify a novel role for the integrated stress response, mediated by EIF2AK4 [eukaryotic initiation factor 2 α-kinase 4, also known as general control nonderepressible 2 kinase (GCN2)], in stimulating autophagy and antigen presentation within dendritic cells during response to YF-17D [83]. In our original systems biology analysis of the YF-17D vaccine, expression of GCN2 in PBMCs on day 7 post-vaccination was shown to be predictive of the later CD8+ T cell response [14]. As a regulator of the integrated stress response, accumulation of uncharged tRNA during amino acid starvation activates GCN2, which results in phosphorylation of eukaryotic initiation factor 2 α (eIF2α). This process leads to reduced activity of the eIF2 complex, resulting in reduced rates of protein synthesis and stress granule formation [84]. However, GCN2’s role in immune responses is not clear. The capability of GCN2 expression to predict CD8+ T cell responses to YF-17D led us to believe that this kinase may be important for priming of the adaptive immune response. Follow up work in mice revealed that YF-17D induced amino acid starvation in dendritic cells, inducing autophagy and antigen cross-presentation in a GCN2-dependent manner [83]. This reveals an unappreciated connection between the ancient nutrient sensing pathway and elicitation of adaptive immunity by dendritic cells.
Another mechanistic discovery enabled by a systems vaccinology is the role that the intestinal microbiota play in stimulating antibody responses to vaccination. This development was prompted by the finding in our systems-level analysis of human immune responses to TIV vaccination that expression of the toll-like receptor TLR5 on day 3 post-vaccination was significantly correlated with the day 28 antibody response [18]. As TLR5 is a receptor specific for flagellin, the protein subunit of bacterial flagellum, its association with antibody response to a viral vaccine was unexpected. The importance of TLR5 sensing of intestinal bacteria during TIV vaccination was confirmed in experiments with antibiotic treated, germ-free, and TLR5−/− mice, all of which experienced significant reduction in antibody responses after TIV vaccination compared to controls [85]. Further work demonstrated that sensing of flagellin by TLR5 promoted plasma cell differentiation directly and by stimulating lymph node macrophages to produce plasma cell growth factors. Interestingly, the loss of intestinal microbiota did not result in reduction of humoral responses to adjuvanted vaccines or the live-attenuated YF-17D vaccine, indicating that the microbiota may be acting as a natural adjuvant in the absence of other sources of immune stimulation. As the microbiome is affected by diet and varies widely across the world [86], gut microbial composition adds another layer of diversity in the human population that should be considered in future studies of immune responses to vaccination. The experimental evaluation of the role of the microbiome in promoting immunity during vaccination in humans is ongoing.
Conclusions
Developments in high-throughput technologies are enabling vaccinologists to investigate the immune responses induced by vaccines at a greater depth than ever before. More importantly, these advances are facilitating the identification of robust molecular and cellular signatures of protective immunity, which can help to generate diagnostic tools that reduce the length and cost of current clinical trials. In this developing field, vaccinologists, immunologists, bioinformaticians, and systems biologists must work hand in hand to advance our understanding of the molecular mechanisms by which vaccines induce protective immunity and help drive development of the next generation of vaccines.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Behbehani AM. The smallpox story: life and death of an old disease. Microbiol Rev. 1983;47:455–509. doi: 10.1128/mr.47.4.455-509.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehrenfeld E, Modlin J, Chumakov K. Future of polio vaccines. Expert Rev Vaccines. 2009;8:899–905. doi: 10.1586/erv.09.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, et al. Comparison of the influenza virus-specific effector and memory B-cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol. 2007;81:215–28. doi: 10.1128/JVI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, Michael NL. Accelerating HIV-1 Vaccine Efficacy Trials. Cell. 2014;159:969–72. doi: 10.1016/j.cell.2014.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funderud A, Aas-Hanssen K, Aksaas AK, Hafte TT, Corthay A, Munthe LA, et al. Isoform-specific regulation of immune cell reactivity by the catalytic subunit of protein kinase A (PKA) Cell Signal. 2009;21:274–81. doi: 10.1016/j.cellsig.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol. 2014;14:361–76. doi: 10.1038/nri3682. [DOI] [PubMed] [Google Scholar]
- 7.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 2010;184:4062–8. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensinger SJ, Tontonoz P. Integration of metabolism and inflammation by lipid-activated nuclear receptors. Nature. 2008;454:470–7. doi: 10.1038/nature07202. [DOI] [PubMed] [Google Scholar]
- 10.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–25. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 11.Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med. 2012;4:193–205. doi: 10.1002/wsbm.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–29. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulendran B. Systems vaccinology: probing humanity’s diverse immune systems with vaccines. Proc Natl Acad Sci U S A. 2014;111:12300–6. doi: 10.1073/pnas.1400476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–31. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–7. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 17.Pulendran B, Oh JZ, Nakaya HI, Ravindran R, Kazmin DA. Immunity to viruses: learning from successful human vaccines. Immunol Rev. 2013;255:243–55. doi: 10.1111/imr.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12:786–95. doi: 10.1038/ni.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Nino D, et al. Early Patterns of Gene Expression Correlate With the Humoral Immune Response to Influenza Vaccination in Humans. J Infect Dis. 2011;203:921–9. doi: 10.1093/infdis/jiq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vahey MT, Wang Z, Kester KE, Cummings J, Heppner DG, Jr, Nau ME, et al. Expression of genes associated with immunoproteasome processing of major histocompatibility complex peptides is indicative of protection with adjuvanted RTS,S malaria vaccine. J Infect Dis. 2010;201:580–9. doi: 10.1086/650310. [DOI] [PubMed] [Google Scholar]
- 21.Reif DM, Motsinger-Reif AA, McKinney BA, Rock MT, Crowe JE, Jr, Moore JH. Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun. 2009;10:112–9. doi: 10.1038/gene.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zak DE, Andersen-Nissen E, Peterson ER, Sato A, Hamilton MK, Borgerding J, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci U S A. 2012;109:E3503–12. doi: 10.1073/pnas.1208972109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E, et al. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–44. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chulay JD, Schneider I, Cosgriff TM, Hoffman SL, Ballou WR, Quakyi IA, et al. Malaria transmitted to humans by mosquitoes infected from cultured Plasmodium falciparum. Am J Trop Med Hyg. 1986;35:66–8. doi: 10.4269/ajtmh.1986.35.66. [DOI] [PubMed] [Google Scholar]
- 27.Waddington CS, Darton TC, Jones C, Haworth K, Peters A, John T, et al. An Outpatient, Ambulant-Design, Controlled Human Infection Model Using Escalating Doses of Salmonella Typhi Challenge Delivered in Sodium Bicarbonate Solution. Clin Infect Dis. 2014;58:1230–40. doi: 10.1093/cid/ciu078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marwick C. Volunteers in typhoid infection study will aid future vaccine development. JAMA. 1998;279:1423–4. doi: 10.1001/jama.279.18.1423. [DOI] [PubMed] [Google Scholar]
- 29.Bodhidatta L, Pitisuttithum P, Chamnanchanant S, Chang KT, Islam D, Bussaratid V, et al. Establishment of a Shigella sonnei human challenge model in Thailand. Vaccine. 2012;30:7040–5. doi: 10.1016/j.vaccine.2012.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardner EM, Gonzalez EW, Nogusa S, Murasko DM. Age-related changes in the immune response to influenza vaccination in a racially diverse, healthy elderly population. Vaccine. 2006;24:1609–14. doi: 10.1016/j.vaccine.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 32.Jackson LA, Janoff EN. Pneumococcal vaccination of elderly adults: new paradigms for protection. Clin Infect Dis. 2008;47:1328–38. doi: 10.1086/592691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutz W, Sanderson W, Scherbov S. The coming acceleration of global population ageing. Nature. 2008;451:716–9. doi: 10.1038/nature06516. [DOI] [PubMed] [Google Scholar]
- 34.Duraisingham SS, Rouphael N, Cavanagh MM, Nakaya HI, Goronzy JJ, Pulendran B. Systems biology of vaccination in the elderly. Curr Top Microbiol Immunol. 2013;363:117–42. doi: 10.1007/82_2012_250. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, et al. Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest. 2011;121:3109–19. doi: 10.1172/JCI57834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagar LE, Gentleman B, Pircher H, McElhaney JE, Watts TH. Influenza-Specific T Cells from Older People Are Enriched in the Late Effector Subset and Their Presence Inversely Correlates with Vaccine Response. PLOS ONE. 2011:6. doi: 10.1371/journal.pone.0023698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang IS, Hong MS, Nolasco H, Park SH, Dan JM, Choi JY, et al. Age-associated change in the frequency of memory CD4(+) T cells impairs long term CD4(+) T cell responses to influenza vaccine. J Immunol. 2004;173:673–81. doi: 10.4049/jimmunol.173.1.673. [DOI] [PubMed] [Google Scholar]
- 38.Orru V, Steri M, Sole G, Sidore C, Virdis F, Dei M, et al. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–56. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padgett DA, Glaser R. How stress influences the immune response. Trends Immunol. 2003;24:444–8. doi: 10.1016/s1471-4906(03)00173-x. [DOI] [PubMed] [Google Scholar]
- 42.Woods JA, Vieira VJ, Keylock KT. Exercise, inflammation, and innate immunity. Neurol Clin. 2006;24:585–99. doi: 10.1016/j.ncl.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor D, Pollard AJ. Characterizing Vaccine Responses Using Host Genomic and Transcriptomic Analysis. Clin Infect Dis. 2013;57:860–9. doi: 10.1093/cid/cit373. [DOI] [PubMed] [Google Scholar]
- 45.Gelder CM, Lambkin R, Hart KW, Fleming D, Williams OM, Bunce M, et al. Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J Infect Dis. 2002;185:114–7. doi: 10.1086/338014. [DOI] [PubMed] [Google Scholar]
- 46.Moss AJ, Gaughran FP, Karasu A, Gilbert AS, Mann AJ, Gelder CM, et al. Correlation between Human Leukocyte Antigen Class II Alleles and HAI Titers Detected Post-Influenza Vaccination. PLOS ONE. 2013:8. doi: 10.1371/journal.pone.0071376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poland GA. Immunogenetic mechanisms of antibody response to measles vaccine: the role of the HLA genes. Vaccine. 1999;17:1719–25. doi: 10.1016/s0264-410x(98)00429-0. [DOI] [PubMed] [Google Scholar]
- 48.Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. Human Leukocyte Antigen Genotypes in the Genetic Control of Adaptive Immune Responses to Smallpox Vaccine. J Infect Dis. 2011;203:1546–55. doi: 10.1093/infdis/jir167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ovsyannikova IG, Pankratz VS, Salk HM, Kennedy RB, Poland GA. HLA alleles associated with the adaptive immune response to smallpox vaccine: a replication study. Hum Genet. 2014;133:1083–92. doi: 10.1007/s00439-014-1449-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li ZK, Nie JJ, Li J, Zhuang H. The effect of HLA on immunological response to hepatitis B vaccine in healthy people: A meta-analysis. Vaccine. 2013;31:4355–61. doi: 10.1016/j.vaccine.2013.06.108. [DOI] [PubMed] [Google Scholar]
- 51.Lauterbach H, Ried C, Epstein AL, Marconi P, Brocker T. Reduced immune responses after vaccination with a recombinant herpes simplex virus type 1 vector in the presence of antiviral immunity. J Gen Virol. 2005;86:2401–10. doi: 10.1099/vir.0.81104-0. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki S, He XS, Holmes TH, Dekker CL, Kemble GW, Arvin AM, et al. Influence of prior influenza vaccination on antibody and B-cell responses. PLOS ONE. 2008;3:e2975. doi: 10.1371/journal.pone.0002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47:401–9. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 54.McElhaney JE, Xie D, Hager WD, Barry MB, Wang Y, Kleppinger A, et al. T cell responses are better correlates of vaccine protection in the elderly. J Immunol. 2006;176:6333–9. doi: 10.4049/jimmunol.176.10.6333. [DOI] [PubMed] [Google Scholar]
- 55.Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat Med. 2004;10:806–10. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- 56.Hoft DF. Tuberculosis vaccine development: goals, immunological design, and evaluation. Lancet. 2008;372:164–75. doi: 10.1016/S0140-6736(08)61036-3. [DOI] [PubMed] [Google Scholar]
- 57.Reyes-Sandoval A, Pearson FE, Todryk S, Ewer K. Potency assays for novel T-cell-inducing vaccines against malaria. Curr Opin Mol Ther. 2009;11:72–80. [PubMed] [Google Scholar]
- 58.Stephenson I, Das RG, Wood JM, Katz JM. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: An international collaborative study. Vaccine. 2007;25:4056–63. doi: 10.1016/j.vaccine.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 59.Frey SE, Bernstein DI, Brady RC, Keitel WA, Sahly HE, Rouphael NG, et al. Phase II trial in adults of concurrent or sequential 2009 pandemic H1N1 and 2009–2010 seasonal trivalent influenza vaccinations. Vaccine. 2015;33:163–73. doi: 10.1016/j.vaccine.2014.10.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greenberg DP, Robertson CA, Noss MJ, Blatter MM, Biedenbender R, Decker MD. Safety and immunogenicity of a quadrivalent inactivated influenza vaccine compared to licensed trivalent inactivated influenza vaccines in adults. Vaccine. 2013;31:770–6. doi: 10.1016/j.vaccine.2012.11.074. [DOI] [PubMed] [Google Scholar]
- 61.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial. New Engl J Med. 2012;366:1275–86. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olotu A, Clement F, Jongert E, Vekemans J, Njuguna P, Ndungu FM, et al. Avidity of Anti-Circumsporozoite Antibodies following Vaccination with RTS,S/AS01(E) in Young Children. PLOS ONE. 2014:9. doi: 10.1371/journal.pone.0115126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pulendran B, Miller J, Querec TD, Akondy R, Moseley N, Laur O, et al. Case of yellow fever vaccine--associated viscerotropic disease with prolonged viremia, robust adaptive immune responses, and polymorphisms in CCR5 and RANTES genes. J Infect Dis. 2008;198:500–7. doi: 10.1086/590187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 65.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–63. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kauffman RE. Clinical trials in children: problems and pitfalls. Paediatr Drugs. 2000;2:411–8. doi: 10.2165/00128072-200002060-00001. [DOI] [PubMed] [Google Scholar]
- 68.Johnstone IM, Titterington DM. Statistical challenges of high-dimensional data. Philos Trans A Math Phys Eng Sci. 2009;367:4237–53. doi: 10.1098/rsta.2009.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- 70.Storey JD, Tibshirani R. Statistical significance for genomewide studies. P Natl Acad Sci USA. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kotsiantis SB, Zaharakis ID, Pintelas PE. Machine learning: a review of classification and combining techniques. Artif Intell Rev. 2006;26:159–90. [Google Scholar]
- 72.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 73.Blum AL, Langley P. Selection of relevant features and examples in machine learning. Artif Intell. 1997;97:245–71. [Google Scholar]
- 74.Inza I, Larranaga P, Etxeberria R, Sierra B. Feature Subset Selection by Bayesian network-based optimization. Artif Intell. 2000;123:157–84. [Google Scholar]
- 75.Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58:234–9. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 76.Jeffery IB, Higgins DG, Culhane AC. Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinform. 2006;7:359. doi: 10.1186/1471-2105-7-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–7. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vastrik I, D’Eustachio P, Schmidt E, Gopinath G, Croft D, de Bono B, et al. Reactome: a knowledge base of biologic pathways and processes (vol 8, pg 39, 2007) Genome Biol. 2009:10. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–64. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–80. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ravindran R, Khan N, Nakaya HI, Li S, Loebbermann J, Maddur MS, et al. Vaccine activation of the nutrient sensor GCN2 in dendritic cells enhances antigen presentation. Science. 2014;343:313–7. doi: 10.1126/science.1246829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–21. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–92. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]