Abstract
The epithelium is the major entry point for many viruses but the processes protecting barrier surfaces against viral infections are incompletely understood. We identify interleukin (IL)-22 produced by group 3 innate lymphoid cells (ILC3s) as an amplifier of interferon (IFN)-λ signaling, a synergism required to curtail replication of rotavirus, the leading cause of childhood gastroenteritis. Cooperation between IL-22 and IFN-λ receptors, both of which are preferentially expressed by intestinal epithelial cells, was required for optimal STAT1 transcription factor activation and expression of interferon-stimulated genes. This data suggests that epithelial cells are protected against virus replication by co-opting two evolutionarily related cytokine networks. These data may inform the design of novel immunotherapies of virus infections that are sensitive to IFNs.
Many viruses such as hepatitis B and C viruses (HBV, HCV), human immunodeficiency virus (HIV), influenza virus, rotavirus, and poliovirus use replication in epithelial cells to establish infections of multicellular hosts but the molecular networks protecting barrier surfaces against viral infections are incompletely understood1,2. It is generally believed that interferons (IFNs), such as type I (i.e., IFN-α, IFN-β) and type III IFNs (i.e., IFN-λ2 and IFN-λ3 in mice, from now on collectively referred to as IFN-λ), play essential roles in antiviral defense. IFNs are produced by virus-infected epithelial cells and induce an anti-viral state in cells receiving an IFN signal, thereby constituting an efficient innate defense mechanism3,4. While type I and type III IFNs and their receptors are only distantly related5, 6, they show virtually identical signaling behaviour in that they activate the STAT1-STAT2 transcription factor signaling pathway, which leads to the formation of the ternary interferon-stimulated gene factor-3 (ISGF3) complex (consisting of STAT1, STAT2 and IRF9) that controls transcription of more than 300 interferon-stimulated genes (ISGs) acting in concert to restrict replication of viruses5, 7, 8, 9, 10, 11.
IFN-λ and the IFN-λ receptor (IFN-λR) share homologies with IL-10 family cytokines and their receptors. In particular, the gene encoding the α chain of the IL-22 receptor (Il22ra1) is the closest relative of the gene encoding the IFN-λR1 chain (Ifnlr1)5, 6. The Il22ra1 and the Ifnlr1 genes are adjacent on mouse chromosome 4 or human chromosome 15, 6 and, both, the IL-22Rα and the IFN-λR1 protein associate with the IL-10Rβ (also known as IL-10R2) chain to form functional, heterodimeric receptor complexes. The close relationship between IFN-λR and IL-22R is interesting because both receptors are preferentially expressed by epithelial cells12, 13, indicating a designated role in controlling epithelial function in response to infections. Although the IFN-λR connects to STAT1 and STAT2 signaling, the IL-22R engages the STAT3 signaling pathway14, and STAT3 signaling has been implicated in virtually all known biological functions of IL-22, in particular in up-regulating expression of antibacterial genes15 and of genes promoting tissue repair16.
Rotavirus infection is a suitable model system to analyze the cytokine neworks required for the protection of the intestinal barrier against virus infection because rotavirus has a preferential tropism for the villous epithelium of the small intestine of infants in humans and mice. Infection with rotavirus, a double-stranded RNA virus of the Reoviridae family is the leading cause of viral gastroenteritis in infants causing annually more than 500,000 fatalities worldwide17. Rotavirus is highly adapted to its host and homologous animal models of rotavirus infection have shown that less than 10 infectious units of virus replicate vigorously and initiate diarrheal disease including viral shedding and efficient spread to uninfected mice. Such efficient replication of virus is in part mediated by efficient machinery of the virus to evade immune recognition and/or to suppress IFN-mediated innate immune responses18, 19, 20. The mucosal type III IFN system21 but not type I IFNs plays an important role in curtailing early rotavirus replication in its homologous host minimizing tissue damage22, 23.
Given the close relationship between IL-22 and IFN-λ and their concerted action on epithelial cells, we have interrogated the role of IL-22 for protection against a murine virus with preferential tropism to small intestinal epithelial cells. We show that group 3 innate lymphoid cells (ILC3)-derived IL-22 is required for control of murine rotavirus infection. IL-1α, an epithelial alarmin released by virus-infected cells, but not IL-23 or IL-1β mediated a high degree of IL-22 expression by ILC3s following rotavirus infection. IL-22-mediated protection against rotavirus-mediated tissue damage did not require STAT3, a transcription factor involved in virtually all previously reported biological activities of IL-22. Instead, IL-22 synergized with IFN-λ for optimal activation of STAT1 and expression of ISGs. Our data identify previously unknown synergies between two cytokines acting on intestinal epithelial cells for the protection of the intestinal barrier against viral infections. Such synergies between these evolutionarily related cytokines might be harnessed for the clinical therapy of chronic virus infections.
Results
IL-22 curtails rotavirus replication in its homologous host
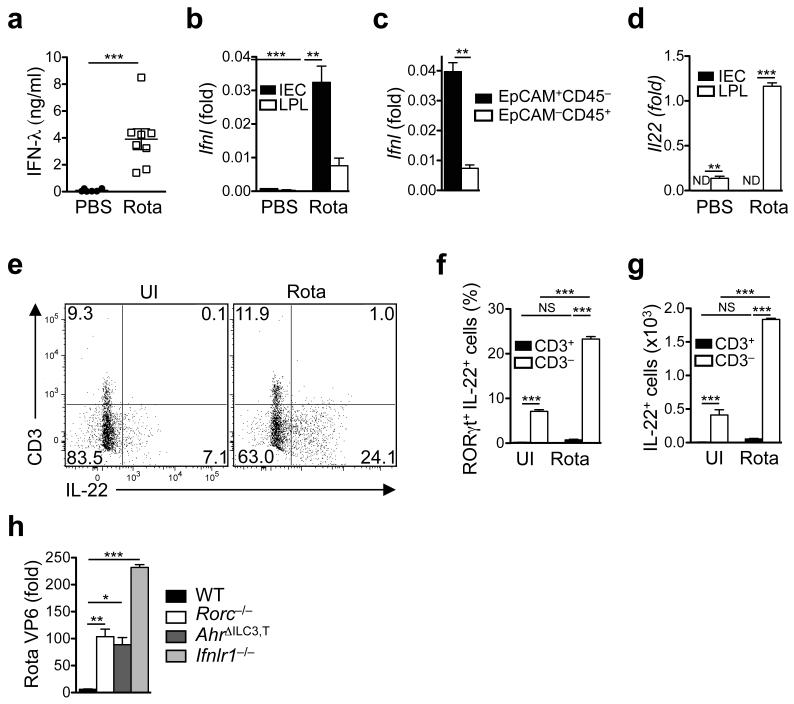
There is a paucity of information about whether amplifiers of IFN signaling exist that could be harnessed for better control of viral infections. The related cytokines IL-22 and IFN-λ act on the same cellular target (i.e. intestinal epithelial cells, IECs), but cooperation between these two mucosal cytokine networks has not been explored. To model the infection of human infants, we infected Il22−/− mice, Ifnlr1−/− mice and wildtype mice either fostered to wildtype mothers (Fig. 1a,b) or to Ifnlr1−/− mothers (Fig. 1c,d) at day 7 of age (hereon referred to as ‘suckling mice’) with 7×102 ID50 (infectious dose 50, see Methods) of homologous murine rotavirus (epizootic diarrhea of infant mice, EDIM) and determined virus load by quantitative RT-PCR from small intestinal tissues (Fig. 1a,c), by measuring the amount of virus shed into the colon lumen using ELISA (Fig. 1b,d) and by visualizing the viral antigen in the small intestine by immunofluorescence staining (Fig. 1e). In extension of previously published work21, Ifnlr1−/− mice had a significantly higher virus burden at day 1 (peak of virus replication; Supplementary Fig. 1) and at day 4 of rotavirus infection (peak of virus shedding; Fig. 1b,d) compared to wildtype mice. Increased virus replication in Ifnlr1−/− mice was likely independent of the expression of a wildtype Mx1 allele, an ISG involved in restricting virus replication, because the Mx1 status did not impact the contol of rotavirus replication (Supplementary Fig. 2a). Strikingly, Il22−/− mice also showed a significant increase in virus load albeit to a lesser extent than Ifnlr1−/− mice (Fig. 1a-d). Thorough analysis of intestinal pathology in infected mice over the entire extension of the small intestinal tube revealed important insights. The pathological lesions following rotavirus infection are discontinuous and areas of histopathological damage are interspersed with healthy tissue. Therefore, it was pivotal to analyze the entire small intestinal tube to assess the extent of tissue damage. In wildtype mice, replicating rotavirus was restricted to the distal third of the small intestine, whereas in both Il22−/− and Ifnlr1−/− mice virus replication was found throughout the full extension of the small intestine (Fig. 1e,f). These data indicate that IL-22 and IFN-λ signaling protected in particular the proximal parts of the small intestine against rotavirus infection.
Fig. 1. Control of rotavirus infection requires IL-22.
Seven day old mice were orally infected with 7×102 ID50 rotavirus and samples were collected at the indicated times following infection. Newborn mice were fostered by wildtype (WT) mothers (a, b) or Ifnlr1−/− mothers (c, d). Virus replication was analyzed by (a, c) RT-qPCR using RNA obtained from small intestinal tissue, (b, d) ELISA of colon homogenates (see Methods), (e) immunofluorescence staining and (f) by determining the distribution and mean number (± SEM) of infected cells per visual field across the small intestine. The size of the pies represents the mean number of infected cells per visual field and their distribution across the entire small intestine: WT (239 ± 18), Il22−/− (958.4 ± 10), Ifnlr1−/− (1187.5 ± 108). (g) H&E staining of small intestinal tissue sections. (h) Clinical score. (i) Body weight at day 19 after infection as the percentage of body weight at day 0. All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-k) Mean ± SEM. (a, b) n ≥ 6. (c-g) n ≥ 4. (i) n ≥ 3. Data are representative of four (a-b), two (c-d) and two (e-g) individual experiments. i, Data combined from two individual experiments. (a-i) One-way ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. Scale bars, 50 μm.
Despite effective virus replication and diarrhea in all mice tested, histological analysis of small intestinal tissues from infected wildtype mice showed only moderate damage of the epithelial barrier in the distal small intestine (Fig. 1g,h). Infected Il22−/− mice and Ifnlr1−/− mice exhibited a more severe pathology characterized by vacuolization, epithelial erosions and villus disruption across the entire small intestine (Fig. 1g,h, Supplementary Fig. 2b). These data indicate that IL-22 and IFN-λ suppress virus replication to an extent that prevents the development of severe epithelial damage. Uninfected mice of all genotypes did not show any signs of pathology (data not shown). In addition, rotavirus-infected Il22−/− and Ifnlr1−/− mice showed retardation of weight gain compared to infected wildtype mice that was not compensated by almost three weeks following infection (Fig. 1i), when virus had become undetectable in all mouse strains investigated. Notably, weight gain of uninfected Il22−/− and Ifnlr1−/− mice did not differ from that of wildtype mice indicating that Il22−/− or Ifnlr1−/− mice have no genetically determined growth retardation.
In contrast to suckling mice, adult wildtype mice infected with 3.5×104 ID50 do not develop clinically overt symptoms of rotavirus infection, although they have low (but detectable) viral titers both in tissues and in feces (Supplementary Fig. 2c,d) indicating that all mice were infected. Adult mice genetically deficient for Il22 or Ifnlr1 also showed impaired control of rotavirus replication (Supplementary Fig. 2c,d). Thus, IL-22 is required to curtail rotavirus replication in both, suckling and adult animals.
Although IL-22 has been implicated in tissue repair following viral infection26, we did not find any significant differences in the proliferation of crypt-resident cells (i.e., stem cells) (Supplementary Fig. 2e,f) and in the upward migration of differentiating IECs (Supplementary Fig. 2g,h) effectively replacing damaged cells following viral infection between infected wildtype, Il22−/− and Ifnlr1−/− mice.
It has previously been noted that mutations in cytokine genes can lead to differences in microbial communities which could impact the course of rotavirus infection. Because distinct microbiota can be acquired through maternal transmission24, we fostered newborn pups either with wildtype or Ifnlr1−/− mothers. No differences in the course of the infection or in the severity of pathology were observed (Fig. 1a,b vs. Fig. 1c,d and data not shown). We also directly examined the small intestinal microbiota of newborn and adult mice using 454 sequencing of 16S rDNA amplicons. In adult Il22−/− mice, differences in colon-resident microbial communities were previously reported25. However, no significant differences in the composition of the small intestinal microbiota could be found between adult wildtype and Il22−/− mice, between litters of adult mice, or between genotypes within litters of adult mice (Supplementary Fig. 3a-c, Supplementary Table 1). Using principal coordinate analysis (PCoA) and analysis of molecular variance (AMOVA), we found the microbiota of suckling mice to be highly litter-specific (Supplementary Fig. 3d-f, Supplementary Table 2). Thus, the course of rotavirus infection in the mouse strains investigated is not determined by differences in microbial communities.
IFN-λ is produced by rotavirus-infected IECs
IFN-λ production is strongly induced following rotavirus infection (Fig. 2a) regardless of the Mx1 status of the mice (Supplementary Fig. 2a), but the cellular sources of IFN-λ production remain obscure. IFN-λ production has been documented for virus-infected epithelial cells21 and for hematopoietic cells (e.g., mononuclear phagocytes, plasmacytoid DCs)27, 28. We purified epithelial cells and hematopoietic cells from uninfected and rotavirus-infected mice. Larger amounts of Ifnl2/3 transcripts (the same primer detected both Ifnl2 and Ifnl3) were found in the epithelial fraction of both suckling (Fig. 2b) and adult rotavirus-infected mice (Supplementary Fig. 4a). Purity of sorted cells was confirmed by differential representation of rotavirus transcripts (Supplementary Fig. 4b) and of tissue-specific genes (Supplementary Fig. 4c,d). As the epithelial fraction contains IECs and intraepithelial leukocytes, we highly purifed IECs and hematopoietic cells based on differential expression of EpCAM and CD45 (Supplementary Fig. 4e) and found Ifnl2/3 transcripts predominantly in EpCAM+CD45– IECs (Fig. 2c). Thus, IECs but not hematopoietic cells are the major source of IFN-λ following rotavirus infection. In order to determine the cell type responding to IFN-λ, we monitored the expression of three ISGs in IECs or hematopoietic cells following rotavirus infection in wildtype and Ifnlr1−/− mice. In IECs (EpCAM+) of the small intestine, ISG expression was entirely IFN-λR-dependent (Supplementary Fig. 4f-h). Thus, IECs are not only the major producers of IFN-λ following rotavirus infection but IECs are also responders to IFN-λ which indicates that the IFN-λ network is an IEC-autonomous, anti-viral defense pathway.
Fig. 2. Rotavirus infection induces production of IFN-λ by intestinal epithelial cells and of IL-22 by ILC3s.
Groups of suckling wildtype mice were infected with rotavirus. Control groups received PBS. (a) IFN-λ production in whole small intestinal tissue of infected suckling mice at day 1 following infection was determined by ELISA. (b, c) Expression of Ifnl (primer detects both Ifnl2 and Ifnl3) was determined by RT-qPCR using a Taqman assay detecting transcripts of both genes. Small intestinal epithelial cells (IEC) and lamina propria leukocytes (LPL) were isolated from suckling mice at day 1 following infection using the EDTA dissociation method (see Methods) (b) or highly purified by sorting epithelial (EpCAM+CD45–) or hematopoietic cells (EpCAM–CD45+) (c). (d) Il22 expression was determined by RT-qPCR using RNA from IEC and LPL isolated by the EDTA dissociation method from suckling mice at day 1 following infection. (e) Flow cytometry analysis of IL-22 and CD3 expression by gated RORγt+ LPL from suckling mice at day 0 (UI, uninfected) and at day 1 following infection. (f) Proportions and (g) absolute numbers of RORγt+IL-22+CD3+ and IL-22+CD3– LPL in uninfected suckling mice at day 1 after infection. (h) Virus load (RT-qPCR of whole small intestinal tissue) from suckling mice of the indicated strains at day 4 after infection. All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-d, f-h) Mean ± SEM. (a) n ≥ 6. (b, d) n ≥ 5. c, n ≥ 3. (f, g) n = 4. (h) n ≥ 5. Data are representative of two (a, c, h) and three (b, d-g) independent experiments. (a, c) Student’s t test. (b, d, f, g) Two-way ANOVA. (h) One-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. ND, not detectable (or not displayable on the scale indicated).
Rotavirus infection enhances IL-22 production by ILC3s
Although IECs were the main source of IFN-λ in rotavirus-infected mice, IL-22 was exclusively expressed by hematopoietic cells of the small intestinal lamina propria in both suckling (Fig. 2d) and adult mice (Supplementary Fig. 5a). Rotavirus infection led to substantially enhanced IL-22 expression in the lamina propria leukocyte population. IL-22 can be produced by group 3 innate lymphoid cells (ILC3s)29, T helper 17 (TH17) cells30, subsets of γδ T cells31 and possibly neutrophils32. The IL-22-producing cells in rotavirus-infected mice expressed the transcription factor RORγt but not CD11b, CD19 (Supplementary Fig. 5b,c) nor CD3 (Fig. 2e) which excludes neutrophils and T cells as substantial IL-22 sources after rotavirus infection. IL-22 production by RORγt+ non-B, non-T cells was indicative of ILC3 subsets. Based on differential CCR6 expression, two major ILC3 populations have been identified33. As the newborn intestine only contains low numbers of CCR6−/lo RORγt+ ILC3s (including the NKp46+ subset of ILC3s)33, the major source of IL-22 in response to rotavirus infection was prenatally emerging CCR6+ ILC3s (Supplementary Fig. 5d). Thus, two evolutionarily related cytokines simultaneously acting on epithelial cells, namely IFN-λ produced by infected IECs and IL-22 produced by CCR6+ ILC3s, curtail rotavirus infection and protect against severe damage to the epithelial barrier.
Following rotavirus infection, both the fraction of ILC3s producing IL-22 (Fig. 2e,f) and the absolute numbers of IL-22-producing ILC3s (Fig. 2g) increased significantly. We tested whether ILC3 deficiency would lead to increased virus replication. Indeed, mice lacking IL-22-producing ILC3s (AhrΔILC3,T and Rorc−/− mice)34, 35 were unable to effectively control rotavirus infection in both suckling (Fig. 2h) and adult mice (Supplementary Fig. 5e), demonstrating that RORγt+ ILC3s are a central component for dampening replication of a virus with preferential tropism for the small intestinal epithelium. In extension of previous data36, we found that rotavirus shedding in mice lacking adaptive immune system components (i.e., Rag2−/− mice lacking all T and B cells and Tcrb−/−Tcrd−/− mice lacking αβ and γδ T cells) was comparable to that observed in wildtype mice (Supplementary Fig. 5e). In contrast, alymphoid Rag2−/−Il2rg−/− mice that have a largely normal mononuclear phagocyte compartment but lack all ILC subsets in addition to conventional lymphocytes showed an increase in rotavirus burden similar to Ifnlr1−/− and Il22−/− mice, assigning an important role to ILCs in curtailing rotavirus infection (Fig. 2h, Supplementary Fig. 5e).
IL-1α enhances IL-22 production by ILC3s
We wondered which virus-induced factors are required for the enhanced expression of IL-22 by ILC3s. IFN-λ signals are not required for IL-22 production because Ifnlr1−/− mice showed no reduction in expression of IL-22 (Supplementary Fig. 5f) or its receptor (Supplementary Fig. 5g). In fact, IL-22 production was enhanced at day 4 following infection in Ifnlr1−/− mice (Supplementary Fig. 5f) correlating with increased expression of the antibacterial lectin RegIIIγ (Supplementary Fig. 5h,i), which is under the control of IL-2215, 29. Previous work assigned an important role to the mononuclear phagocyte-derived cytokines IL-2337 and IL-1β38 in controlling IL-22 production by ILC3s. However, the production of IL-23 (Fig. 3a) and IL-1β (Fig. 3b) was not significantly enhanced following rotavirus infection, and transcripts of these cytokines were only mildly elevated in lamina propria leukocytes but not in IECs (Supplementary Fig. 1e,f and 6a,b). In contrast, rotavirus infection significantly enhanced expression of the alarmin IL-1α by IECs, both on the protein (Fig. 3c) and on the mRNA level (Fig. 3d, Supplementary Fig. 1d). Lamina propria leukocytes (LPL) contained few Il1a transcripts (Fig. 3d). The amount of IL-1α protein available after rotavirus infection was more than 10-fold larger than that of IL-23 or IL-1β (Fig. 3a-c). Mice deficient for the Il23a gene (encoding the p19 subunit of IL-23) showed normal IL-22 (Fig. 3e) and IFN-λ expression (Supplementary Fig. 6c) and had rotavirus titers (day 4) comparable to wildtype mice (Fig. 3f), whereas Il1r1−/− mice lacking both IL-1α and IL-1β signalling showed significantly enhanced virus replication (Fig. 3f). Reduced virus control in Il1r1−/− mice was likely due to reduced expression of Il22 in lamina propria leukocytes (Fig. 3e) because expression of IFN-λ was not reduced (Supplementary Fig. 6c).
Fig. 3. IL-1α produced by intestinal epithelial cells controls rotavirus-induced IL-22 production by ILC3s.
Suckling mice from the indicated mouse strains were infected with rotavirus. (a-c) Cytokine production in small intestinal tissue of uninfected (UI) and infected (day 1) suckling wildtype mice was determined by ELISA. (d, e) Il1a (d) or Il22 (e) expression by small intestinal LPL and IECs (d) or small intestinal tissue (e) of uninfected and infected (day 1) suckling wildtype mice was determined by RT-qPCR. (f) Virus load in the small intestine of suckling mice was determined by RT-qPCR at day 4 after infection. (g-j) Mice were treated with the indicated neutralizing antibodies at day −1 (g-j) and day 1 (h-j) of infection. (g) IL-22 production by RORγt+ LPL was determined by flow cytometry at day 1 following infection. (h) Virus load in colonic tissue at day 4 after infection was determined by ELISA. (i) H&E staining of small intestinal tissue sections. (j) Clinical score. (k, l) Small intestinal lamina propria CD3–, CD19–, CD11b–, KLRG1–, NK1.1–, IL-7R+, Kit+ cells from wildtype animals were highly purified by sorting and cultured in the presence of IL-7, SCF and IL-1α or IL-1β for 3 days. (k) Il22 transcripts in cultured ILC3s were measured by RT-qPCR. (l) IL-22 production was measured by ELISA in culture supernatant. All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a, c) Student’s t test. Data are representative of two (a-c, h-l) or three (d-e, g) independent experiments. (d, e) Two-way ANOVA. (f, j-l) One-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001. NS, not significant.
To identify the relevant type of IL-1 required for IL-22 production and for curtailing rotavirus replication, we injected mice with neutralizing antibodies specific for either IL-1α or IL-1β prior to rotavirus infection. Neutralization of IL-1α abrogated rotavirus-induced IL-22 expression (Fig. 3g, Supplementary Fig. 6d-f) and led to enhanced rotavirus replication (Supplementary Fig. 6g) and shedding (Fig. 3h) and substantially aggravated epithelial pathology (Fig. 3i,j). Neutralization of IL-1β had no such effect. The effectiveness of the antibody treatment was monitored by evaluating IL-1-induced genes (Supplementary Fig. 6h,i). Ex vivo, IL-1α was as efficient as IL-1β in inducing IL-22 production from ILC3s (Fig. 3k,l). Thus, an epithelial alarmin, IL-1α produced and released in response to rotavirus infection is required for enhanced IL-22 production by RORγt+ ILC3s and for curtailment of rotavirus replication.
IL-22 and IFN-λ synergize for optimal expression of ISGs
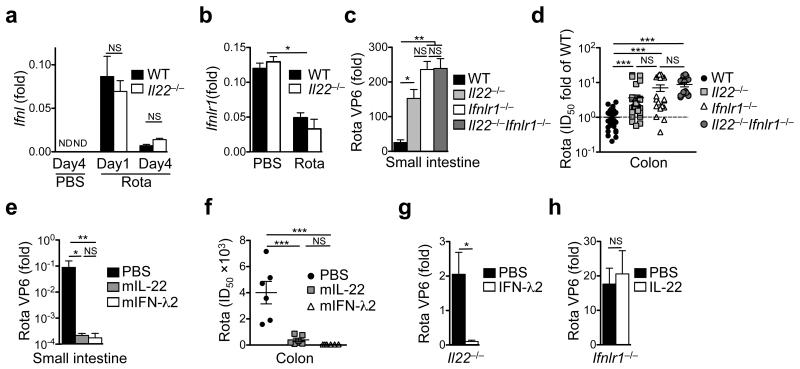
How does IL-22 limit virus replication? IL-22 was not required for the expression of IFN-λ (Fig. 4a) or its receptor (Fig. 4b). Although Ifnl2/3 expression was comparable between wildtype and Il22−/− mice at day 1 following infection, Il22−/− mice produced 3-5-fold more IFN-λ at day 4 following rotavirus infection likely because of increased virus titers. However, the amount of Ifnl2/3 transcripts at day 4 was 5-8-fold lower than at day 1 (Fig. 4a, Supplementary Fig. 1b). Rotavirus infection led to significant downregulation of the IFN-λ receptor mRNA regardless of the presence of IL-22 (Fig. 4b, Supplementary Fig. 1i), whereas mRNA expression of the IL-22 receptor was unchanged (Supplementary Fig. 5g). Thus, IFN-λ signals might become limiting during rotavirus infection perhaps because of the downregulation of IFN-λR expression by IECs or of other perturbations of the IFN-λ signaling path19, 20, 23. We compared virus control in Il22−/− mice, Ifnlr1−/− mice and in mice doubly deficient for IL-22 and the IFN-λ receptor (Il22−/−Ifnlr1−/− mice). Double knockout mice did not have significantly higher virus titers than Ifnlr1−/− or Il22−/− mice (Fig. 4c,d). The absence of any synergistic or additive effects of combined Ifnlr1 and Il22 deficiency in comparison to Ifnlr1 deficiency could indicate that IL-22-mediated virus control requires a functional IFN-λ signalling pathway. To experimentally address this, we exogenously applied IL-22 or IFN-λ to mice that were infected at day 12 after birth and analyzed virus replication. Alhtough injection of IL-22 or IFN-λ into wildtype mice (Fig. 4e,f) or of IFN-λ into Il22−/− mice (Fig. 4g) led to a significant reduction in virus titers, injection of IL-22 into Ifnlr1−/− mice did not affect rotavirus replication (Fig. 4h), demonstrating that the impact of IL-22 on rotavirus replication requires a functional IFN-λ signalling pathway.
Fig. 4. Control of rotavirus replication by IL-22 depends on IFN-λ receptor signaling.
Mice of the indicated strains were infected with rotavirus and samples were collected at the indicated time points after infection. (a, b) Ifnl (a) and Ifnlr1 (b) expression in the small intestine of mock and rotavirus infected suckling mice. (c, d) Virus load was determined by RT-qPCR (c) or ELISA (d) at day 4 after infection of suckling mice. Pooled ELISA data from two independent experiments were normalised to fold ID50 of wildtype mice (d). (e-h) Two week old suckling mice of the indicated strains were subcutaneously injected with PBS or the indicated cytokines 8 hours before infection with rotavirus. Injections were repeated on days 1 and 2 after infection. Virus load was determined at day 3 of rotavirus infection by RT-qPCR of RNA obtained from small intestinal tissue (e, g, h) or by ELISA from colon homogenates (f). All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-h) Mean ± SEM. (a-c) n ≥ 5. (d) n ≥ 12. (e, f) n ≥ 6. (g, h) n ≥ 4. (a-f) One-way ANOVA. (g, h) Student’s t test. Data are representative of (a, b) three and (c, e-h) two independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. ND, not detectable (or not displayable on the scale indicated).
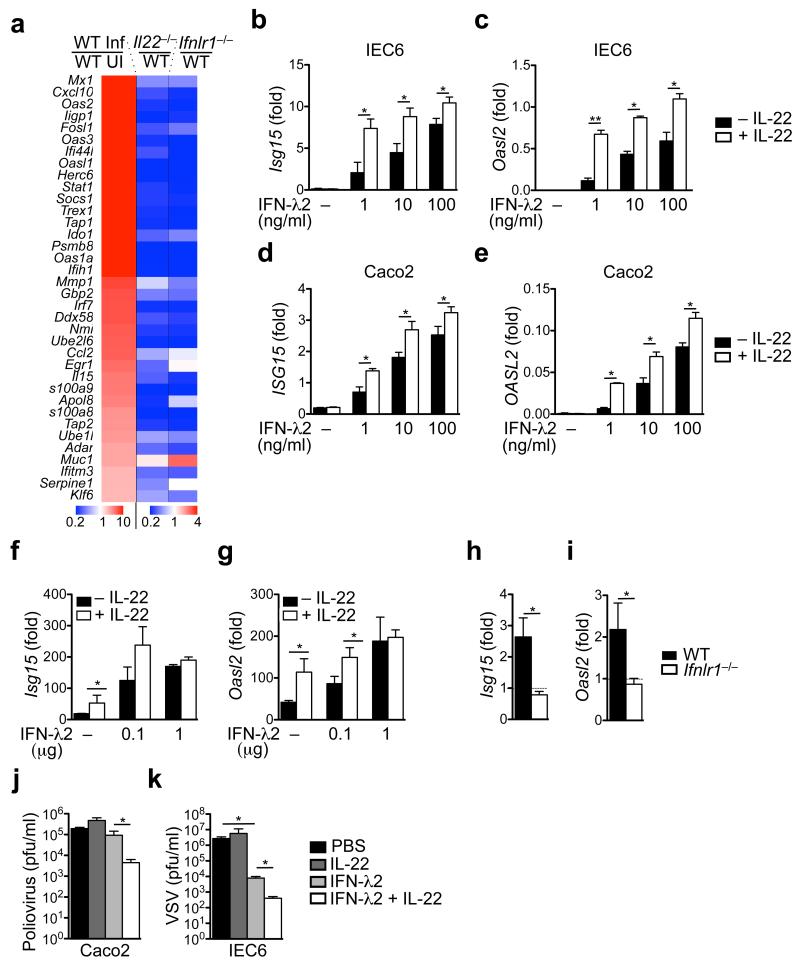
As IL-22 and IFN-λ act on IECs, we transcriptionally profiled epithelial cells isolated from day 1 (Fig. 5a, Supplementary Fig. 7a) and day 4 (Supplementary Fig. 7b) rotavirus-infected wildtype, Il22−/− and Ifnlr1−/− mice. IL-22 target genes (e.g., Reg3g, Reg3b) were not up-regulated in Il22−/− mice, but they were overexpressed in Ifnlr1−/− mice at day 4 of infection (Supplementary Fig. 7b), reflecting increased IL-22 expression in Ifnlr1−/− mice (Supplementary Fig. 5f). ISGs were not or only very weakly induced in Ifnlr1−/− mice (Fig. 5a, Supplementary Fig. 7a-d). Upregulation of ISGs was also reduced in Il22−/− mice (Fig. 5a, Supplementary Fig. 7a-d). In general, upregulation of ISGs was more profoundly impaired in Ifnlr1−/− mice in comparison to Il22−/− mice. However, the various ISG families differed in the extent to which they depended on IFN-λR, IL-22 or both. This probably resulted from the complex mechanisms regulating ISG induction and from the fact that the various ISGs contain different numbers of interferon-stimulated response elements (ISRE) and/or IFN-γ-activated sites (GAS) sequences in their promoters39. Furthermore, we detected a 16-fold induction of Stat1 mRNA expression upon rotavirus infection, which is impaired in Il22−/− mice (0.2 times) and even stronger in Ifnlr1−/− mice (0.12 times) in comparison to wildtype mice (Fig. 5a, Supplemetary Fig. 7a). This might explain why IL-22 seems to be crucial for the induction of some ISGs to an extent similar to IFN-λR. The positive feedback loop of STAT1 activation and newly synthesized protein is required for the full induction of ISGs and is impaired in Il22−/− mice39.
Fig. 5. Cooperation between IL-22 and IFN-λ is required for inducing an efficient antiviral state in IECs.
(a) Gene expression analysis of IECs isolated from suckling animals sacrificed at day 4 after infection. Heat map visualizing relative expression of selected genes by comparing infected wildtype (WT Inf) with uninfected wildtype (UI) mice and infected Il22−/− with infected wildtype mice as well as infected Ifnlr1−/− with infected wildtype mice. (b-e) The intestinal epithelial cell lines IEC6 (b, c) and Caco2 (d, e) were stimulated for 16 hours with the indicated amounts of mouse (IEC6) or human (Caco2) the indicated doses of IFN-λ2 alone or in combination with 100 ng/ml of mouse or human IL-22. The expression of the indicated ISGs was evaluated by RT-qPCR. (f, g) Suckling wildtype mice were injected subcutaneously with indicated amounts of IFN-λ2 and/or 1 μg of IL-22. Three hours after injection, Isg15 (f) and Oasl2 (g) expression in IECs was analyzed by RT-qPCR. (h, i) Adult mice were injected subcutaneously with 1 μg of IL-22 and expression of ISGs was analyzed by RT-qPCR using RNA obtained from IECs isolated from the small intestines 3 hours after cytokine injection. (j, k) Caco2 (j) cells or IEC6 cells (k) were treated with 20 ng/ml of IFN-λ2 and/or 100 ng/ml of IL-22 before infection with poliovirus (j) or VSV (k). All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-k) Mean ± SEM. (a) n ≥ 5. (b-e) n = 4. (f, g) n ≥ 3. (h, i) n = 3. (j, k) n ≥ 4. (b-k) Student’s t test. Data are representative of (a-g, j, k) three and (h, i) two independent experiments. *P < 0.05; **P < 0.01. NS, not significant.
We experimentally tested whether IL-22 can synergize with IFN-λ for optimal ISG induction by assessing the expression of six representative ISGs (i.e., Isg15, Oasl2, Gbp1, Mx1, Eif2ak2 and Rsad2) using a murine (IEC6) and a human (Caco2) intestinal epithelial cell line that are responsive to IL-22 and IFN-λ. Although IL-22 treatment alone did not induce ISG expression, it potently enhanced IFN-λ-induced ISG expression and this effect was most significant when IFN-λ concentrations were limiting (Fig. 5b-e, Supplementary Fig. 7e-l). Similar results were obtained when wildtype mice were treated in vivo with IL-22 and IFN-λ before analyzing expression of Isg15 and Oasl2 in IECs (Fig. 5f,g). In vivo, IL-22 alone was able to induce robust ISG expression in adult mice (Fig. 5f,g). This required constitutive, low level IFN-λ expression by and signaling within IECs (Supplementary Fig. 4a), because it was absent when IL-22 was injected into Ifnlr1−/− mice (Fig. 5h,i). Importantly, IL-22 and IFN-λ acted in synergy to curtail replication of two other viruses that can infect epithelial cells, namely poliovirus (Fig. 5j) and vesicular stomatitis virus (Fig. 5k). Thus, IL-22 limits virus replication in epithelial cells by potentiating IFN-λ-dependent ISG expression in IECs.
IL-22-mediated virus control is independent of STAT3
Virtually all biological effects of IL-22 recognized to date are believed to be mediated by inducing STAT3 signalling downstream of the IL-22R in epithelial cells14, 16. To address the role of epithelial STAT3 signalling in the context of rotavirus infection, we generated mice with a deletion of the Stat3 gene in all intestinal epithelial cells (Stat3fl/flVil1-Cre, hereafter Stat3ΔIEC) (Supplementary Fig. 8a,b). In extension of our data demonstrating normal proliferation and migration of stem cells in infected Il22−/− mice (Supplementary Fig. 2e-h), Stat3ΔIEC mice showed no deviation in rotavirus titers (Fig. 6a) and normal expression of Isg15 (Fig. 6b, Supplementary Fig. 8c), although expression of recognized STAT3 target genes (e.g., Reg3g, Birc5, Pla2g2a) was significantly reduced during steady-state (Supplementary Fig. 8d-f) and after rotavirus infection (Fig. 6c-h). Thus, the IL-22-induced and STAT3-controlled gene expression program in epithelial cells is dispensable for IL-22-mediated suppression of virus replication.
Fig. 6. IL-22 and IFN-λ synergize for phosphorylation of STAT1.
(a-h) Wildtype and Stat3ΔIEC mice were infected with rotavirus and samples collected at day 4 after infection. Virus load (a) and the expression of STAT1-dependent (b) or STAT3-dependent genes (c-h) was determined by RT-qPCR of RNA from IECs. (i) IEC6 cells were stimulated with the indicated cytokines for 30 min. Phosphorylation of the indicated STAT proteins was determined by flow cytometry (open histograms). Unstimulated IEC6 cells are shown as a control (grey histograms). (j) Wildtype mice were subcutaneously injected with 1 μg of IFN-λ2 and/or 1 μg of IL-22. One hour later, STAT phosphorylation in small intestinal tissues was assessed by immunoblotting. (k) Phospho-STAT (pSTAT) analysis in intestinal epithelial cell lines IEC6 and Caco2 stimulated with 20 ng/ml IFN-λ2 and/or 100 ng/ml IL-22 for 30 min. All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-h) Mean ± SEM; n ≥ 4. Data are representative of two independent experiments. (a) Student’s t test. (b-h) Two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
IL-22 and IFN-λ synergize for optimal STAT1 activation
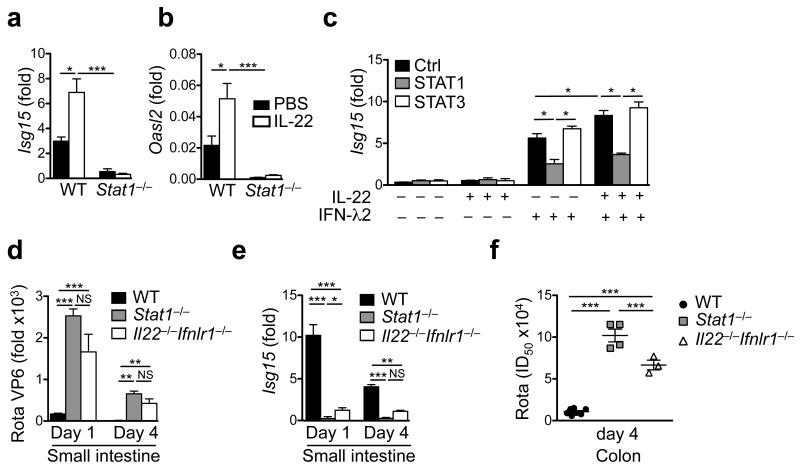
Previous data using hepatoma cell lines had indicated that IL-22 may also connect to the STAT1 signaling pathway7, 40, 41, providing a testable hypothesis for how IL-22 may cooperate with IFN-λ signaling. We stimulated IECs with graded doses of IFN-λ in the presence or absence of IL-22 and analyzed phosphorylation of STAT proteins. In cell lines, IL-22 or low doses of IFN-λ induced only weak phosphorylation of STAT1, whereas high doses of IFN-λ led to robust STAT1 phosphorylation (Fig. 6i). Remarkably, combination of low dose IFN-λ with IL-22 stimulation led to STAT1 phosphorylation comparable to high dose IFN-λ stimulation (Fig. 6i). Application of IL-22 or IFN-λ in vivo led to weak STAT1 phosphorylation (pSTAT1) in IECs (Fig. 6j, Supplementary Fig. 8g). Combined application of IL-22 and IFN-λ led to synergistically enhanced pSTAT1 formation (Fig. 6j) and high level ISG mRNA expression. Similar results were obtained in vitro with two independent IEC lines (Fig. 6k, Supplementary Fig. 8h,i). Although phosphorylation of STAT3 was only induced by IL-22 (Fig. 6i-k, Supplementary Fig. 8g-i), pSTAT2 formation required the presence of IFN-λ and could not be induced by IL-22 (Fig. 6k). IL-22 injections failed to enhance ISG expression in Stat1−/− mice (Fig. 7a,b), whereas Stat3ΔIEC mice showed normal ISG expression following rotavirus infection (Fig. 6b). We also tested the expression of suppressor of cytokine signaling (Socs) genes 1 and 3, which are inhibitory regulators of STAT signaling. Socs1 and Socs3 expression was not increased in Il22−/− mice (Supplementary Fig. 8j) ruling out that enhanced replication of rotavirus in the absence of IL-22 is due to enhanced expression of negative regulators of STAT1 signaling. Socs3 expression was not altered in Ifnlr1−/− mice, whereas Socs1 was reduced, consistent with the finding that Socs1 expression can be induced by STAT1 signaling42. Thus, our data demonstrate that IL-22 and IFN-λ synergize for STAT1 phosphorylation which is required to curtail virus replication in IECs and to limit tissue damage.
Fig. 7. Enhancement of IFN-λ-dependent ISG expression by IL-22 requires STAT1 signaling.
(a, b) Groups of adult mice of the indicated strains were treated with 1 μg of IL-22. Three hours later, gene expression of the indicated genes was determined by RT-qPCR using RNA isolated from small intestinal tissue. (c) The indicated STAT proteins were knocked down in IEC6 cells using siRNAs. Cells were stimulated with 100 ng/ml IL-22, 100 ng/ml IFN-λ2 or both for 16 hours. Isg15 expression was evaluated by RT-qPCR. (d-f) Suckling mice of the indicated strains were infected with rotavirus. (d) Rotavirus replication in the small intestine was analyzed by RT-qPCR. (e) Analysis of Isg15 expression by small intestinal IECs by RT-qPCR. (f) Virus shedding into colon tissue was analyzed by ELISA. All RT-qPCR data are shown as fold (2−ΔCt) relative to Hprt. (a-f) Mean ± SEM. (a-c) n ≥ 3. (d-f) n ≥ 4. Data are representative of two (a, b, d-f) and four (c) independent experiments. (a-f) Two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
To more definitively address the role of STAT1 for enhanced expression of ISGs following stimulation with IFN-λ and IL-22 we used siRNA-mediated knock-down of STAT1 or STAT3 in IECs (Supplementary Fig. 8k) and determined expression of ISGs. Combining IL-22 and IFN-λ led to significantly higher mRNA expression of ISGs than stimulation with IFN-λ alone, and this synergy was entirely dependent on STAT1 but not STAT3 (Fig. 7c, Supplementary Fig. 8k). We reasoned that if STAT1 signaling is the central signaling path mediating cooperation between IFN-λ and IL-22, then the phenotype of Stat1−/− and Il22−/− Ifnlr1−/− mice should be largely comparable. Rotavirus replication (Fig. 7d) and Isg15 induction (Fig. 7e) were largely comparable between Stat1−/− mice and Il22−/−Ifnlr1−/− mice. However, Stat1−/− mice showed significantly more virus shed via the feces (Fig. 7f) suggesting additional STAT1-mediated effects independent of IL-22 and IFN-λ. Thus, cooperation between the mucosal cytokines IL-22 and IFN-λ for optimal induction of ISGs and for curtailment of rotavirus replication can be explained to a large extent by the cooperation of these cytokines for optimal activation of STAT1.
Discussion
Border surfaces such as those of the intestinal epithelium are major entry ports for viruses and they need multi-layered and fail-safe mechanisms of protection. Here, we have identified a close cooperation of two, evolutionarily related mucosal cytokine networks (i.e., IFN-λ and IL-22)5, 6 that synergize to curtail mucosal virus infections which included rotavirus infection, the leading cause of childhood gastroenteritis worldwide. Our data demonstrate that IFN-λ production by IECs alone, albeit required21, is not sufficient to limit rotavirus-afflicted damage to IECs. Instead, the concerted action of IL-22 provided by ILC3s and IFN-λ is required to effectively control rotavirus replication. The related receptors for IL-22 and IFN-λ are virtually exclusively expressed by epithelial cells and simultaneous engagement of both receptors on IECs was required for effective induction of ISGs that confer an antiviral state to IECs. It is interesting that type I IFNs only make a minor contribution to immunity to homologous rotavirus infection while they are required for immunity to infections with heterologous rotaviruses (e.g., rhesus macaque rotavirus infection in mice)22, 43. These findings reflect low expression of the type I IFN receptor by IECs and strong host adaptation of rotavirus strains and existence of immune evasion strategies targeting the IFN pathways19,20,23. It has been demonstrated that IFN-λ but not type I IFNs controlled persistent enteric infection with another important and health-relevant intestinal virus, norovirus44,45. Collectively these data identify IFN-λ as an important antiviral defense strategy at the intestinal barrier.
Following rotavirus infection, IECs produced and released IL-1α, an epithelial alarmin that enhanced IL-22 production by CCR6+ ILC3s. Previously IL-2337 and IL-1β38 expressed by hematopoietic cells were shown to be major regulators of IL-22 expression during steady-state and in the context of inflammatory responses. However, during rotavirus infection neither IL-23 nor IL-1β were required to sustain IL-22 expression. Instead, only neutralization of IL-1α inhibited IL-22 production of ILC3s after rotavirus infection. Another IL-1 family cytokine, IL-18 is produced by epithelial cells and IL-18 release following Toxoplasma gondii was required for the detrimental high IL-22 production following infection46. Collectively these data add to the emerging concept that IL-1 family cytokines produced by epithelial cells (i.e., IL-1α, IL-18) are an important determinant of IL-22 production by ILC3s.
IL-22 has previously been implicated in regulating expression of antimicrobial peptides in IECs required for protection against bacterial infections15, 47 and promoting tissue protection and repair following various types of epithelial insults such as dextran sodium sulfate-mediated colitis16, irradiation48, graft-versus-host disease49 and influenza infection26. Virtually all of the biological functions assigned to IL-22 have been linked to STAT3 signaling in epithelial cells, although it was known that IL-22 can lead to weak STAT1 activation in several cell lines7, 40, 41. However, the biological consequences of such IL-22-driven STAT1 activation were not explored. Recent data have shown that exogenous application of IL-22 to rotavirus-infected mice led to a reduction in virus load, an effect independent of type I and type II interferons50. However, the mechanism by which IL-22 restrains virus replication remained unknown and it was not addressed if endogenously produced IL-22 has any role in restricting rotavirus infection
Our data now demonstrate cooperation between IL-22 and IFN-λ for synergistic activation of STAT1 is required for optimal transcription of ISGs and for limiting replication of mucosal viruses such as rotavirus. Collectively, our data assign an important and previously unappreciated role to IL-22 for curtailing virus infections. The cooperation between an epithelial cell-autonomous antiviral defense system (type III IFN) with an evolutionarily related but hematopoietic-cell derived cytokine (IL-22), both of which act on epithelial cells might document a second layer of virus control that became necessary throughout co-evolution of viruses and vertebrate hosts to fend off viral immunoevasion strategies such as downregulation of the IFN-λR and interference with STAT1 signaling by rotavirus18. Interestingly, rotavirus infections in humans and mice occur preferentially in infants. Therefore, our data contribute to understanding immunity in early life when adaptive immune system components are not yet fully operative. These results represent a significant advance in our understanding of how mucosal cytokine networks cooperate for efficient antiviral immunity to limit damage at the epithelial barrier.
Methods
Mice
Conventional (specific pathogen-free) C57BL/6 mice were purchased from Janvier Laboratories. Tcrb−/− (ref. 51) and Tcrd−/− mice52 were purchased from Jackson Laboratories and crossed in our animal facility to obtain Tcrb−/−Tcrd−/− mice. Mice with a floxed Stat3 allele53 were kindly provided by Veronika Sexl (University of Veterinary Medicine Vienna) with permission of Valeria Poli (Universita di Torino, Torino, Italy). Vil1-Cre mice54 were kindly provided by Marc Stemmler (Max-Planck-Institute of Immunobiology & Epigenetics, Freiburg, Germany) with permission of Sylvie Robine (Institute Curie, Paris, France). Mice with an IEC-specific deletion of Stat3 (Stat3ΔIEC) were generated by breeding Stat3fl/fl mice with Vil1-Cre mice. Rag2−/− and Rag2−/−Il2rg−/− mice were provided by Caro Johner (Max-Planck-Institute of Immunobiology & Epigenetics, Freiburg, Germany). Il22−/− mice55, B6.A2G-Mx1 wild-type mice carrying intact Mx1 alleles (WT), B6.A2G-Mx1-Ifnlr1−/− mice lacking functional type III IFN receptors (Ifnlr1−/−)56, B6.A2G-Mx1-Ifnlr1−/−Il22−/− double-knockout mice (Ifnlr1−/−Il22−/−), and AhrΔILC3,T mice34 and Stat1−/− mice57 were bred in our animal facility. Rorc(γt)-CreTG mice58 were a gift of Dan Littman (Skirball Institute of Biomolecular Medicine, NY, USA). Il1r1−/− mice59 and Il23a−/− mice60 were kindly provided by Bernhard Ryffel and Christoph Hoelscher, respectively. Animals were housed in accordance with the guidelines defined by the Federation for Laboratory Animal Science Associations (www.felasa.eu/recommendations) and the national animal welfare body Gesellschaft für Versuchstierkunde (www.gv-solas.de/index.html), and experiments were performed in compliance with the German animal protection law (TierSchG) and approved by the local animal welfare committee of the University of Freiburg.
Cell lines and reagents
Human epithelial colorectal adenocarcinoma cell line Caco2, human colon carcinoma cell line HT29 and African green monkey kidney cell line Vero and Madin-Darby Canine Kidney MDCK were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% FCS (Sigma), glutamine (Invitrogen), 4.5 g/l glucose (Sigma) and penicillin/streptomycin. The rat intestinal epithelial cell line IEC6 was kindly provided by Mathias Hornef (Hannover Medical School, Hannover, Germany)21. All cell lines were maintained at 37°C and in a 5% CO2 humidified atmosphere. Recombinant murine and human IFN-λ2 and IL-22 were purchased from Peprotech and used at the concentrations indicated. Cytokines were diluted in phosphate-buffered saline (PBS).
Isolation of lamina propria leukocytes and intestinal epithelial cells
Isolation of lamina propria leukocytes was performed as described previously61. In brief, intestines were isolated, cut open longitudinally and washed briefly in PBS. Dissociation of epithelial cells was performed by shaking at 100 rpm/min at 37°C in HBSS containing 5 mM EDTA and 10 mM Hepes for 20 min. Epithelial cells were collected and spun down for 5 min at 2000 rpm/min at 4°C and cell pellets were resuspended in Trizol using a FastPrep®-24 Instrument (MP Biomedicals). To obtain highly purified epithelial cells, the cell pellets were digested for 20 min using Dispase (5 U/ml; BD), Collagenase D (0.5 mg/ml; Roche) and DNaseA (0.5 mg/ml; Sigma-Aldrich). Cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI), CD45 and EpCAM antibodies and sorted using a BD FACSAria III cell sorter (BD Biosciences). The remaining lamina propria fraction was cut into pieces of 1 mm2 before performing enzymatic digestion using the same enzyme mix as for the digestion of epithelial cells. Lymphocyte enrichment was performed by Percoll gradient centrifugation (Sigma-Aldrich).
In vitro stimulation and intracellular staining
For cytokine staining, cells were stimulated at 37°C for 4 h in RPMI containing 10 μg/ml brefeldin A. Cells were permeabilized with Cytofix/Cytoperm (BD Biosciences) and stained for IL-22. For intracellular RORγt staining, cells were permeabilized with Foxp3 staining solutions according to the manufacturer’s protocol (eBioscience) and stained with APC-conjugated RORγt antibody. For detection of intracellular phosphorylated STAT1 and STAT3 proteins, IEC6 cells were harvested and fixed with 4% paraformaldehyde, washed with PBS and resuspended in ice-cold methanol at 4°C for 20 min. After washing, cells were incubated with Fc-block and antibodies against pSTAT1 and pSTAT3 for 60 min at RT.
Antibodies and ELISA kits
The following fluorophore-conjugated antibodies reactive to mouse antigens were purchased from eBioscience: CD3 (145-2C11), CD4 (L3T4), CD11b (M1/70), CD19 (1D3), CD45.2 (104), CD45 (30F11), CD117 (2B8), CD127 (A7R34), KLRG1 (2F1), NK1.1 (PK136), IL-22 (1H8PWSR), RORγt (B2D), BrdU (PRB-1) and from BD Biosciences: EpCAM (G8.8), E-cadherin (36), pY701STAT1 (4a), pY705STAT3 (4), purified CD16/CD32 (2.4G2). Polyclonal goat anti-rotavirus NCDV antibody was purchased from Meridian LS. Secondary donkey anti-rabbit AF488 and anti-goat AF555 or AF488 antibodies were purchased from Invitrogen. Neutralizing antibodies against murine IL-1α (ALF-161) and murine IL-1β (B122) were purchased from BioXCell. For determination of IFN-λ, IL-1α, IL-1β, IL-22 and IL-23 proteins ELISA kits were purchased from eBioscience.
Flow cytometry and cell sorting
After immunostaining, single-cell suspensions of lamina propria lymphocytes were acquired using a FACS Canto II flow cytometer and the FACS Diva software (BD Biosciences). For data analysis, FlowJo V9.2 software (TreeStar) was used. For cell sorting of intestinal epithelial cells and lymphocytes, full small intestines from rotavirus infected as well as control suckling mice were cut into pieces of 1 mm2 and subjected to enzymatic digestion. After blocking of Fc receptors with CD16/CD32 antibodies, single-cell suspensions were incubated with fluorescent conjugated antibodies against CD45 and EpCAM. After washing, cells were incubated with DAPI for exclusion of dead cells and sorted (purity >98%) using a BD FACSAria III cell sorter (BD Biosciences).
Immunohistology
Small intestines were fixed with 4% paraformaldehyde for 2 hours on ice and washed with PBS. Later, the tissue was incubated with 30% sucrose at 4°C overnight, embedded in optimum cutting temperature compound (Sakura, Finetek) and frozen in liquid nitrogen. For rotavirus detection, after blocking with 10% donkey serum, sections were incubated with anti-rotavirus and APC-conjugated E-cadherin antibodies followed by secondary anti-goat AF488 antibody. For BrdU pulse-chase assays, mice were i.p. injected with 50 mg/kg body weight of BrdU (Sigma). Mice were sacrificed at the times indicated in figure legends. Frozen sections were treated with 3N HCl for one hour and incubated for two hours with anti-BrdU-FITC antibodies. Before mounting with mounting medium (PermaFluor, Thermo Scientific) sections were incubated with DAPI for nuclear counterstaining. Tissue sections were visualized using an ApoTome-equipped Axioplan 2 microscope connected to an AxioCam digital Camera (Carl Zeiss MicroImaging, Inc.) Alternatively, small intestines were fixed overnight with 4% paraformaldehyde at room temperature and embedded in paraffin. After cutting and deparaffinization, H&E staining was performed according to Mayer’s protocol with reagents from Roth. Epithelial vacuolization (graded 0-3 to indicate whether there was no damage (0) or damage was seen in a third, 2/3 (2) or 3/3 (3) of the villi in a visual field), villus disruption (graded 0 or 1 to indicate absence (0) or presence (1) of broken or damaged appearance of the villus) and breakage of epithelial barrier (graded 0 or 1 to indicate absence (0) or presence (1) of loss of epithelial cells) were recorded in blinded fashion in distal, middle and proximal tissue sections from intestines of four mice per experimental group. A representative image to illustrate the histopathological changes is displayed in Supplementary Fig. 2b. The number of infected cells per visual field was determined in parallel. The clinical score was determined as follows: Histopathological score × (infected cells per visual field/100).
RNA isolation and quantitative real time PCR
RNA was isolated with Trizol (Invitrogen) reagent according to the manufacturer’s instructions. 1 μg RNA was reverse-transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Real Time PCR was performed using the SYBR®Green Master Mix (Applied Biosystems) or TaqMan Universal Master Mix including gene specific probes (Applied Biosystems) and run on an ABIPrism 7900 sequence detector using the ΔΔCt method from the SDS 2.4 software (Applied Biosystems). The relative expression of tested genes was calculated using the formula 2−ΔCt, where ΔCt = Ct target gene - Ct endogenous control gene62. Hprt was used as endogenous control housekeeping gene. The following TaqMan assays were used for mouse samples: Hprt: Mm00446968_m1; Ifnl2/3: Mm04204156_gH, and for human cell lines: Hprt: Hs99999909_m1. All primers were purchased from Sigma. The primers used are summarized in Supplementary Table 4.
Virus infection and monitoring
Murine rotavirus strain EDIM was kindly provided by Mathias Hornef (Hannover Medical School, Hannover, Germany). Virus stocks were prepared and titrated as described earlier21. Briefly, pooled intestinal tissue with contents collected from orally infected suckling mice at day 4 post infection was homogenized in PBS. The infectious dose 50 (ID50) was determined by infecting groups of 7 day old suckling mice (n=6) with 5 μl of 10-fold serial dilutions of the virus stock (from 10−2 to 10−9). Mice infected with each dilution were kept in separated cages and sacrificed 24 hours later for determination of rotavirus presence in full small intestine by qPCR using two different set of primers. The ID50 of the rotavirus stock was calculated to be 1.7×105 using the Reed-Muench method. Unless explicitly stated otherwise, 7 days old suckling mice were infected by oral application of 7×102 ID50 of rotavirus strain EDIM. Adult mice (4-6 weeks old) were orally infected with 3.5×104 ID50 rotavirus. Age-matched animals were used for individual experiments. To determine viral antigen concentration in colon homogenates or stool samples, samples were homogenized in the dilution buffer supplied with RIDASCREEN Rotavirus ELISA Kit (R-Biopharm) and ELISA was performed according to the manufacturer’s protocol. Two-fold serial dilutions of our rotavirus stock (standard curve) were analyzed side-by-side with mouse samples by ELISA in order to calculate the amount of rotavirus detected by ELISA in colon homogenates and feces relative to the ID50. Vesicular stomatitis virus (VSV) serotype Indiana stocks were grown and titrated on MDCK cells. Plaque assays were performed under 1:1 mixture of 3% AVICEL (Thermo Fisher Scientific), 2x concentrated DMEM medium (Invitrogen) and 0.1% BSA. Cell culture infection was performed at room temperature with virus diluted in 0.1% BSA PBS for 1h. Poliovirus type I Mahoney strain was kindly provided by Satoshi Koike (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Virus stocks were grown and titrated on Vero cells. Plaque assays were performed under 1:1 mixture of 3% AVICEL (Thermo Fisher Scientific), 2x concentrated DMEM medium (Invitrogen) and 0.1% BSA. Cell culture infection was performed at room temperature for with virus diluted in 0.1% BSA PBS for 1h.
Microbiota analysis
16S rDNA libraries were constructed and sequenced as described previously63, with modifications. 16S rDNA libraries were constructed following the 454 Amplicon Library Preparation Manual as of April 2014 (Roche, pp. 2-4), with each reaction containing 20 ng of DNA. 454 Lib-L fusion primers contained multiplex identifiers (MIDs) and the template-specific parts 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 541R (5′-WTTACCGCGGCTGCTGG-3′). Positive and no-template negative controls were included for each MID. PCR conditions included the following steps: 94°C for 3 min, 33 cycles of 15 s at 94°C, 45 s at 55°C and 1 min at 72°C, followed by final elongation for 8 min at 72°C. PCR products were purified by gel electrophoresis (1.5% agarose gels), followed by extraction of 500-700 bp fragments using a QIAquick gel extraction kit (QIAGEN). Following fluorometric quantitation with a Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen), groups of 5 samples with different MIDs were pooled in equal amounts. These pools were prepared for sequencing using GS FLX Titanium chemistry emPCR as described in the emPCR Amplification Method Manual - Lib L SV (XL+) (May 2011), modified as suggested in the 454 Long Fragment Protocol TCB11001 (2011). Emulsion PCR products were sequenced from the 3′ end using Titanium chemistry on a GS FLX+ instrument. Raw sequencing data were pre-processed with the shotgun pipeline of GS Run Processing Software version 2.9 to extract FASTA and quality files.
Data were processed using UPARSE64 as provided by USEARCH version 7.0.1090, following the pipeline suggested in the UPARSE manual64. During quality filtering, sequences were trimmed to 350 bp and quality-filtered to a maximum expected error of 0.5. One mouse (IL22_KO3_M16) was excluded from further analysis due to low quality of sequencing results. Sequences were clustered into operational taxonomic units (OTUs) at the default OTU radius of 3%.
The representative sequences of the resulting 314 OTUs (Supplementary Table 3) were run through the command-line version of the Ribosomal Database Project (RDP) Classifier65 version 2.7, and classifications with a minimum bootstrap value of 0.8 were retained. Sequences classified as chloroplast were culled. For representative sequences not identified to class level, BLAST-based classification was attempted: The representative sequences were BLASTed against the NCBI Nucleotide database (nt), and hit sequences with an alignment over the full length of the representative sequence which also were 100% identical to this query sequence were identified. Among these full-length identical hits, all individual nt accession numbers were counted; multiple hits to the same entry were excluded. OTUs with representative sequences for which at least 50 identical hits were identified were retained, and where possible were assigned a classification based on the hits not marked as “uncultured”; all other OTUs not identified to class level were excluded. For identification to species level, representative sequences that were identified to genus with at least 98% bootstrap support were BLASTed against two different reference datasets, the RDP database66 (release 11, update 2) and the “The All-Species Living Tree” Project 16S rRNA database LTPs11167. For both databases, the top BLAST hits carrying species annotations were further examined: The corresponding species name was retained if the identity to the query sequence was at least 97%, the aligned region covered at least 97% of query sequence length, no hit sequence with another species annotation matched at the same BLAST expectation value, and the species annotation did not conflict with the genus as identified by RDP classifier (except if the same conflict also occurred between the LTPs111 sequence of the species type and its RDP classifier annotation). If the same species name was retained from TaxCollector-based and LTPs-based analyses, this species name was added to the classification of the representative sequence (see Supplementary Data Set).
Further analyses were based on the mothur pipeline (mothur version 1.33.3)68: The dataset was normalised by randomly subsampling the sequence set for each mouse to 3,163 sequences, the number found in the sample with the lowest sequence count. OTU-based statistics including rarefaction curves, Good’s coverage and AMOVA comparisons were calculated with mothur (see Supplementary Fig. 3a,d and Supplementary Tables 1-3). PCoA analyses were performed using the capscale function of the R package vegan69 with Jaccard index distances calculated within mothur.
Immunoblot
Tissue was homogenized in lysis buffer containing 100 mM NaCl, 50 mM Tris (pH 7.5), 1 mM EDTA, 0.1% TX-100, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM vanadate, and 1x protease inhibitor cocktail tablets (Roche Diagnostics GmbH). Samples were lysed on ice at 4°C for 30 min and centrifuged for 5 min at 15,000 rpm at 4°C. Protein concentration was determined using the Bradford assay (Bio-Rad, Munich, Germany). 20 μg of protein was separated on sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred on nitrocellulose membrane. The membranes were blocked with 3% bovine serum albumin (BSA) in TRIS-buffered saline with 0.05% Tween-20 (TBST). Proteins were detected with primary antibodies specific for phospho-STAT1 (Tyr701, Cell Signaling Technology), STAT1 (Transduction Laboratories, BD Biosciences Pharmingen), phospho-STAT3 (Tyr705, Cell Signaling Technology), STAT3 (Santa Cruz Biotechnology) and β-actin (Sigma-Aldrich Chemie GmbH). Fluorescent secondary goat anti-mouse (IRDye 680) or anti-rabbit (IRDye 800) antibodies (Li-Cor Biosciences) were used with the Odyssey infrared imaging system. Alternatively, horseradish peroxidase-labeled secondary antibodies (Jackson Immunoresearch) were used with the chemiluminescence detection system from Pierce.
siRNA-mediated knock-down of gene expression
IEC6 cells at 50% confluence were transfected with Silencer® Select Pre-designed siRNA (Cat# 4390816) for STAT1 (ID: s129044), STAT3 (ID: s129046) or negative control Scrambled Stealth™ (Life Technologies) at a final concentration of 200 nM using Lipofectamine® 2000 (Life Technologies) during 72 h. After transfection, cells were used for cytokine stimulation and gene expression analyzed by flow cytometry, RT-PCR or Western blot.
Statistical analysis
GraphPad Prism 6.0 software was used for data analysis. Statistical significance was determined by two-tailed t-test for two groups or one-way ANOVA (with Tukey’s multiple comparisons test) for three or more groups with the assumption of normal distribution of data and equal sample variance. Two-way ANOVA was used when one measurement was performed on samples with two nominal variables (e.g., infected vs. uninfected and different genotypes or cell types). Sample sizes were selected on the basis of preliminary results to ensure an adequate power. All experimental data were included into the statistical analysis.
Supplementary Material
Acknowledgments
We thank G. Häcker for support; the members of the Diefenbach laboratory for valuable discussions and to A. Triantafyllopoulou for critical reading of the manuscript. We are thankful to N. Ghilardi for providing Il23a−/− mice. We thank K. Oberle, S. Woltemate and Z. Fiebig for excellent technical assistance. We are grateful to M. Follo, K. Geiger and J. Bodinek-Wersing for cell sorting. The work was supported by an ERC Starting Grant #311377 to A.D., the DFG Priority Program 1656 “Intestinal Microbiota” (DI764/3 to A.D. and SU133/9 to S.S.), GRK1104 to A.D. and F.G., GSC-4, Spemann Graduate School of Biology and Medicine to T.M., SFB900/Z1 to S.S. and the Cluster of Excellence “Inflammation at Interfaces” (Borstel-Kiel-Lübeck-Plön) EXC306 to C.H.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Accession codes
Microbiota 16S rDNA amplicon data were submitted to the European Nucleotide Archive under project accession number PRJEB8991.
References
- 1.Moon C, Stappenbeck TS. Viral interactions with the host and microbiota in the intestine. Curr Opin Immunol. 2012;24(4):405–410. doi: 10.1016/j.coi.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14(7):654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katze MG, He Y, Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat Rev Immunol. 2002;2(9):675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 4.Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev. 2013;255(1):25–39. doi: 10.1111/imr.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, et al. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4(1):69–77. doi: 10.1038/ni875. [DOI] [PubMed] [Google Scholar]
- 6.Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4(1):63–68. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 7.Dumoutier L, Tounsi A, Michiels T, Sommereyns C, Kotenko SV, Renauld JC. Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-lambda 1: similarities with type I interferon signaling. J Biol Chem. 2004;279(31):32269–32274. doi: 10.1074/jbc.M404789200. [DOI] [PubMed] [Google Scholar]
- 8.Ank N, West H, Bartholdy C, Eriksson K, Thomsen AR, Paludan SR. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J Virol. 2006;80(9):4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyle SE, Schreckhise H, Khuu-Duong K, Henderson K, Rosler R, Storey H, et al. Interleukin-29 uses a type 1 interferon-like program to promote antiviral responses in human hepatocytes. Hepatology. 2006;44(4):896–906. doi: 10.1002/hep.21312. [DOI] [PubMed] [Google Scholar]
- 10.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8(7):559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotenko SV. IFN-lambdas. Curr Opin Immunol. 2011;23(5):583–590. doi: 10.1016/j.coi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R. IL-22 increases the innate immunity of tissues. Immunity. 2004;21(2):241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS pathogens. 2008;4(3):e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutz S, Ouyang W. Regulation of interleukin-10 and interleukin-22 expression in T helper cells. Curr Opin Immunol. 2011;23(5):605–612. doi: 10.1016/j.coi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14(3):282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 16.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Warntjen M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206(7):1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramig RF. Pathogenesis of intestinal and systemic rotavirus infection. J Virol. 2004;78(19):10213–10220. doi: 10.1128/JVI.78.19.10213-10220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen A, Rott L, Phan N, Mukherjee G, Greenberg HB. Rotavirus NSP1 protein inhibits interferon-mediated STAT1 activation. J Virol. 2014;88(1):41–53. doi: 10.1128/JVI.01501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway G, Coulson BS. Innate cellular responses to rotavirus infection. J Gen Virol. 2013;94(Pt 6):1151–1160. doi: 10.1099/vir.0.051276-0. [DOI] [PubMed] [Google Scholar]
- 20.Arnold MM, Sen A, Greenberg HB, Patton JT. The battle between rotavirus and its host for control of the interferon signaling pathway. PLoS pathogens. 2013;9(1):e1003064. doi: 10.1371/journal.ppat.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pott J, Mahlakoiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, et al. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci USA. 2011;108(19):7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng N, Kim B, Fenaux M, Nguyen H, Vo P, Omary MB, et al. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J Virol. 2008;82(15):7578–7590. doi: 10.1128/JVI.00391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen A, Rothenberg ME, Mukherjee G, Feng N, Kalisky T, Nair N, et al. Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc Natl Acad Sci USA. 2012;109(50):20667–20672. doi: 10.1073/pnas.1212188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209(8):1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190(10):5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P, Thakar MS, Ouyang W, Malarkannan S. IL-22 from conventional NK cells is epithelial regenerative and inflammation protective during influenza infection. Mucosal Immunol. 2013;6(1):69–82. doi: 10.1038/mi.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lauterbach H, Bathke B, Gilles S, Traidl-Hoffmann C, Luber CA, Fejer G, et al. Mouse CD8alpha+ DCs and human BDCA3+ DCs are major producers of IFN-lambda in response to poly IC. J Exp Med. 2010;207(12):2703–2717. doi: 10.1084/jem.20092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol. 2012;189(6):2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10(1):83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203(10):2271–2279. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31(2):321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 32.Zindl CL, Lai JF, Lee YK, Maynard CL, Harbour SN, Ouyang W, et al. IL-22-producing neutrophils contribute to antimicrobial defense and restitution of colonic epithelial integrity during colitis. Proc Natl Acad Sci USA. 2013;110(31):12768–12773. doi: 10.1073/pnas.1300318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494(7436):261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 34.Kiss EA, Vonarbourg C, Kopfmann S, Hobeika E, Finke D, Esser C, et al. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 2011;334(6062):1561–1565. doi: 10.1126/science.1214914. [DOI] [PubMed] [Google Scholar]
- 35.Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5(1):64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 36.Franco MA, Greenberg HB. Immunity to rotavirus infection in mice. J Infect Dis. 1999;179(Suppl 3):S466–469. doi: 10.1086/314805. [DOI] [PubMed] [Google Scholar]
- 37.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36(2):276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynders A, Yessaad N, Vu Manh TP, Dalod M, Fenis A, Aubry C, et al. Identity, regulation and in vivo function of gut NKp46+RORgammat+ and NKp46+RORgammat− lymphoid cells. EMBO J. 2011;30(14):2934–2947. doi: 10.1038/emboj.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1(6):519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmann M, Ulziibat S, Hardle L, Pfeilschifter J, Muhl H. IFNalpha converts IL-22 into a cytokine efficiently activating STAT1 and its downstream targets. Biochem Pharmacol. 2013;85(3):396–403. doi: 10.1016/j.bcp.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 41.Lejeune D, Dumoutier L, Constantinescu S, Kruijer W, Schuringa JJ, Renauld JC. Interleukin-22 (IL-22) activates the JAK/STAT, ERK, JNK, and p38 MAP kinase pathways in a rat hepatoma cell line. Pathways that are shared with and distinct from IL-10. J Biol Chem. 2002;277(37):33676–33682. doi: 10.1074/jbc.M204204200. [DOI] [PubMed] [Google Scholar]
- 42.Dai X, Sayama K, Yamasaki K, Tohyama M, Shirakata Y, Hanakawa Y, et al. SOCS1-negative feedback of STAT1 activation is a key pathway in the dsRNA-induced innate immune response of human keratinocytes. J Invest Dermatol. 2006;126(7):1574–1581. doi: 10.1038/sj.jid.5700294. [DOI] [PubMed] [Google Scholar]
- 43.Angel J, Franco MA, Greenberg HB, Bass D. Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J Interferon Cytokine Res. 1999;19(6):655–659. doi: 10.1089/107999099313802. [DOI] [PubMed] [Google Scholar]
- 44.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347(6219):269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015;347(6219):266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munoz M, Eidenschenk C, Ota N, Wong K, Lohmann U, Kuhl AA, et al. Interleukin-22 Induces Interleukin-18 Expression from Epithelial Cells during Intestinal Infection. Immunity. 2015;42(2):321–331. doi: 10.1016/j.immuni.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 47.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14(3):275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudakov JA, Hanash AM, Jenq RR, Young LF, Ghosh A, Singer NV, et al. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 2012;336(6077):91–95. doi: 10.1126/science.1218004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanash AM, Dudakov JA, Hua G, O’Connor MH, Young LF, Singer NV, et al. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 2012;37(2):339–350. doi: 10.1016/j.immuni.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346(6211):861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.